The Impact of Human DNA Glycosylases on the Activity of DNA Polymerase β toward Various Base Excision Repair Intermediates
Abstract
1. Introduction
2. Results and Discussion
2.1. Interaction of Polβ with Model DNA Substrates
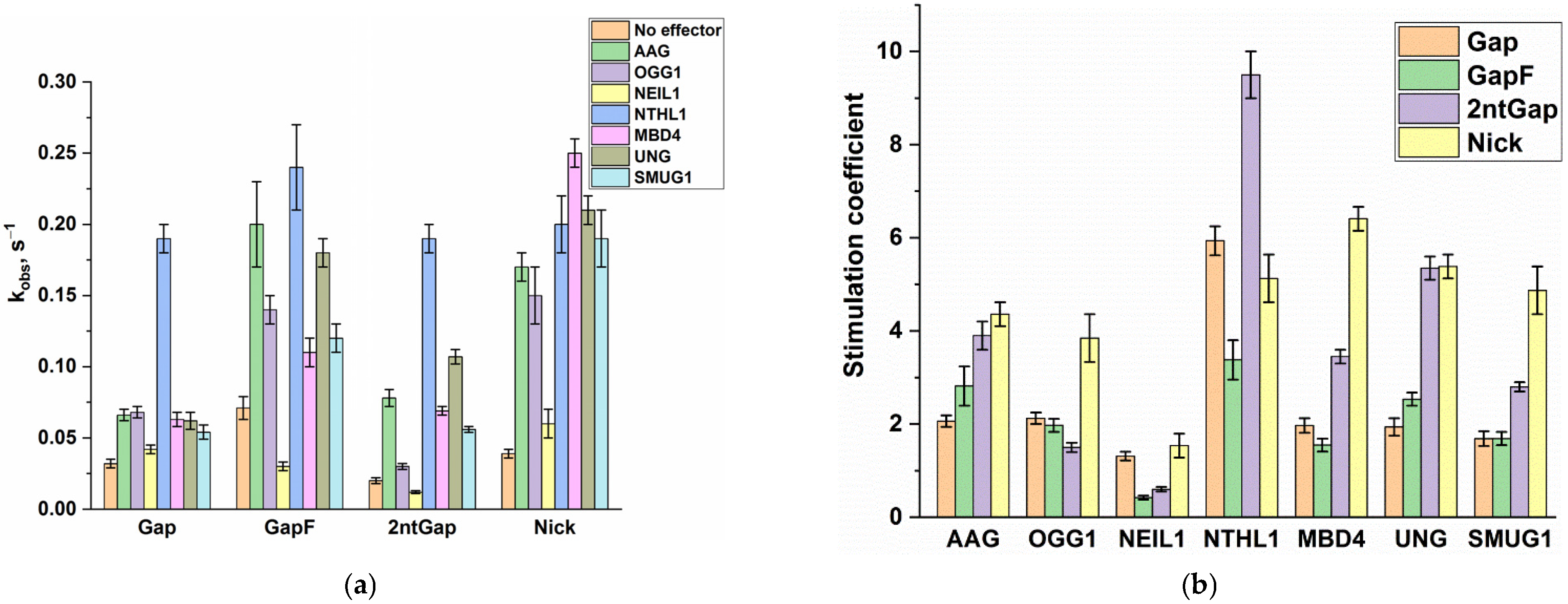
2.2. The Impact of DNA Glycosylases on Polβ Polymerase Activity as Revealed by Polyacrylamide Gel Electrophoresis (PAGE)
3. Materials and Methods
3.1. Protein Expression and Purification
3.2. Oligodeoxynucleotides
3.3. Polβ Polymerase Activity Assays Using PAGE Analysis
3.4. The Stopped-Flow Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Friedberg, E.C.; Walker, G.C.; Siede, W.; Wood, R.D.; Schultz, R.A.; Ellenberger, T. DNA Repair and Mutagenesis; ASM Press: Washington, DC, USA, 2005. [Google Scholar] [CrossRef]
- Lombard, D.B.; Chua, K.F.; Mostoslavsky, R.; Franco, S.; Gostissa, M.; Alt, F.W. DNA repair, genome stability, and aging. Cell 2005, 120, 497–512. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008, 9, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Hořejší, Z.; Koed, K.; Krämer, A.; Tort, F.; Zieger, K.; Guldberg, P.; Sehested, M.; Nesland, J.M.; Lukas, C.; et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005, 434, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Dianov, G.L.; Hübscher, U. Mammalian base excision repair: The forgotten archangel. Nucleic Acids Res. 2013, 41, 3483–3490. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Mosbaugh, D.W.; Bennett, S.E. Uracil-Excision DNA Repair. Prog. Nucleic Acid Res. Mol. Biol. 1994, 48, 315–370. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Kim, K. Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science 1995, 269, 699–702. [Google Scholar] [CrossRef]
