Novel Functional Features of cGMP Substrate Proteins IRAG1 and IRAG2
Abstract
1. Introduction
2. Functional Features of IRAG1
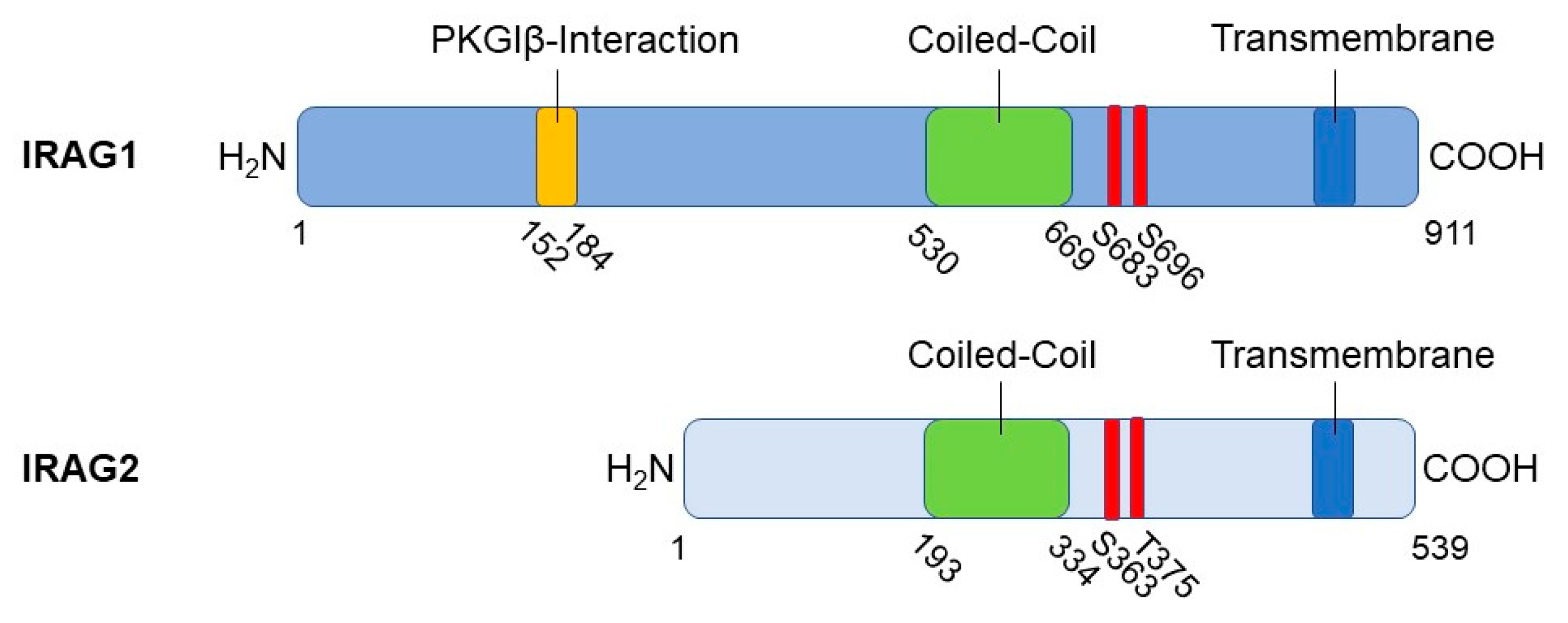
2.1. Structure, Interaction Partners, and Cellular Functions of IRAG1
2.2. Expression Pattern and Localization of IRAG1
2.3. Impact of IRAG1 on Ca2+ Signaling
2.4. (Patho-)Physiological Functions of IRAG1
2.4.1. IRAG1 and the Gastrointestinal System
2.4.2. IRAG1 and (Cardio-)Vascular System
2.4.3. IRAG1 and Cancer
2.4.4. Further (Patho-)Physiological Functions of IRAG1
2.5. Polymorphisms of IRAG1 Gene
2.6. Significance of IRAG1 as a Diagnostical/Prognostic Marker
3. Functional Features of IRAG2
3.1. Structure of IRAG2
3.2. Expression Pattern and Localization of IRAG2
3.3. Cellular Functions of IRAG2
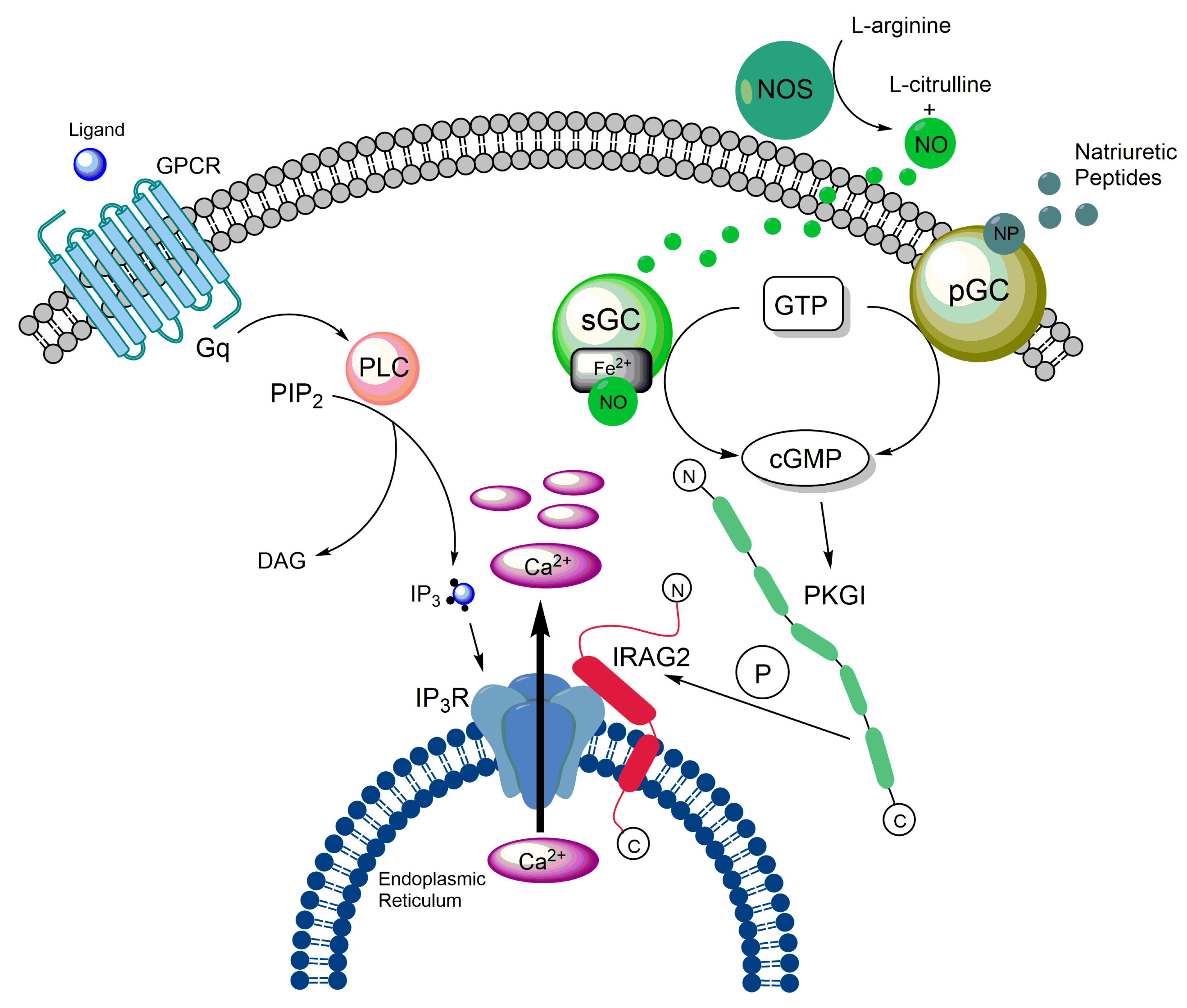
3.4. Impact of IRAG2 on Ca2+ Signaling
3.5. IRAG2 as a Substrate of cGMP-Dependent Protein Kinase I
3.6. (Patho-)Physiological Functions of IRAG2
3.6.1. Function of IRAG2 in Intestinal Type 2 Immunity
3.6.2. Function of IRAG2 on HCN Channels
3.6.3. Potential Role of IRAG2 in Taste-Signal Transduction
3.6.4. Function of IRAG2 in Exocrine Pancreatic Acinar Cells
3.6.5. Function of IRAG2 in Platelets
3.7. Significance of IRAG2 as a Prognostic Marker of Cancer
3.8. Significance of IRAG2 Polymorphisms
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ammendola, A.; Geiselhöringer, A.; Hofmann, F.; Schlossmann, J. Molecular Determinants of the Interaction between the Inositol 1,4,5-Trisphosphate Receptor-associated cGMP Kinase Substrate (IRAG) and cGMP Kinase Iβ. J. Biol. Chem. 2001, 276, 24153–24159. [Google Scholar] [CrossRef] [PubMed]
- Schlossmann, J.; Ammendola, A.; Ashman, K.; Zong, X.; Huber, A.; Neubauer, G.; Wang, G.-X.; Allescher, H.-D.; Korth, M.; Wilm, M.; et al. Regulation of intracellular calcium by a signalling complex of IRAG, IP3 receptor and cGMP kinase Iβ. Nature 2000, 404, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Geiselhöringer, A.; Werner, M.; Sigl, K.; Smital, P.; Wörner, R.; Acheo, L.; Stieber, J.; Weinmeister, P.; Feil, R.; Feil, S.; et al. IRAG is essential for relaxation of receptor-triggered smooth muscle contraction by cGMP kinase. EMBO J. 2004, 23, 4222–4231. [Google Scholar] [CrossRef] [PubMed]
