Putative Wound Healing Induction Functions of Exosomes Isolated from IMMUNEPOTENT CRP
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. IMMUNEPOTENT CRP Protein Characterization
4.2. Exosome Proteomics Characterization
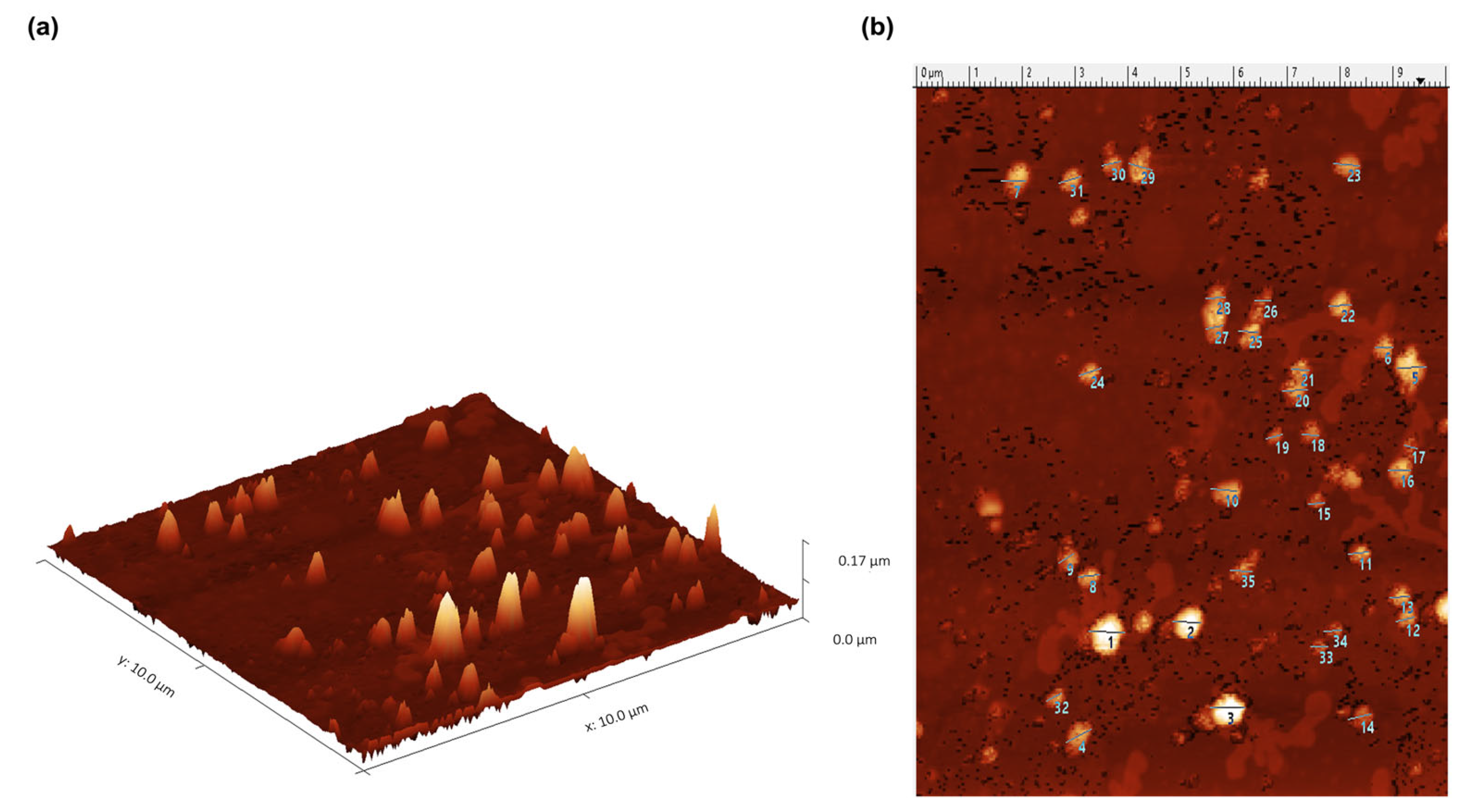
4.3. Exosome Shape and Size Characterization through Atomic Force Microscopy
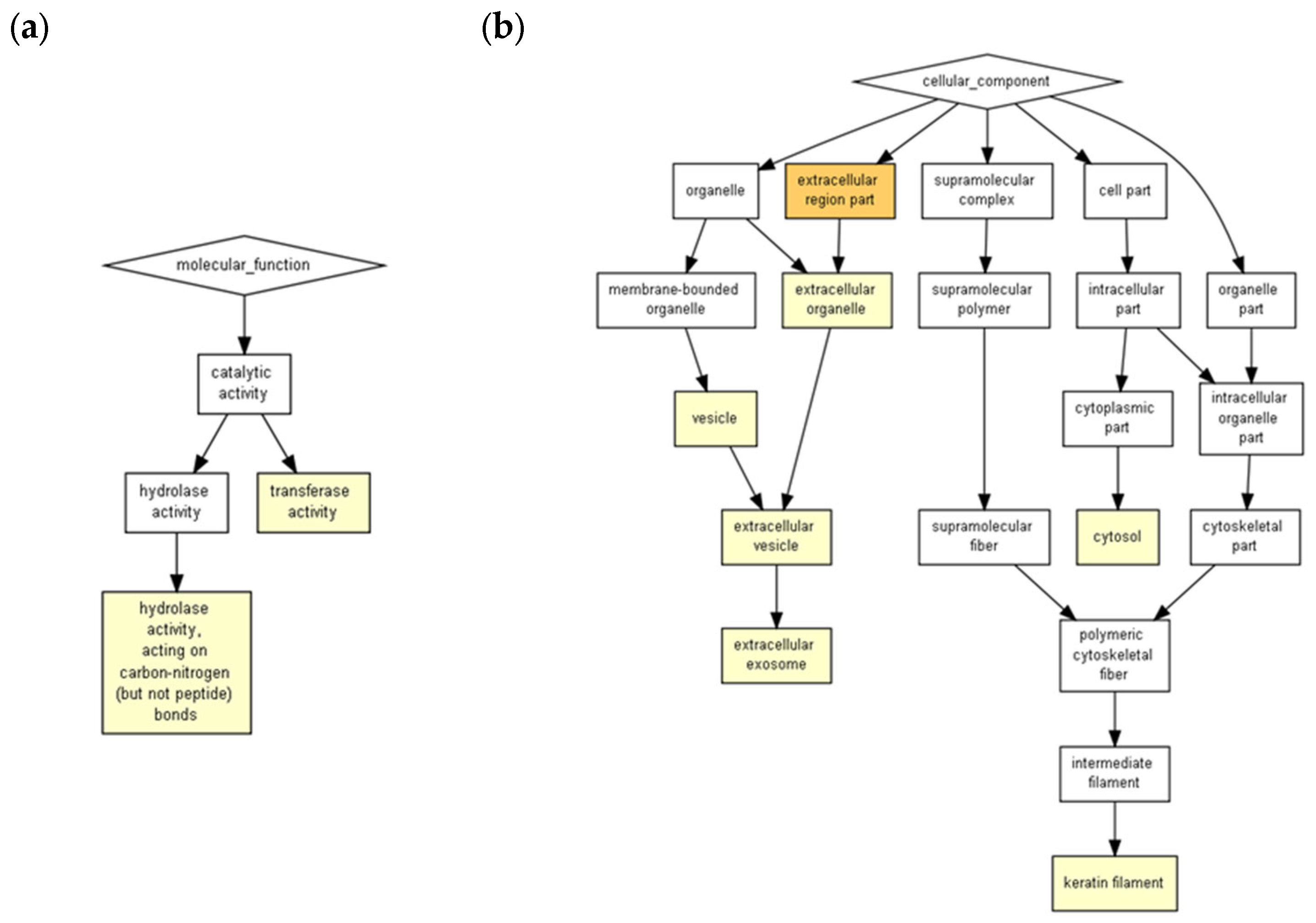
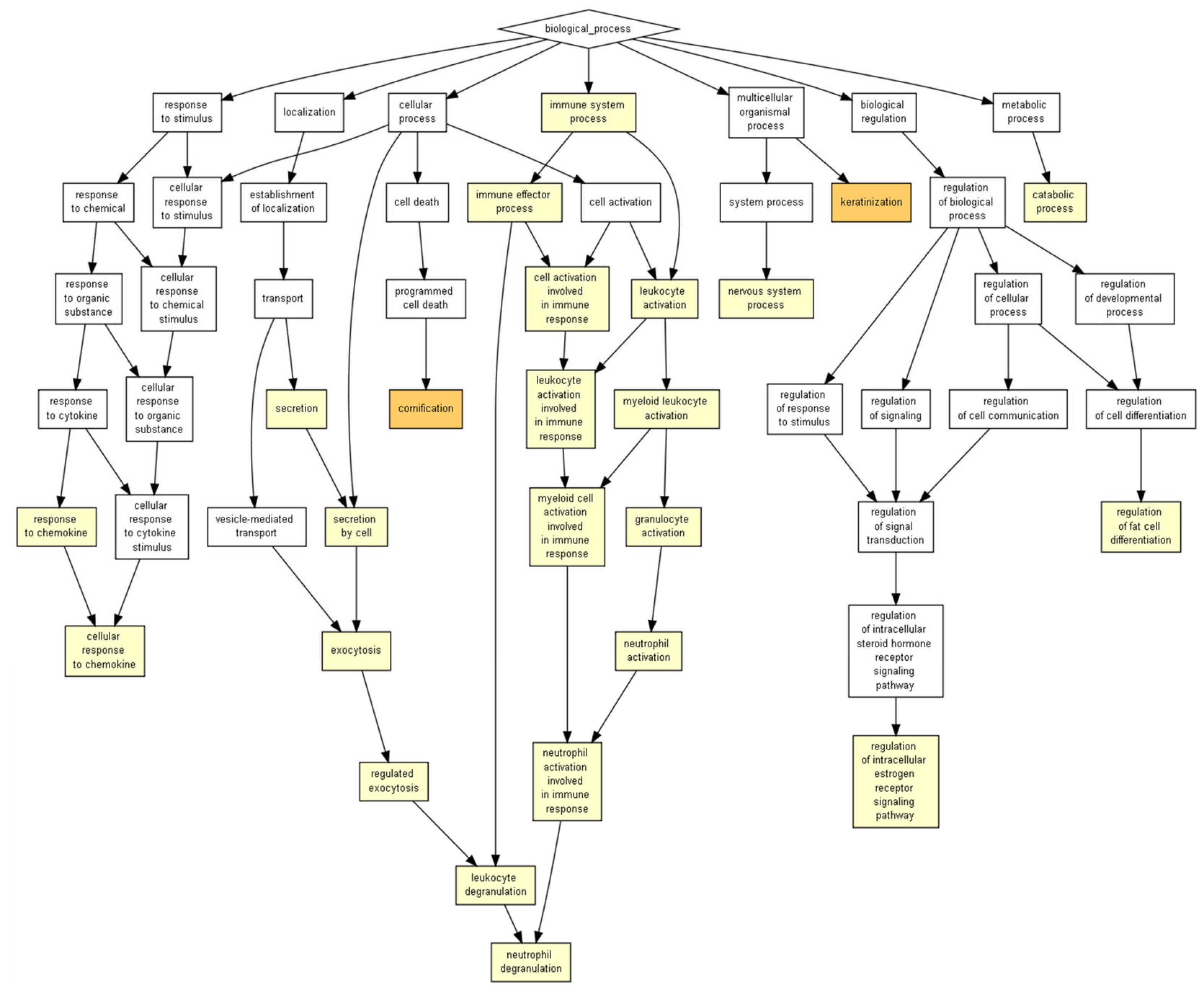