- Piersen, C.E.; Prasad, R.; Wilson, S.H.; Lloyd, R.S. Evidence for an imino intermediate in the DNA polymerase β deoxyribose phosphate excision reaction. J. Biol. Chem. 1996, 271, 17811–17815. [Google Scholar] [CrossRef]
- Srivastava, D.K.; Berg, B.J.V.; Prasad, R.; Molina, J.T.; Beard, W.A.; Tomkinson, A.E.; Wilson, S.H. Mammalian abasic site base excision repair: Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 1998, 273, 21203–21209. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Kim, K.; Bogenhagen, D.F. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: An alternative pathway of base excision DNA repair. Mol. Cell. Biol. 1994, 14, 6187–6197. [Google Scholar] [CrossRef]
- Lindahl, T.; Satoh, M.S.; Dianov, G. Enzymes acting at strand interruptions in DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995, 347, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Frosina, G.; Fortini, P.; Rossi, O.; Carrozzino, F.; Raspaglio, G.; Cox, L.S.; Lane, D.P.; Abbondandolo, A.; Dogliotti, E. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 1996, 271, 9573–9578. [Google Scholar] [CrossRef] [PubMed]
- Klungland, A.; Lindahl, T. Second pathway for completion of human DNA base excision-repair: Reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 1997, 16, 3341–3348. [Google Scholar] [CrossRef] [PubMed]
- Moor, N.A.; Lavrik, O.I. Protein–Protein Interactions in DNA Base Excision Repair. Biochemistry 2018, 83, 411–422. [Google Scholar] [CrossRef]