- Desch, M.; Sigl, K.; Hieke, B.; Salb, K.; Kees, F.; Bernhard, D.; Jochim, A.; Spiessberger, B.; Höcherl, K.; Feil, R.; et al. IRAG determines nitric oxide- and atrial natriuretic peptide-mediated smooth muscle relaxation. Cardiovasc. Res. 2010, 86, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Antl, M.; Von Bruhl, M.-L.; Eiglsperger, C.; Werner, M.; Konrad, I.; Kocher, T.; Wilm, M.; Hofmann, F.; Massberg, S.; Schlossmann, J.; et al. IRAG mediates NO/cGMP-dependent inhibition of platelet aggregation and thrombus formation. Blood 2007, 109, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Shaughnessy, J.D.; Largaespada, D.A.; Tian, E.; Fletcher, C.F.; Cho, B.C.; Vyas, P.; Jenkins, N.A.; Copeland, N.G. Mrvil, a common MRV integration site in BXH2 myeloid leukemias, encodes a protein with homology to a lymphoid-restricted membrane protein Jaw1. Oncogene 1999, 18, 2069–2084. [Google Scholar] [CrossRef]
- Behrens, T.W.; Jagadeesh, J.; Scherle, P.; Kearns, G.; Yewdell, J.; Staudt, L.M. Jaw1, A lymphoid-restricted membrane protein localized to the endoplasmic reticulum. J. Immunol. 1994, 153, 682–690. [Google Scholar] [CrossRef]
- Shindo, Y.; Kim, M.-R.; Miura, H.; Yuuki, T.; Kanda, T.; Hino, A.; Kusakabe, Y. Lrmp/Jaw1 is Expressed in Sweet, Bitter, and Umami Receptor-Expressing Cells. Chem. Senses 2010, 35, 171–177. [Google Scholar] [CrossRef]
- Prüschenk, S.; Majer, M.; Schreiber, R.; Schlossmann, J. IRAG2 Interacts with IP3-Receptor Types 1, 2, and 3 and Regulates Intracellular Ca2+ in Murine Pancreatic Acinar Cells. Int. J. Mol. Sci. 2021, 22, 13409. [Google Scholar] [CrossRef]
- Tedoldi, S.; Paterson, J.; Cordell, J.; Tan, S.-Y.; Jones, M.; Manek, S.; Tos, A.D.; Roberton, H.; Masir, N.; Natkunam, Y.; et al. Jaw1/LRMP, a germinal centre-associated marker for the immunohistological study of B-cell lymphomas. J. Pathol. 2006, 209, 454–463. [Google Scholar] [CrossRef]
- Duarte, N.; Lundholm, M.; Holmberg, D. The Idd6.2 diabetes susceptibility region controls defective expression of the Lrmp gene in nonobese diabetic (NOD) mice. Immunogenetics 2007, 59, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Grimm, C.H.; Rogner, U.C.; Avner, P. LrmpandBcat1are Candidates for the Type I Diabetes Susceptibility Locus Idd6. Autoimmunity 2003, 36, 241–246. [Google Scholar] [CrossRef] [PubMed]