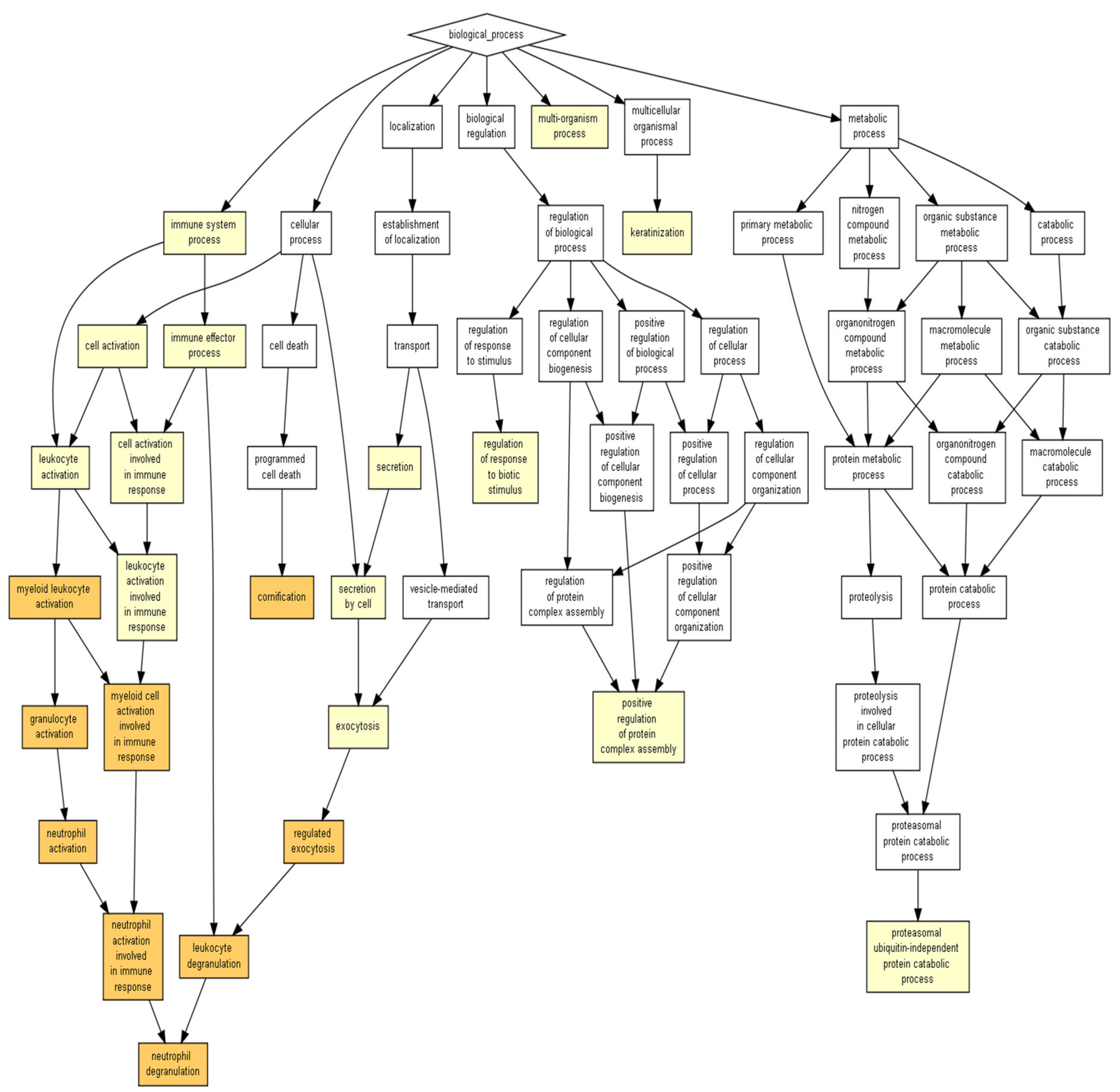
4.4. Biological Pathway Functions Prediction
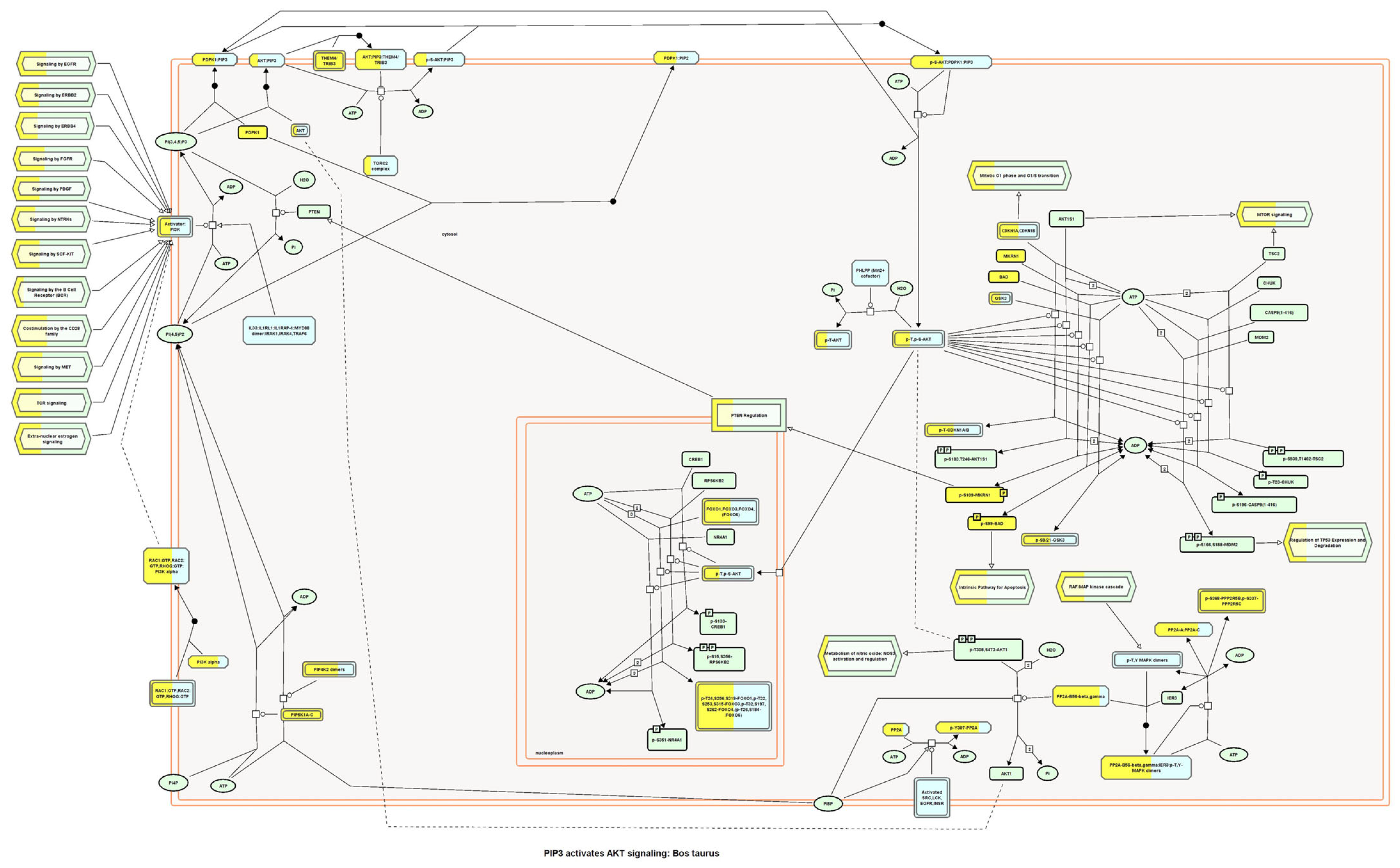
4.5. Evaluation of Wound-Healing-Related Signaling Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gurunathan, S.; Kang, M.-H.; Kim, J.-H. A comprehensive review on factors influences biogenesis, functions, therapeutic and clinical implications of exosomes. Int. J. Nanomed. 2021, 16, 1281–1312. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, Q.; Tang, J.; Xiong, L.; Li, L. Extracellular vesicles: Emerging therapeutics in cutaneous lesions. Int. J. Nanomed. 2021, 16, 6183–6202. [Google Scholar] [CrossRef] [PubMed]
- ExoCarta. List of Top 100 Proteins That Are often Identified in Exosomes. ExoCarta. 2022. Available online: http://www.exocarta.org/ (accessed on 10 March 2022).