- Endutkin, A.V.; Yudkina, A.V.; Sidorenko, V.S.; Zharkov, D.O. Transient protein–protein complexes in base excision repair. J. Biomol. Struct. Dyn. 2019, 37, 4407–4418. [Google Scholar] [CrossRef]
- Prasad, R.; Beard, W.A.; Batra, V.K.; Liu, Y.; Shock, D.D.; Wilson, S.H. A review of recent experiments on step-to-step “hand-off” of the DNA intermediates in mammalian base excision repair pathways. Mol. Biol. 2011, 45, 536–550. [Google Scholar] [CrossRef]
- Akbari, M.; Otterlei, M.; Diaz-Peña, J.; Aas, P.A.; Kavli, B.; Liabakk, N.B.; Hagen, L.; Imai, K.; Durandy, A.; Slupphaug, G.; et al. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 2004, 32, 5486–5498. [Google Scholar] [CrossRef]
- Braithwaite, E.K.; Kedar, P.S.; Stumpo, D.J.; Bertocci, B.; Freedman, J.H.; Samson, L.D.; Wilson, S.H. DNA polymerases β and λ mediate overlapping and independent roles in base excision repair in mouse embryonic fibroblasts. PLoS ONE 2010, 5, e12229. [Google Scholar] [CrossRef]
- Yudkina, A.V.; Endutkin, A.V.; Diatlova, E.A.; Moor, N.A.; Vokhtantsev, I.P.; Grin, I.R.; Zharkov, D.O. Displacement of slow-turnover DNA glycosylases by molecular traffic on DNA. Genes 2020, 11, 866. [Google Scholar] [CrossRef]
- Wiederhold, L.; Leppard, J.B.; Kedar, P.; Karimi-Busheri, F.; Rasouli-Nia, A.; Weinfeld, M.; Tomkinson, A.E.; Izumi, T.; Prasad, R.; Wilson, S.H.; et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004, 15, 209–220. [Google Scholar] [CrossRef]