- von Werder, A.; Mayr, M.; Schneider, G.; Oesterle, D.; Fritsch, R.M.; Seidler, B.; Schlossmann, J.; Hofmann, F.; Schemann, M.; Allescher, H.D.; et al. Truncated IRAG variants modulate cGMP-mediated inhibition of human colonic smooth muscle cell contraction. Am. J. Physiol. Cell Physiol. 2011, 301, C1445–C1457. [Google Scholar] [CrossRef] [PubMed]
- Casteel, D.E.; Boss, G.R.; Pilz, R.B. Identification of the Interface between cGMP-dependent Protein Kinase Iβ and Its Interaction Partners TFII-I and IRAG Reveals a Common Interaction Motif. J. Biol. Chem. 2005, 280, 38211–38218. [Google Scholar] [CrossRef]
- Casteel, D.E.; Zhang, T.; Zhuang, S.; Pilz, R.B. cGMP-dependent protein kinase anchoring by IRAG regulates its nuclear translocation and transcriptional activity. Cell. Signal. 2008, 20, 1392–1399. [Google Scholar] [CrossRef]
- Majer, M.; Prueschenk, S.; Schlossmann, J. Loss of PKGIβ/IRAG1 Signaling Causes Anemia-Associated Splenomegaly. Int. J. Mol. Sci. 2021, 22, 5458. [Google Scholar] [CrossRef]
- Biswas, S.; Kojonazarov, B.; Hadzic, S.; Majer, M.; Bajraktari, G.; Novoyatleva, T.; Ghofrani, H.A.; Grimminger, F.; Seeger, W.; Weissmann, N.; et al. IRAG1 Deficient Mice Develop PKG1β Dependent Pulmonary Hypertension. Cells 2020, 9, 2280. [Google Scholar] [CrossRef]
- Peters, C.H.; Myers, M.E.; Juchno, J.; Haimbaugh, C.; Bichraoui, H.; Du, Y.; Bankston, J.R.; Walker, L.A.; Proenza, C. Isoform-specific regulation of HCN4 channels by a family of endoplasmic reticulum proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 18079–18090. [Google Scholar] [CrossRef]
- Geiselhöringer, A.; Gaisa, M.; Hofmann, F.; Schlossmann, J. Distribution of IRAG and cGKI-isoforms in murine tissues. FEBS Lett. 2004, 575, 19–22. [Google Scholar] [CrossRef]
- Ali, S.; Solano, A.S.; Gonzales, A.L.; Thakore, P.; Krishnan, V.; Yamasaki, E.; Earley, S. Nitric oxide signals through IRAG to inhibit TRPM4 channels and dilate cerebral arteries. Function 2021, 2, zqab051. [Google Scholar] [CrossRef]