- Kwok, Z.H.; Wang, C.; Jin, Y. Extracellular vesicle transportation and uptake by recipient cells: A critical process to regulate human diseases. Processes 2021, 9, 273. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Kosaric, N.; Bonham, C.A.; Gurtner, G.C. Wound Healing: A Cellular Perspective. Physiol. Rev. 2019, 99, 665–706. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, V.; Samal, S.K.; Mohanty, S.; Nayak, S.K. Recent advancements in biopolymer and metal nanoparticle-based materials in diabetic wound healing management. Int. J. Biol. Macromol. 2019, 122, 137–148. [Google Scholar] [CrossRef]
- Gianino, E.; Miller, C.; Gilmore, J. Smart wound dressings for diabetic chronic wounds. Bioengineering 2018, 5, 51. [Google Scholar] [CrossRef]
- Li, B.; Luan, S.; Chen, J.; Zhou, Y.; Wang, T.; Li, Z.; Fu, Y.; Zhai, A.; Bi, C. The MSC-derived exosomal lncRNA H19 promotes wound healing in diabetic foot ulcers by upregulating pten via MicroRNA-152-3p. Mol. Ther. Nucleic Acids 2020, 19, 814–826. [Google Scholar] [CrossRef]
- Zhang, W.; Bai, X.; Zhao, B.; Li, Y.; Zhang, Y.; Li, Z.; Wang, X.; Luo, L.; Han, F.; Zhang, J.; et al. Cell-free therapy based on adipose tissue stem cell-derived exosomes promotes wound healing via the PI3K/Akt signaling pathway. Exp. Cell Res. 2018, 370, 333–342. [Google Scholar] [CrossRef]
- Wang, J.; Wu, H.; Peng, Y.; Zhao, Y.; Qin, Y.; Zhang, Y.; Xiao, Z. Hypoxia adipose stem cell-derived exosomes promote high-quality healing of diabetic wound involves activation of PI3K/AKT PATHWAYS. J. Nanobiotechnol. 2021, 19, 202. [Google Scholar] [CrossRef]
- Franco-Molina, M.A.; Mendoza-Gamboa, E.; Mir, D.F.; Sierra-Rivera, C.A.; Zapata-Benavides, P.; Vera-García, M.E.; Tamez-Guerra, R.; Rodríguez-Padilla, C. Anti-inflammatory and antioxidant effects of IMMUNEPOTENT CRP in Lipopolysaccharide (LPS)-stimulated human macrophages. Afr. J. Microbiol. Res. 2011, 5, 3726–3736. [Google Scholar] [CrossRef]
- Franco-Molina, M.A.; Mendoza-Gamboa, E.; Coronado-Cerda, E.E.; Zarate-Triviño, D.; Arizpe-Coronado, J.E.; Zapata-Benavides, P.; Ramos Zayas, Y.; Tamez-Guerra, R.; Rodríguez-Padilla, C. Clinical trial evaluating the effectiveness of biocompound immunepotent CRP in the third-molar extraction. Biotechnol. Biotechnol. Equip. 2016, 31, 182–186. [Google Scholar] [CrossRef]
- Faustman, D.L.; Davis, M. Stem Cells in the spleen: Therapeutic potential for Sjogren’s syndrome, type I diabetes, and other disorders. Int. J. Biochem. Cell Biol. 2010, 42, 1576–1579. [Google Scholar] [CrossRef]
- Franco-Molina, M.A.; Santana-Krímskaya, S.E.; Coronado-Cerda, E.E.; Hernández-Luna, C.E.; Zarate-Triviño, D.G.; Zapata-Benavides, P.; Mendoza-Gamboa, E.; Rodríguez-Salazar, M.C.; Tamez-Guerra, R.; Rodríguez-Padilla, C. Increase of the antitumour efficacy of the biocompound IMMUNEPOTENT CRP by enzymatic treatment. Biotechnol. Biotechnol. Equip. 2018, 31, 1028–1035. [Google Scholar] [CrossRef]
- Franco-Molina, M.A.; Santana-Krímskaya, S.E.; Zarate-Triviño, D.G.; Zapata-Benavides, P.; Hernández-Martínez, S.P.; Cervantes-Wong, F.; Rodríguez-Padilla, C. Bovine Dialyzable Leukocyte Extract IMMUNEPOTENT CRP: Evaluation of Biological Activity of the Modified Product. Appl. Sci. 2021, 11, 3505. [Google Scholar] [CrossRef]
- Gazze, S.A.; Thomas, S.J.; Garcia-Parra, J.; James, D.W.; Rees, P.; Marsh-Durban, V.; Corteling, R.; Gonzalez, D.; Conlan, R.S.; Francis, L.W. High content, quantitative AFM analysis of the scalable biomechanical properties of extracellular vesicles. Nanoscale 2021, 13, 6129–6141. [Google Scholar] [CrossRef]