- Hegde, M.L.; Hegde, P.M.; Arijit, D.; Boldogh, I.; Mitra, S. Human DNA glycosylase NEIL1’s interactions with downstream repair proteins is critical for efficient repair of oxidized DNA base damage and enhanced cell survival. Biomolecules 2012, 2, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Wiederhold, L.; Leppard, J.B.; Kedar, P.; Prasad, R.; Wang, H.; Boldogh, I.; Karimi-Busheri, F.; Weinfeld, M.; Tomkinson, A.E.; et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair. 2006, 5, 1439–1448. [Google Scholar] [CrossRef]
- Aspinwall, R.; Rothwell, D.G.; Roldan-Arjona, T.; Anselmino, C.; Ward, C.J.; Cheadle, J.P.; Sampson, J.R.; Lindahl, T.; Harris, P.C.; Hickson, I.D. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl. Acad. Sci. USA 1997, 94, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, C.A.; Tsai, M.D. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase β. Biochemistry 2002, 41, 11226–11235. [Google Scholar] [CrossRef] [PubMed]
- Arndt, J.W.; Gong, W.; Zhong, X.; Showalter, A.K.; Liu, J.; Dunlap, C.A.; Lin, Z.; Paxson, C.; Tsai, M.D.; Chan, M.K. Insight into the catalytic mechanism of DNA polymerase β: Structures of intermediate complexes. Biochemistry 2001, 40, 5368–5375. [Google Scholar] [CrossRef]
- Bakhtina, M.; Roettger, M.P.; Kumar, S.; Tsai, M.D. A unified kinetic mechanism applicable to multiple DNA polymerases. Biochemistry 2007, 46, 5463–5472. [Google Scholar] [CrossRef] [PubMed]
- Doublié, S.; Sawaya, M.R.; Ellenberger, T. An open and closed case for all polymerases. Structure 1999, 7, R31–R35. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Prasad, R.; Wilson, S.H.; Kraut, J.; Pelletier, H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: Evidence for an induced fit mechanism. Biochemistry 1997, 36, 11205–11215. [Google Scholar] [CrossRef] [PubMed]