- Fritsch, R.M.; Saur, D.; Kurjak, M.; Oesterle, D.; Schlossmann, J.; Geiselhöringer, A.; Hofmann, F.; Allescher, H.-D. InsP3R-associated cGMP Kinase Substrate (IRAG) Is Essential for Nitric Oxide-induced Inhibition of Calcium Signaling in Human Colonic Smooth Muscle. J. Biol. Chem. 2004, 279, 12551–12559. [Google Scholar] [CrossRef] [PubMed]
- Masuda, W.; Betzenhauser, M.J.; Yule, D.I. InsP3R-associated cGMP Kinase Substrate Determines Inositol 1,4,5-Trisphosphate Receptor Susceptibility to Phosphoregulation by Cyclic Nucleotide-dependent Kinases. J. Biol. Chem. 2010, 285, 37927–37938. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, A.; Klatt, P.; Massberg, S.; Ny, L.; Sausbier, M.; Hirneiß, C.; Wang, G.; Korth, M.; Aszódi, A.; Andersson, K.; et al. Defective smooth muscle regulation in cGMP kinase I-deficient mice. EMBO J. 1998, 17, 3045–3051. [Google Scholar] [CrossRef] [PubMed]
- Frei, E.; Huster, M.; Smital, P.; Schlossmann, J.; Hofmann, F.; Wegener, J.W. Calcium-dependent and calcium-independent inhibition of contraction by cGMP/cGKI in intestinal smooth muscle. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 297, G834–G839. [Google Scholar] [CrossRef] [PubMed]
- Koehler, K.; Hmida, D.; Schlossmann, J.; Landgraf, D.; Reisch, N.; Schuelke, M.; Huebner, A. Homozygous mutation in murine retrovirus integration site 1 gene associated with a non-syndromic form of isolated familial achalasia. Neurogastroenterol. Motil. 2020, 32, e13923. [Google Scholar] [CrossRef]
- Palmieri, O.; Mazza, T.; Bassotti, G.; Merla, A.; Tolone, S.; Biagini, T.; Cuttitta, A.; Bossa, F.; Martino, G.; Latiano, T.; et al. microRNA-mRNA network model in patients with achalasia. Neurogastroenterol. Motil. 2020, 32, e13764. [Google Scholar] [CrossRef] [PubMed]
- Angermeier, E.; Domes, K.; Lukowski, R.; Schlossmann, J.; Rathkolb, B.; de Angelis, M.H.; Hofmann, F. Iron deficiency anemia in cyclic GMP kinase knockout mice. Haematologica 2016, 101, e48–e51. [Google Scholar] [CrossRef]
- Föller, M.; Feil, S.; Ghoreschi, K.; Koka, S.; Gerling, A.; Thunemann, M.; Hofmann, F.; Schuler, B.; Vogel, J.; Pichler, B.; et al. Anemia and splenomegaly in cGKI-deficient mice. Proc. Natl. Acad. Sci. USA 2008, 105, 6771–6776. [Google Scholar] [CrossRef]