- Zhang, Q.; Higginbotham, J.N.; Jeppesen, D.K.; Yang, Y.-P.; Li, W.; McKinley, E.T.; Graves-Deal, R.; Ping, J.; Britain, C.M.; Dorsett, K.A.; et al. Transfer of functional cargo in Exomeres. Cell Rep. 2019, 27, 940–954. [Google Scholar] [CrossRef]
- Anand, S.; Samuel, M.; Mathivanan, S. Exomeres: A new member of extracellular vesicles family. Subcell. Biochem. 2021, 97, 89–97. [Google Scholar] [CrossRef]
- Hu, N.; Cai, Z.; Jiang, X.; Wang, C.; Tang, T.; Xu, T.; Chen, H.; Li, X.; Du, X.; Cui, W. Hypoxia-pretreated ADSC-derived exosome-embedded hydrogels promote angiogenesis and accelerate diabetic wound healing. Acta Biomater. 2022, 157, 175–186. [Google Scholar] [CrossRef]
- Wang, K.; Dong, R.; Tang, J.; Li, H.; Dang, J.; Zhang, Z.; Yu, Z.; Guo, B.; Yi, C. Exosomes laden self-healing injectable hydrogel enhances diabetic wound healing via regulating macrophage polarization to accelerate angiogenesis. Chem. Eng. J. 2022, 430, 132664. [Google Scholar] [CrossRef]
- Geng, X.; Qi, Y.; Liu, X.; Shi, Y.; Li, H.; Zhao, L. A multifunctional antibacterial and self-healing hydrogel laden with bone marrow mesenchymal stem cell-derived exosomes for accelerating diabetic wound healing. Biomater. Adv. 2022, 133, 112613. [Google Scholar] [CrossRef] [PubMed]
- Barbera, S.; Lugano, R.; Pedalina, A.; Mongiat, M.; Santucci, A.; Tosi, G.M.; Dimberg, A.; Galvagni, F.; Orlandini, M. The C-type lectin CD93 controls endothelial cell migration via activation of the Rho family of small GTPases. Matrix Biol. 2021, 99, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Abreu-Blanco, M.T.; Verboon, J.M.; Parkhurst, S.M. Coordination of rho family GTPase activities to orchestrate cytoskeleton responses during Cell Wound Repair. Curr. Biol. 2014, 24, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Franco-Molina, M.A.; Mendoza-Gamboa, E.; Zapata-Benavides, P.; Vera-García, M.E.; Castillo-Tello, P.; García de la Fuente, A.; Mendoza, R.D.; Garza, R.G.; Támez-Guerra, R.S.; Rodríguez-Padilla, C. IMMUNEPOTENT CRP (bovine dialyzable leukocyte extract) adjuvant immunotherapy: A phase I study in non-small cell lung cancer patients. Cytotherapy 2008, 10, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Mansbridge, J.N.; Knapp, M. Changes in keratinocyte maturation during wound healing. J. Investig. Dermatol. 1987, 89, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; He, R.; Ren, J.; Zhang, W.; Wang, K.; Zhu, L.; Liang, T. Exosomal HMGB1 derived from hypoxia-conditioned bone marrow mesenchymal stem cells increases angiogenesis via the JNK/HIF-1α pathway. FEBS Open Bio 2021, 11, 1364–1373. [Google Scholar] [CrossRef]
- Gosain, A.; Jones, S.B.; Shankar, R.; Gamelli, R.L.; DiPietro, L.A. Norepinephrine modulates the inflammatory and proliferative phases of wound healing. J. Trauma Inj. Infect. Crit. Care 2006, 60, 736–744. [Google Scholar] [CrossRef]
- Dilhari, A.; Pathirage, S.; Gunasekara, C.; Fernando, N.; Weerasekara, D.; McBain, A.; Weerasekera, M. salmonella enterica serovar Paratyphi a isolated from a hard-to heal diabetic ulcer: A case report. J. Wound Care 2020, 29, 12–15. [Google Scholar] [CrossRef]
- Pavlov, K.O.; Fedoseev, V.F.; Tyulyubayev, V.S.; Igumnova, T.V.; Malyavsky, I.Y. Salmonella necrotizing infection on the background of diabetes mellitus (clinical, pathological and anatomical case). Wounds Wound Infect. Prof. B.M. Kostyuchenok J. 2019, 6, 44–47. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, N.; Hu, L.; Xi, Y.; Mi, W.; Ma, Y. Integrin β1 in adipose derived stem cells accelerates wound healing via activating PI3K/akt pathway. Tissue Eng. Regen. Med. 2020, 17, 183–192. [Google Scholar] [CrossRef]