- Balbo, P.B.; Wang, E.C.W.; Tsai, M.D. Kinetic mechanism of active site assembly and chemical catalysis of DNA polymerase β. Biochemistry 2011, 50, 9865–9875. [Google Scholar] [CrossRef]
- Demple, B.; Harrison, L. Repair of oxidative damage to DNA: Enzymology and biology. Annu. Rev. Biochem. 1994, 63, 915–948. [Google Scholar] [CrossRef]
- Weinfeld, M.; Mani, R.S.; Abdou, I.; Aceytuno, R.D.; Glover, J.N.M. Tidying up loose ends: The role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem. Sci. 2011, 36, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Kladova, O.A.; Alekseeva, I.V.; Saparbaev, M.; Fedorova, O.S.; Kuznetsov, N.A. Modulation of the apurinic/apyrimidinic endonuclease activity of human APE1 and of its natural polymorphic variants by base excision repair proteins. Int. J. Mol. Sci. 2020, 21, 7147. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, A.A.; Iakovlev, D.A.; Misovets, I.V.; Ishchenko, A.A.; Saparbaev, M.K.; Kuznetsov, N.A.; Fedorova, O.S. Pre-steady-state kinetic analysis of damage recognition by human single-strand selective monofunctional uracil-DNA glycosylase SMUG1. Mol. Biosyst. 2017, 13, 2638–2649. [Google Scholar] [CrossRef] [PubMed]
- Yakovlev, D.A.; Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Search for Modified DNA Sites with the Human Methyl-CpG-Binding Enzyme MBD4. Acta Nat. 2017, 9, 26–37. [Google Scholar] [CrossRef]
- Kuznetsov, N.A.; Koval, V.V.; Zharkov, D.O.; Nevinsky, G.A.; Douglas, K.T.; Fedorova, O.S. Kinetics of substrate recognition and cleavage by human 8-oxoguanine-DNA glycosylase. Nucleic Acids Res. 2005, 33, 3919–3931. [Google Scholar] [CrossRef]
- Kuznetsov, N.A.; Kiryutin, A.S.; Kuznetsova, A.A.; Panov, M.S.; Barsukova, M.O.; Yurkovskaya, A.V.; Fedorova, O.S. The formation of catalytically competent enzyme–substrate complex is not a bottleneck in lesion excision by human alkyladenine DNA glycosylase. J. Biomol. Struct. Dyn. 2017, 35, 950–967. [Google Scholar] [CrossRef]
- Kladova, O.A.; Grin, I.R.; Fedorova, O.S.; Kuznetsov, N.A.; Zharkov, D.O. Conformational Dynamics of Damage Processing by Human DNA Glycosylase NEIL1. J. Mol. Biol. 2019, 431, 1098–1112. [Google Scholar] [CrossRef]
- Kladova, O.A.; Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Mutational and kinetic analysis of lesion recognition by Escherichia coli endonuclease VIII. Genes 2017, 8, 140. [Google Scholar] [CrossRef]
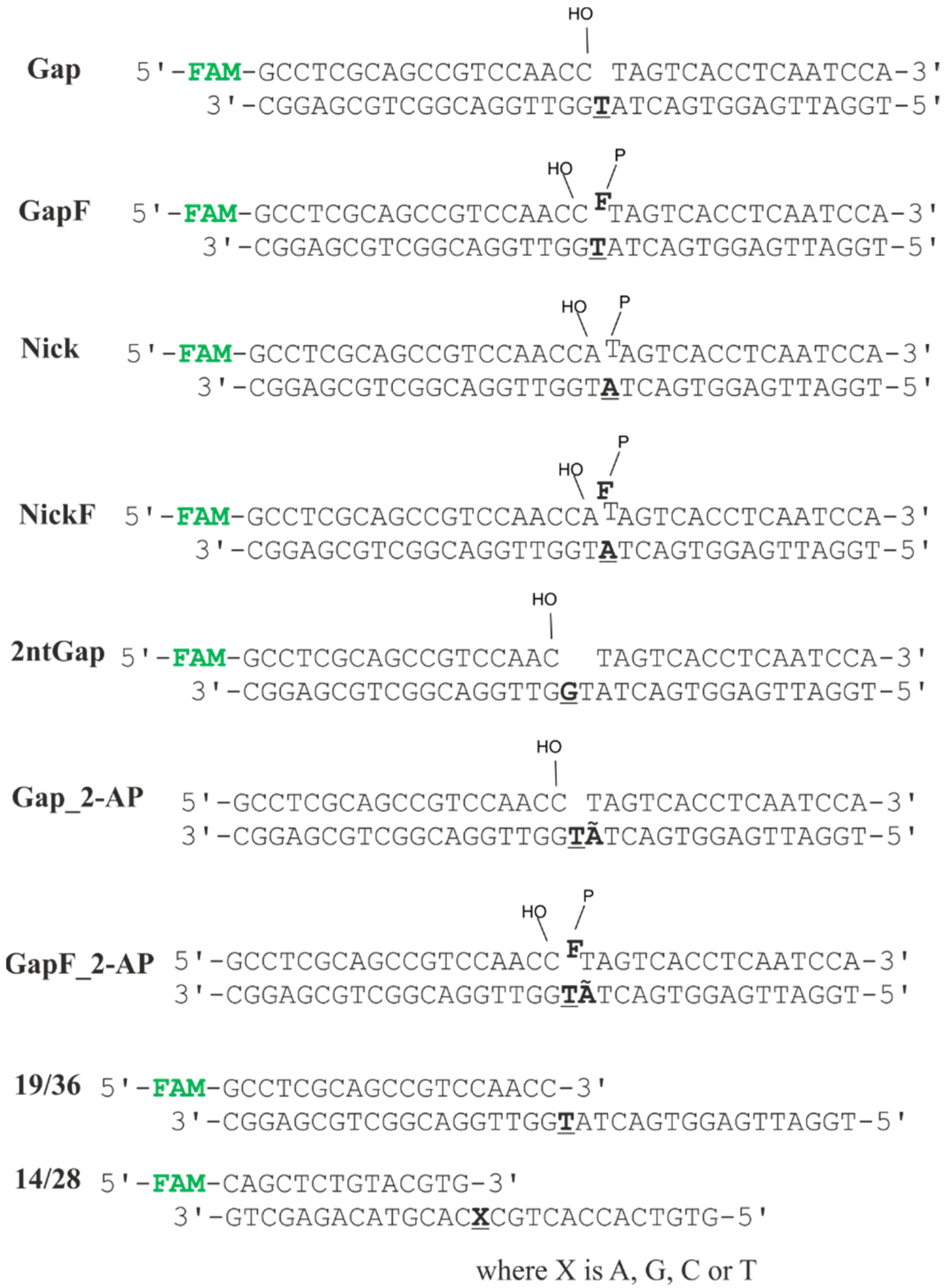
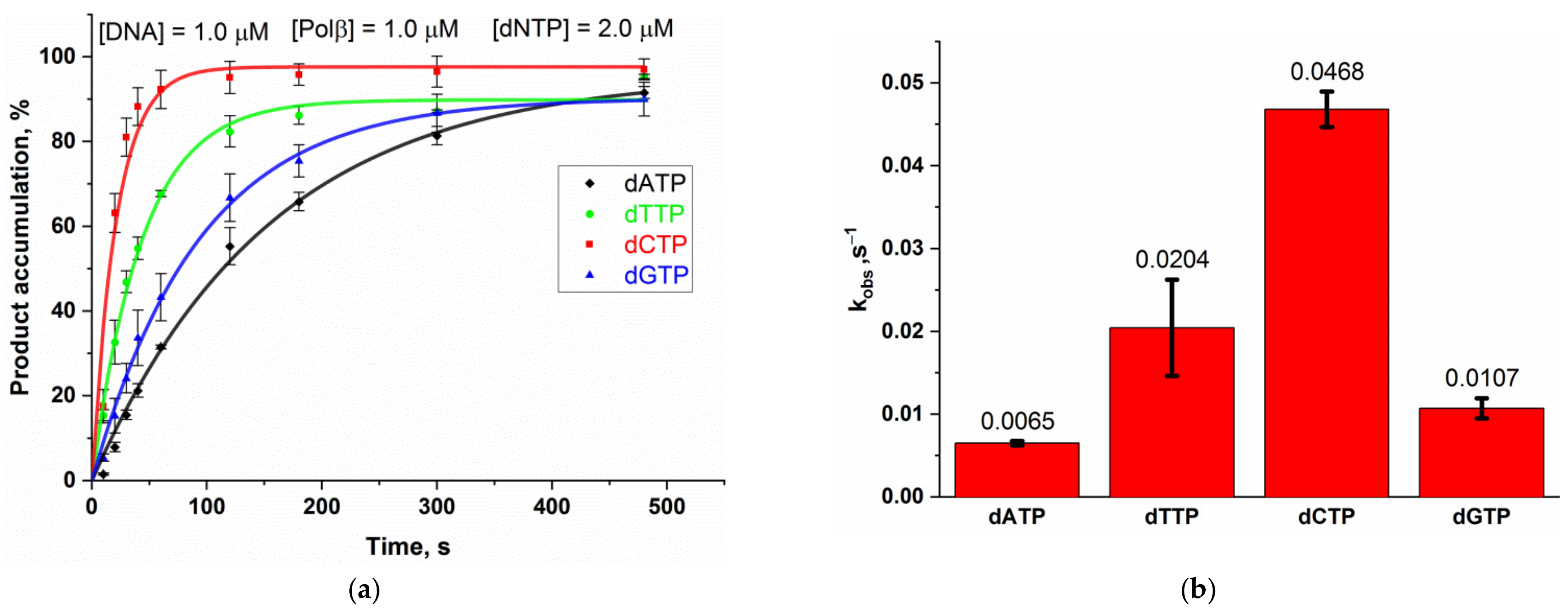
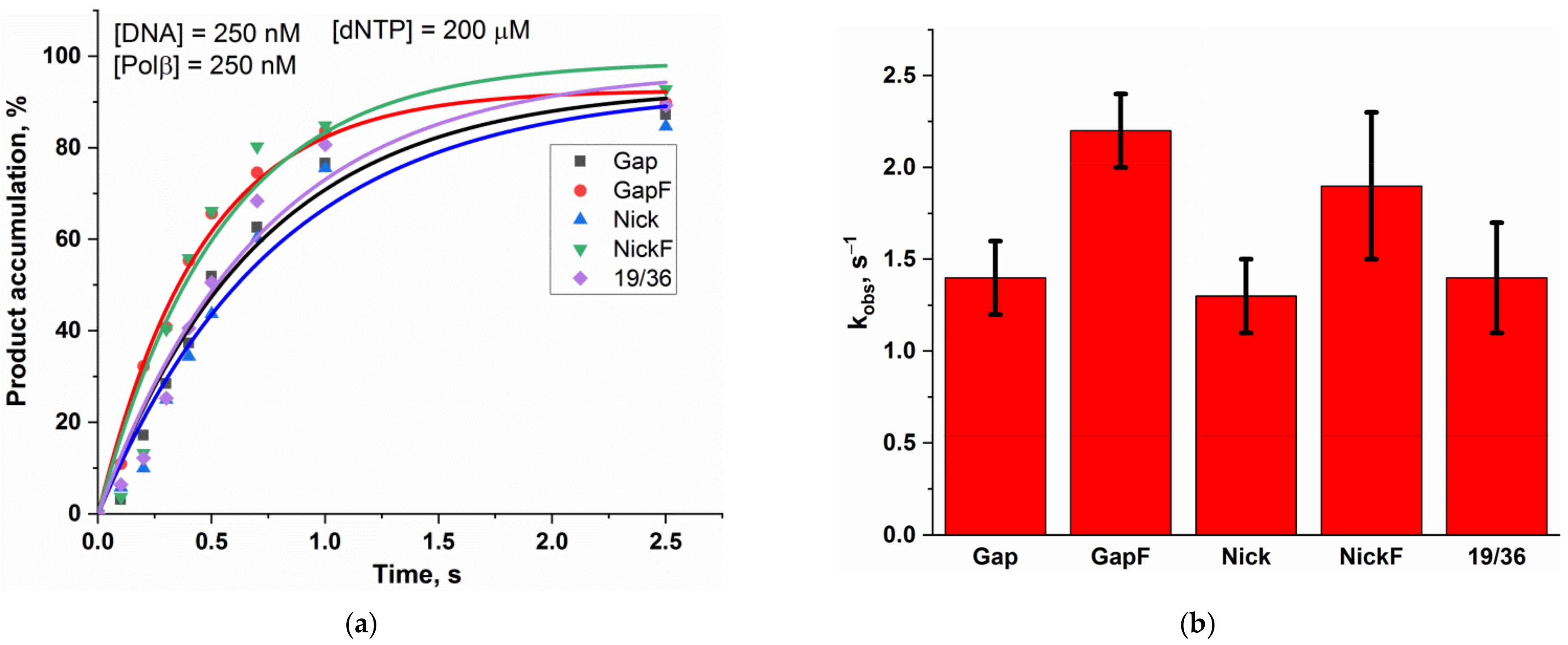

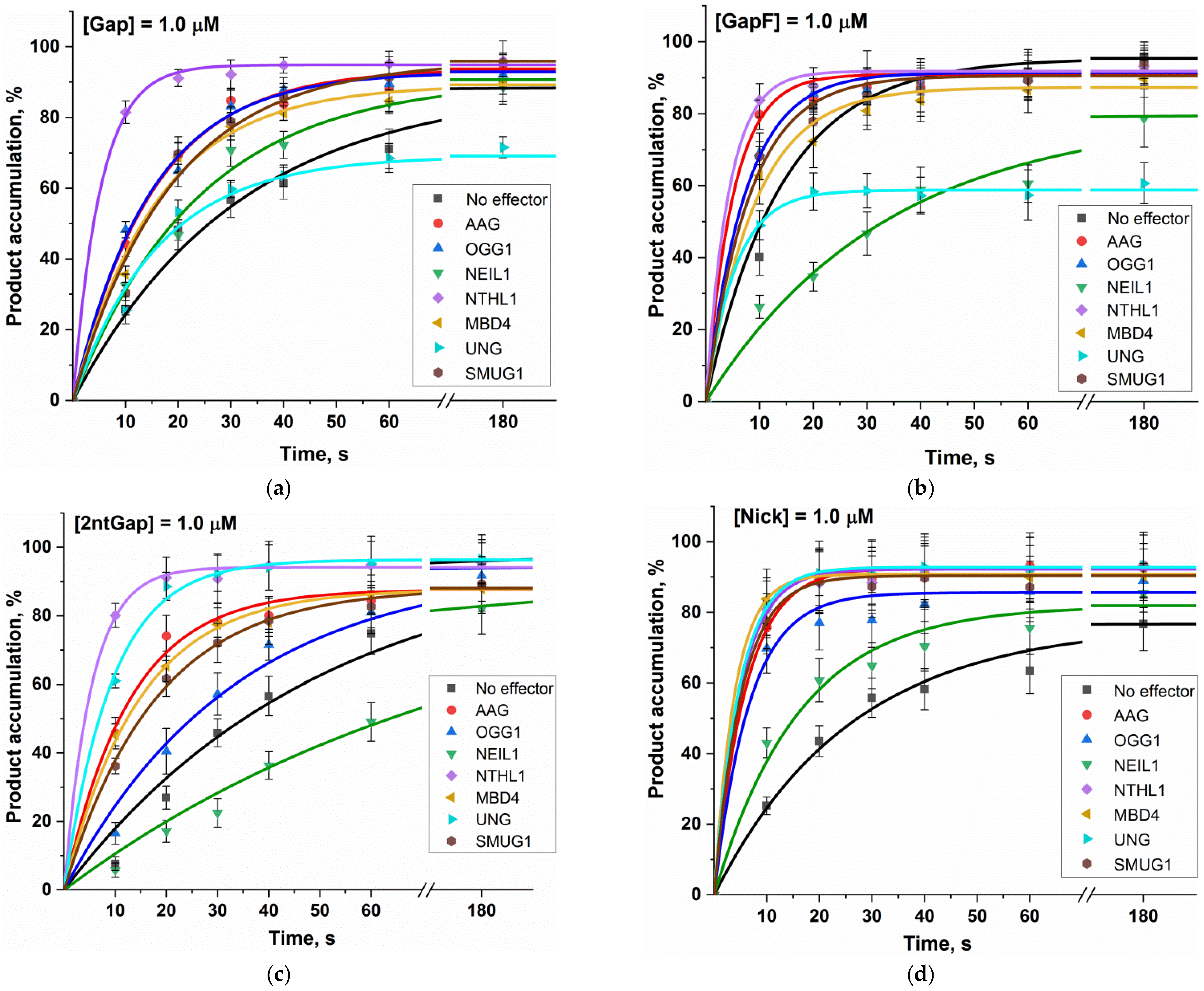
| Enzyme | Substrate Specificity | Structural Superfamily | Mono- (M) or Bi- (B) Functional * | Known Interaction with Polβ |
|---|---|---|---|---|
| UNG | U in single-stranded (ss) and double-stranded (ds) DNA | α/β-fold | M | Interaction has been revealed by an immunoprecipitation assay [18]. |
| SMUG1 | U in ss- and dsDNA | α/β-fold | M | – |
| TDG | T, U, 3,N4-ethenoC, and oxidized/deaminated derivatives of 5-methylC opposite to G in dsDNA | α/β-fold | M | – |
| MBD4 | T and U opposite to G in dsDNA | HhH | M | – |
| NTHL1 | Oxidized pyrimidines in dsDNA | HhH | B | – |
| MYH | A and 2-OH-A opposite to G or 8-oxoguanine in dsDNA | HhH | M | – |