- Li, G.; Li, Y.Y.; Sun, J.E.; Lin, W.H.; Zhou, R.X. Ilk–PI3K/AKT pathway participates in cutaneous wound contraction by regulating fibroblast migration and differentiation to myofibroblast. Lab. Investig. 2016, 96, 741–751. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, X.; Yang, S.; Song, X.; Sun, N.; Chen, C.; Zhang, Y.; Yao, D.; Huang, J.; Wang, J.; et al. Kanglexin accelerates diabetic wound healing by promoting angiogenesis via FGFR1/Erk Signaling. Biomed. Pharmacother. 2020, 132, 110933. [Google Scholar] [CrossRef]
- Emmerson, E.; Campbell, L.; Davies FC, J.; Ross, N.L.; Ashcroft, G.S.; Krust, A.; Chambon, P.; Hardman, M.J. Insulin-like growth factor-1 promotes wound healing in estrogen-deprived mice: New insights into cutaneous IGF-1R/ERA Cross talk. J. Investig. Dermatol. 2012, 132, 2838–2848. [Google Scholar] [CrossRef]
- Xiao, C.; Roland, J.T.; Nam, K.T.; Goldenring, J.R.; Zavros, Y. Sonic hedgehog induces gastric re-epithelialization, angiogenesis and formation of granulation tissue that may have clinical implications during ulcer healing after helicobacter pylori eradication. Gastroenterology 2011, 140, S-477. [Google Scholar] [CrossRef]
- Jia, Y.; Yang, J.; Lu, T.; Pu, X.; Chen, Q.; Ji, L.; Luo, C. Repair of spinal cord injury in rats via exosomes from bone mesenchymal stem cells requires sonic hedgehog. Regen. Ther. 2021, 18, 309–315. [Google Scholar] [CrossRef]
- Wu, X.; Li, L.; Iliuk, A.; Tao, A. Highly Efficient Phosphoproteome Capture and Analysis from Urinary Extracellular Vesicles. J. Proteome Res. 2018, 170, 3308–3316. [Google Scholar] [CrossRef]

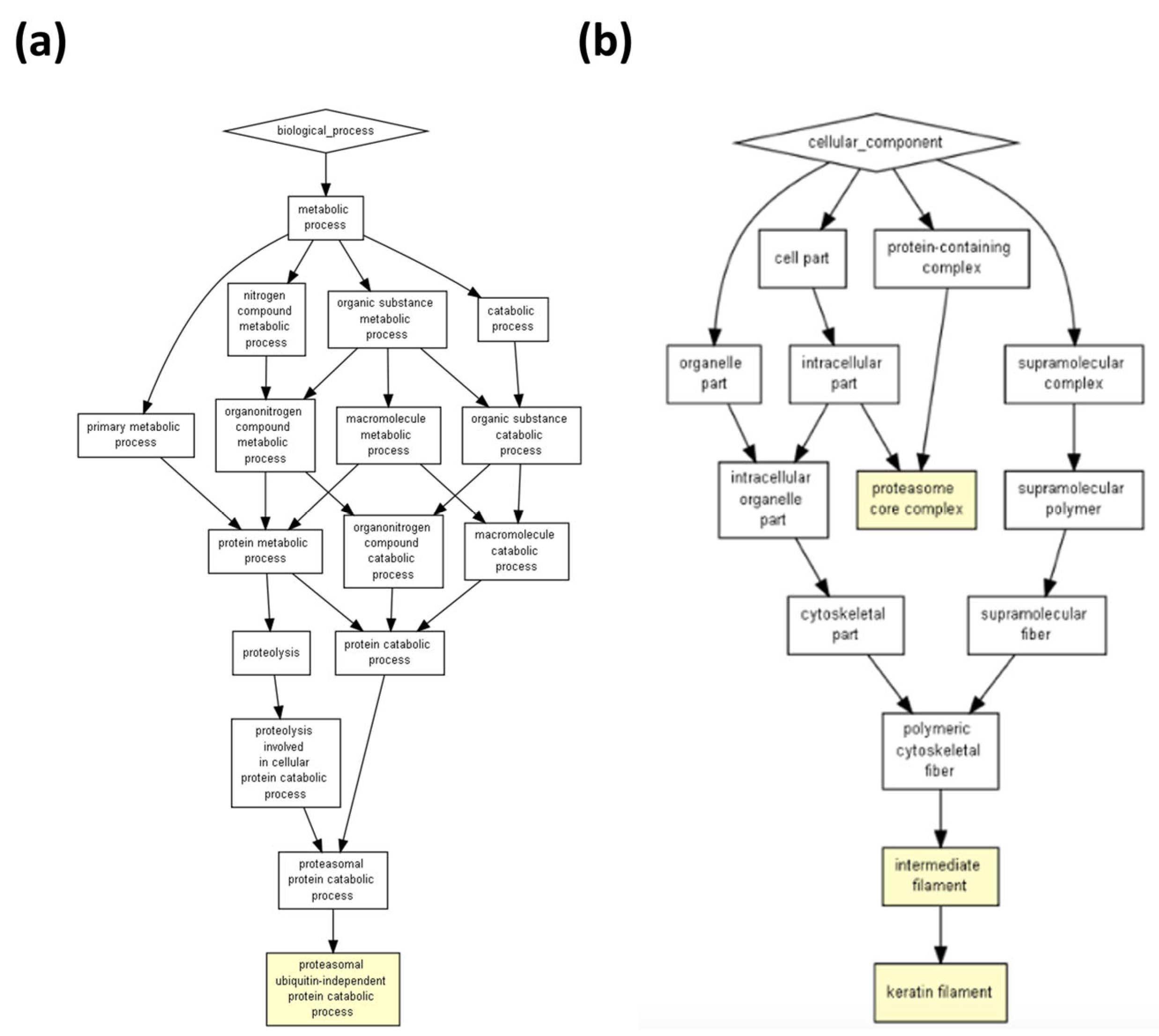

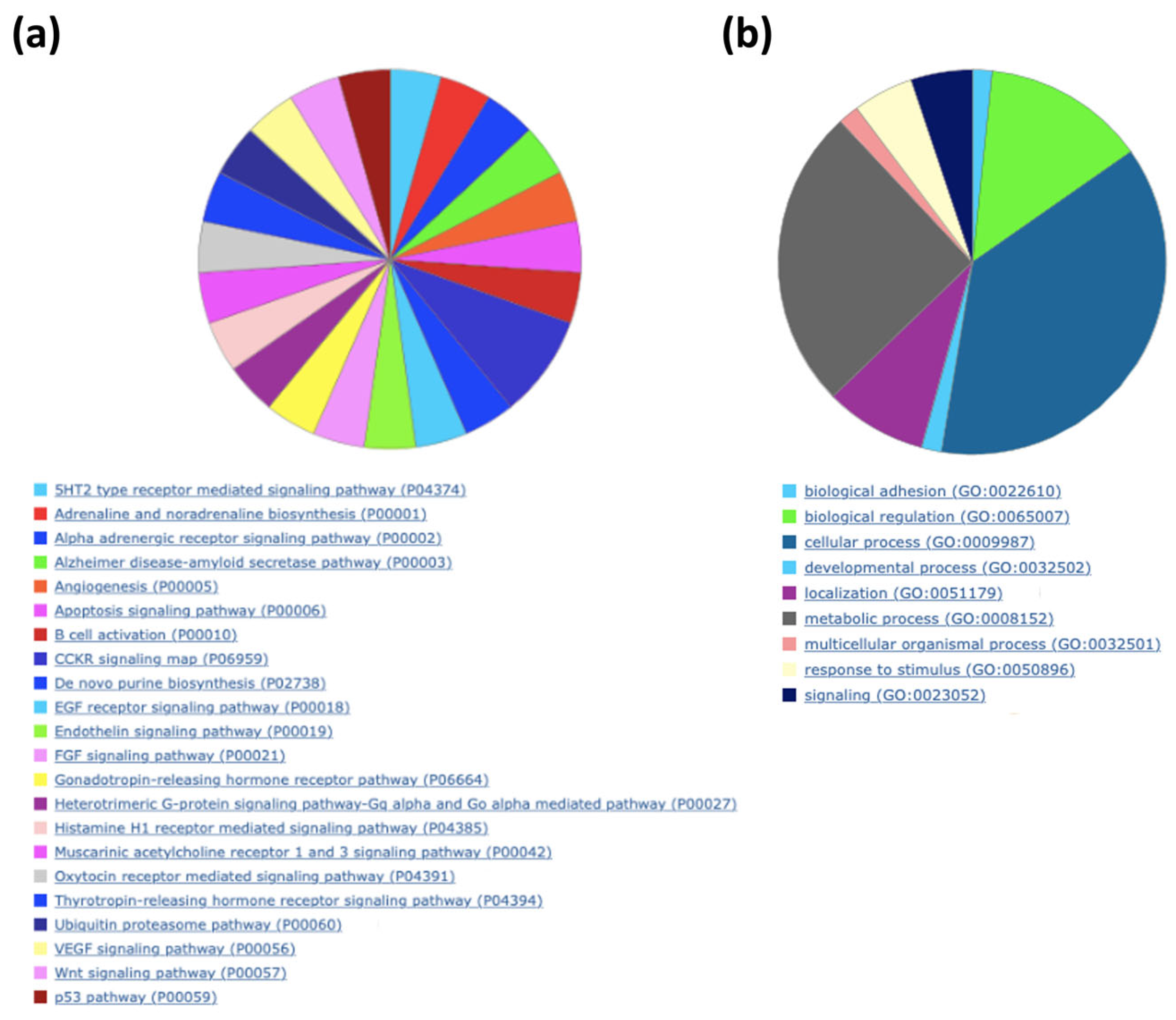

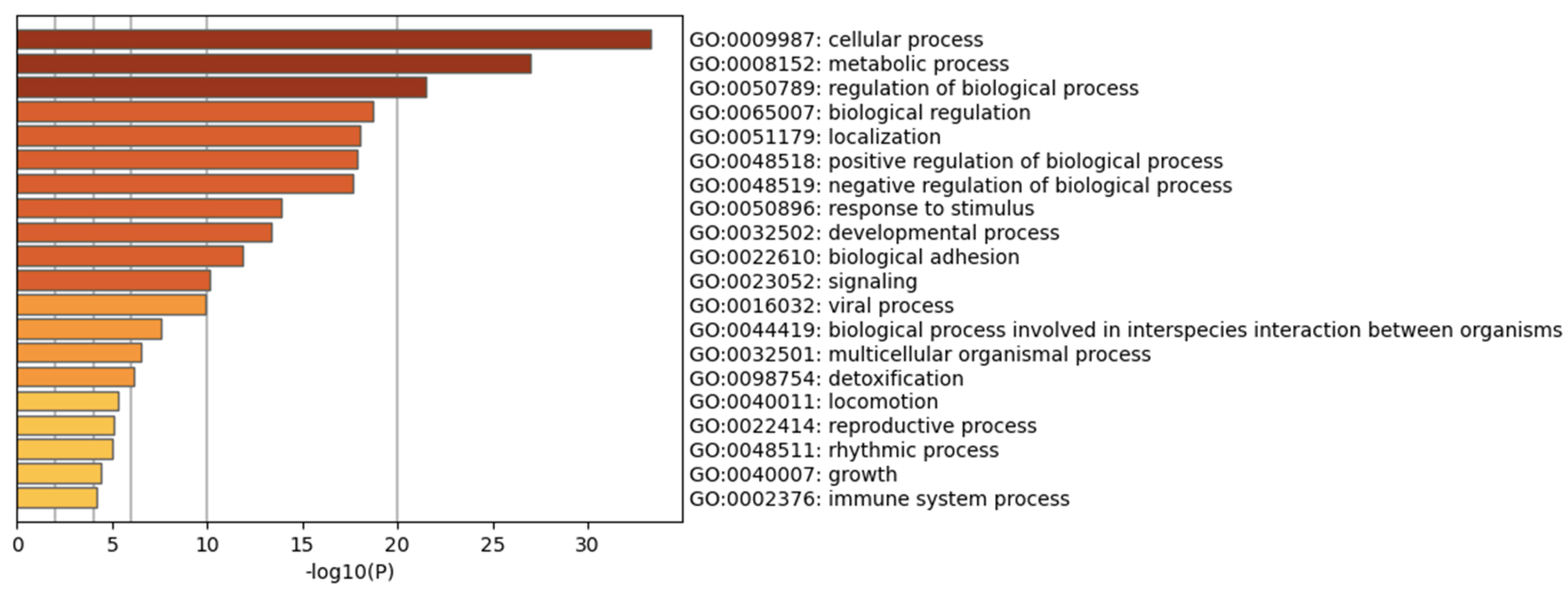


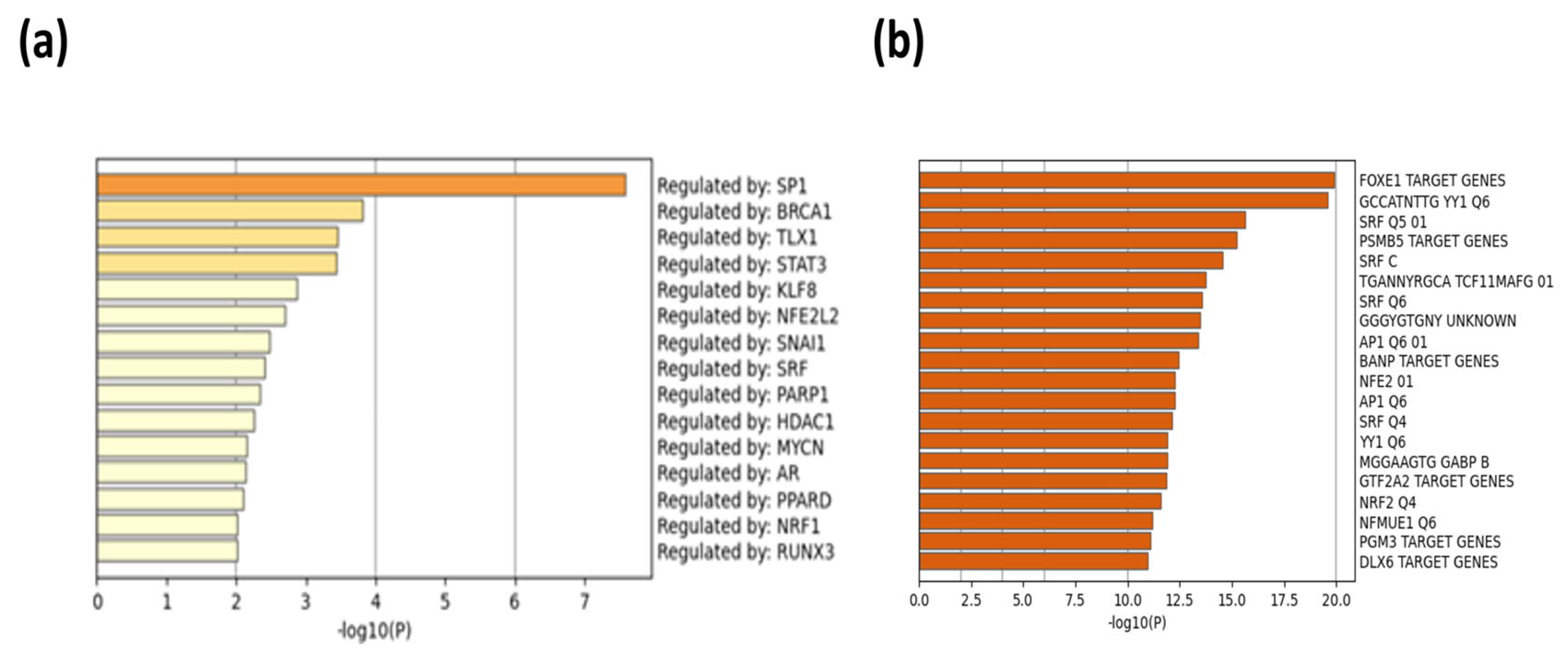
| Description | p-Value | FDR Q-Value | Enrichment (N, B, n, b) * |
|---|---|---|---|
| Keratinization | 0.000004 | 0.0301 | 1.61 (1283, 40, 757, 38) |
| Cornification | 0.00000598 | 0.0225 | 1.58 (1283, 44, 757, 41) |
| Exocytosis | 0.0000369 | 0.0923 | 1.22 (1283, 160, 891, 136) |
| Leukocyte degranulation | 0.0000521 | 0.098 | 1.25 (1283, 123, 891, 107) |
| Regulated exocytosis | 0.0000541 | 0.0813 | 1.22 (1283, 154, 891, 131) |
| Immune effector process | 0.0000609 | 0.0763 | 1.18 (1283, 169, 973, 151) |
| Granulocyte activation | 0.0000674 | 0.0724 | 1.25 (1283, 122, 891, 106) |
| Neutrophil activation | 0.0000674 | 0.0634 | 1.25 (1283, 122, 891, 106) |
| Myeloid cell activation involved in immune response | 0.0000675 | 0.0564 | 1.25 (1283, 122, 891, 106) |
| Response to chemokine | 0.000073 | 0.0549 | 28.51 (1283, 3, 45, 3) |
| Cellular response to chemokine | 0.000073 | 0.0499 | 28.51 (1283, 3, 45, 3) |
| Neutrophil degranulation | 0.0000846 | 0.053 | 1.25 (1283, 121, 891, 105) |
| Neutrophil activation involved in immune response | 0.0000846 | 0.0489 | 1.25 (1283, 121, 891, 105) |
| Secretion by cell | 0.0000888 | 0.0477 | 1.21 (1283, 173, 882, 144) |
| Myeloid leukocyte activation | 0.0000952 | 0.0477 | 1.24 (1283, 125, 891, 108) |
| Leukocyte activation involved in immune response | 0.000106 | 0.0499 | 1.20 (1283, 131, 973, 119) |
| Cell activation involved in immune response | 0.000106 | 0.047 | 1.20 (1283, 131, 973, 119) |