| OGG1 | 8-oxoguanine and FapyG opposite to C in dsDNA | HhH | B | Interaction has been detected by an immunoprecipitation assay [19]. It has been shown that Polβ can displace OGG1 from DNA [20]. |
| AAG | Ring-alkylated purines, hypoxanthine, and 1,N6-ethenoA in ss and dsDNA | FMT_C | M | Interaction has been registered by an immunoprecipitation assay [19]. |
| NEIL1 | Oxidized pyrimidines and purines, ring-open N7-alkylated G modifications, and psoralen cross-links in ss- and dsDNA | H2TH | B | Interaction has been revealed by far-western [21] and immunoprecipitation analyses [22]. Amino acid residues 312–349 of NEIL1 and an N-terminal part of Polβ (residues 1–140) are reported to be critical for this interaction [22]. It has been found that Polβ can displace NEIL1 from DNA [20]. |
| NEIL2 | Oxidized pyrimidines and purines in bubble DNA | H2TH | B | Interaction has been revealed by far-western and immunoprecipitation analyses. It has been shown that the N-terminal domain of NEIL2 (amino acid residues 1–198) interacts with an N-terminal part of Polβ (residues 1–140) [23]. |
| NEIL3 | Oxidized pyrimidines and purines in ssDNA | H2TH | B | – |
| k1 (SFF), s−1 | k2 (SFF), s−1 | kobs (PAGE-RQF), s−1 | |
|---|---|---|---|
| Gap_2-AP | 58 ± 1 | 1.44 ± 0.01 | 1.4 ± 0.2 |
| GapF_2-AP | 73 ± 1 | 3.96 ± 0.02 | 2.2 ± 0.2 |
| No Effector | AAG | OGG1 | NEIL1 | NTHL1 | MBD4 | UNG | SMUG1 | |
|---|---|---|---|---|---|---|---|---|
| Gap | 0.032 ± 0.003 | 0.066 ± 0.004 | 0.068 ± 0.004 | 0.042 ± 0.003 | 0.19 ± 0.01 | 0.063 ± 0.005 | 0.062 ± 0.006 | 0.054 ± 0.005 |
| GapF | 0.071 ± 0.008 | 0.20 ± 0.03 | 0.14 ± 0.01 | 0.030 ± 0.003 | 0.24 ± 0.03 | 0.11 ± 0.01 | 0.18 ± 0.01 | 0.12 ± 0.01 |
| 2ntGap | 0.020 ± 0.002 | 0.078 ± 0.006 | 0.030 ± 0.002 | 0.012 ± 0.001 | 0.19 ± 0.01 | 0.069 ± 0.003 | 0.107 ± 0.005 | 0.056 ± 0.002 |
| Nick | 0.039 ± 0.003 | 0.17 ± 0.01 | 0.15 ± 0.02 | 0.06 ± 0.01 | 0.20 ± 0.02 | 0.25 ± 0.01 | 0.21 ± 0.01 | 0.19 ± 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakman, A.S.; Boichenko, S.S.; Kuznetsova, A.A.; Ishchenko, A.A.; Saparbaev, M.; Kuznetsov, N.A. The Impact of Human DNA Glycosylases on the Activity of DNA Polymerase β toward Various Base Excision Repair Intermediates. Int. J. Mol. Sci. 2023, 24, 9594. https://doi.org/10.3390/ijms24119594
Bakman AS, Boichenko SS, Kuznetsova AA, Ishchenko AA, Saparbaev M, Kuznetsov NA. The Impact of Human DNA Glycosylases on the Activity of DNA Polymerase β toward Various Base Excision Repair Intermediates. International Journal of Molecular Sciences. 2023; 24(11):9594. https://doi.org/10.3390/ijms24119594
Chicago/Turabian StyleBakman, Artemiy S., Stanislav S. Boichenko, Aleksandra A. Kuznetsova, Alexander A. Ishchenko, Murat Saparbaev, and Nikita A. Kuznetsov. 2023. "The Impact of Human DNA Glycosylases on the Activity of DNA Polymerase β toward Various Base Excision Repair Intermediates" International Journal of Molecular Sciences 24, no. 11: 9594. https://doi.org/10.3390/ijms24119594
APA StyleBakman, A. S., Boichenko, S. S., Kuznetsova, A. A., Ishchenko, A. A., Saparbaev, M., & Kuznetsov, N. A. (2023). The Impact of Human DNA Glycosylases on the Activity of DNA Polymerase β toward Various Base Excision Repair Intermediates. International Journal of Molecular Sciences, 24(11), 9594. https://doi.org/10.3390/ijms24119594