| Leukocyte activation | 0.000144 | 0.0603 | 1.18 (1283, 153, 973, 137) |
| Nervous system process | 0.000164 | 0.065 | 2.77 (1283, 36, 232, 18) |
| Regulation of intracellular estrogen receptor Signaling pathway | 0.00027 | 0.101 | 106.92 (1283, 4, 6, 2) |
| Secretion | 0.000384 | 0.137 | 1.19 (1283, 178, 882, 146) |
| PANTHER Pathways | # | # | Expected | Fold Enrichment | Raw p Value | FDR |
|---|---|---|---|---|---|---|
| Glycolysis | 20 | 9 | 1.35 | 6.69 | 0.0000484 | 0.00129 |
| Cytoskeletal regulation by Rho GTPase | 86 | 33 | 5.79 | 5.7 | 2.24 × 1013 | 1.79 × 1011 |
| Parkinson’s disease | 101 | 29 | 6.79 | 4.27 | 1.82 × 109 | 7.28 × 108 |
| Huntington’s disease | 152 | 39 | 10.22 | 3.81 | 5.37 × 1011 | 2.87 × 109 |
| Integrin signaling pathway | 200 | 39 | 13.45 | 2.9 | 4.26 × 108 | 0.00000136 |
| Inflammation mediated by chemokine and cytokine signaling pathways | 261 | 33 | 17.56 | 1.88 | 0.00146 | 0.0334 |
| Unclassified | 17,971 | 1103 | 1,208.89 | 0.91 | 1.64 × 1014 | 2.62 × 1012 |
| Description | p-Value | FDR q-Value | Enrichment (N, B, n, b) |
|---|---|---|---|
| Granulocyte activation | 0.00000222 | 0.0167 | 1.32 (1283, 122, 764, 96) |
| Neutrophil activation | 0.00000222 | 0.00835 | 1.32 (1283, 122, 764, 96) |
| Myeloid leukocyte activation | 0.00000234 | 0.00587 | 1.32 (1283, 125, 764, 98) |
| Cornification | 0.00000305 | 0.00573 | 1.53 (1283, 44, 764, 40) |
| Neutrophil degranulation | 0.00000305 | 0.00459 | 1.32 (1283, 121, 764, 95) |
| Neutrophil activation involved in immune response | 0.00000305 | 0.00383 | 1.32 (1283, 121, 764, 95) |
| Leukocyte degranulation | 0.00000439 | 0.00472 | 1.31 (1283, 123, 764, 96) |
| Myeloid cell activation involved in immune response | 0.00000599 | 0.00563 | 1.31 (1283, 122, 764, 95) |
| Regulated exocytosis | 0.00000966 | 0.00807 | 1.26 (1283, 154, 764, 116) |
| Leukocyte activation involved in immune response | 0.0000164 | 0.0123 | 1.28 (1283, 131, 764, 100) |
| Cell activation involved in immune response | 0.0000164 | 0.0112 | 1.28 (1283, 131, 764, 100) |
| Keratinization | 0.0000178 | 0.0111 | 1.51 (1283, 40, 764, 36) |
| Leukocyte activation | 0.0000287 | 0.0166 | 1.25 (1283, 153, 764, 114) |
| Cell activation | 0.0000751 | 0.0403 | 1.23 (1283, 161, 764, 118) |
| Immune effector process | 0.0000887 | 0.0445 | 1.22 (1283, 169, 764, 123) |
| Multi-organism process | 0.0000919 | 0.0432 | 1.19 (1283, 217, 764, 154) |
| Exocytosis | 0.0000964 | 0.0426 | 1.23 (1283, 160, 764, 117) |
| Secretion by cell | 0.000261 | 0.109 | 1.20 (1283, 173, 764, 124) |
| Secretion | 0.000561 | 0.222 | 1.19 (1283, 178, 764, 126) |
| Proteasomal ubiquitin-independent protein catabolic process | 0.000671 | 0.252 | 1.68 (1283, 14, 764, 14) |
| Regulation of response to biotic stimulus | 0.000671 | 0.24 | 1.68 (1283, 14, 764, 14) |
| Positive regulation of protein complex assembly | 0.000784 | 0.268 | 1.36 (1283, 52, 764, 42) |
| Immune system process | 0.000933 | 0.305 | 1.14 (1283, 287, 764, 194) |
| Description | p-Value | FDR q-Value | Enrichment (N, B, n, b) |
|---|---|---|---|
| Cornification | 0.00000183 | 0.0138 | 1.98 (1283, 44, 457, 31) |
| Keratinization | 0.00000738 | 0.0277 | 1.97 (1283, 40, 457, 28) |
| Catabolic process | 0.000202 | 0.507 | 1.30 (1283, 225, 457, 104) |
| Response to corticosteroid | 0.000537 | 1 | 2.25 (1283, 15, 457, 12) |
| Cellular catabolic process | 0.000566 | 0.851 | 1.28 (1283, 212, 457, 97) |
| Epidermis development | 0.000945 | 1 | 2.34 (1283, 12, 457, 10) |
| GO | Category | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|---|
| R-HSA-6798695 | Reactome Gene Sets | Neutrophil degranulation | 94 | 11.75 | −52.85 | −48.5 |
| R-HSA-8953897 | Reactome Gene Sets | Cellular responses to stimuli | 111 | 13.88 | −48.49 | −44.45 |
| R-HSA-194315 | Reactome Gene Sets | Signaling by Rho GTPases | 98 | 12.25 | −41.21 | −37.59 |
| WP3888 | WikiPathways | VEGFA-VEGFR2 signaling pathway | 72 | 9.00 | −35.01 | −31.51 |
| GO:0097435 | GO Biological Processes | Supramolecular fiber organization | 76 | 9.50 | −33.32 | −29.88 |
| R-HAS-6809371 | Reactome Gene Sets | Formation of the cornified envelope | 40 | 5.00 | −30.75 | −27.58 |
| R-HSA-72766 | Reactome Gene Sets | Translation | 52 | 6.50 | −27.18 | −24.11 |
| R-HSA-5653656 | Reactome Gene Sets | Vesicle-mediated transport | 76 | 9.50 | −25.95 | −23.02 |
| GO:1903311 | GO Biological Processes | Regulation of mRNA metabolic process | 47 | 5.88 | −22.52 | −19.70 |
| GO:0006457 | GO Biological Processes | Protein folding | 38 | 4.75 | −20.58 | −17.87 |
| hsa05132 | KEGG Pathway | Salmonella infection | 41 | 5.12 | −20.15 | −17.51 |
| GO:0043254 | GO Biological Processes | Regulation of protein-containing complex assembly | 50 | 6.25 | −19.00 | −16.43 |
| GO:0043086 | GO Biological Processes | Negative regulation of catalytic activity | 71 | 8.88 | −18.67 | −16.11 |
| GO:0060341 | GO Biological Processes | Regulation of cellular localization | 70 | 8.75 | −18.02 | −15.51 |
| GO:0051129 | GO Biological Processes | Negative regulation of cellular component organization | 64 | 8.00 | −17.61 | −15.16 |
| R-HAS-75153 | Reactome Gene Sets | Apoptotic execution phase | 19 | 2.38 | −16.53 | −14.20 |
| R-HSA-3371497 | Reactome Gene Sets | HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand | 19 | 2.38 | −16.00 | −13.72 |
| GO:0022613 | GO Biological Processes | Ribonucleoprotein complex biogenesis | 48 | 6.00 | −15.74 | −13.47 |
| Hsa04810 | KEGG Pathway | Regulation of actin cytoskeleton | 33 | 4.12 | −15.21 | −13.00 |
| M71 | Canonical Pathway | PID ILK Pathway | 17 | 2.12 | −15.14 | −12.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García Coronado, P.L.; Franco Molina, M.A.; Zárate Triviño, D.G.; Menchaca Arredondo, J.L.; Zapata Benavides, P.; Rodriguez Padilla, C. Putative Wound Healing Induction Functions of Exosomes Isolated from IMMUNEPOTENT CRP. Int. J. Mol. Sci. 2023, 24, 8971. https://doi.org/10.3390/ijms24108971
García Coronado PL, Franco Molina MA, Zárate Triviño DG, Menchaca Arredondo JL, Zapata Benavides P, Rodriguez Padilla C. Putative Wound Healing Induction Functions of Exosomes Isolated from IMMUNEPOTENT CRP. International Journal of Molecular Sciences. 2023; 24(10):8971. https://doi.org/10.3390/ijms24108971
Chicago/Turabian StyleGarcía Coronado, Paola Leonor, Moisés Armides Franco Molina, Diana Ginette Zárate Triviño, Jorge Luis Menchaca Arredondo, Pablo Zapata Benavides, and Cristina Rodriguez Padilla. 2023. "Putative Wound Healing Induction Functions of Exosomes Isolated from IMMUNEPOTENT CRP" International Journal of Molecular Sciences 24, no. 10: 8971. https://doi.org/10.3390/ijms24108971
APA StyleGarcía Coronado, P. L., Franco Molina, M. A., Zárate Triviño, D. G., Menchaca Arredondo, J. L., Zapata Benavides, P., & Rodriguez Padilla, C. (2023). Putative Wound Healing Induction Functions of Exosomes Isolated from IMMUNEPOTENT CRP. International Journal of Molecular Sciences, 24(10), 8971. https://doi.org/10.3390/ijms24108971