- Singh, A.K.; Spieβberger, B.; Zheng, W.; Xiao, F.; Lukowski, R.; Wegener, J.W.; Weinmeister, P.; Saur, D.; Klein, S.; Schemann, M.; et al. Neuronal cGMP kinase I is essential for stimulation of duodenal bicarbonate secretion by luminal acid. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012, 26, 1745–1754. [Google Scholar] [CrossRef]
- Desch, M.; Schinner, E.; Kees, F.; Hofmann, F.; Seifert, R.; Schlossmann, J. Cyclic cytidine 3′,5′-monophosphate (cCMP) signals via cGMP kinase I. FEBS Lett. 2010, 584, 3979–3984. [Google Scholar] [CrossRef]
- Santoro, C.; Giugliano, T.; Kraemer, M.; Torella, A.; Schwitalla, J.C.; Cirillo, M.; Melis, D.; Berlit, P.; Nigro, V.; Perrotta, S.; et al. Whole exome sequencing identifies MRVI1 as a susceptibility gene for moyamoya syndrome in neurofibromatosis type 1. PLoS ONE 2018, 13, e0200446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Qin, F.; Li, X.; Du, X.; Li, T. Identification of novel proteins for lacunar stroke by integrating genome-wide association data and human brain proteomes. BMC Med. 2022, 20, 211. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.J.; Cummins, C.; Radhakrishnan, R.S. Sildenafil Recovers Burn-Induced Cardiomyopathy. Cells 2020, 9, 1393. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.H.; Singh, R.K.; Bankston, J.R.; Proenza, C. Regulation of HCN Channels by Protein Interactions. Front. Physiol. 2022, 13, 928507. [Google Scholar] [CrossRef]
- Massberg, S.; Sausbier, M.; Klatt, P.; Bauer, M.; Pfeifer, A.; Siess, W.; Fässler, R.; Ruth, P.; Krombach, F.; Hofmann, F. Increased Adhesion and Aggregation of Platelets Lacking Cyclic Guanosine 3′,5′-Monophosphate Kinase I. J. Exp. Med. 1999, 189, 1255–1264. [Google Scholar] [CrossRef]
- Schinner, E.; Salb, K.; Schlossmann, J. Signaling via IRAG is essential for NO/cGMP-dependent inhibition of platelet activation. Platelets 2011, 22, 217–227. [Google Scholar] [CrossRef]
- Wilson, L.S.; Elbatarny, H.S.; Crawley, S.W.; Bennett, B.M.; Maurice, D.H. Compartmentation and compartment-specific regulation of PDE5 by protein kinase G allows selective cGMP-mediated regulation of platelet functions. Proc. Natl. Acad. Sci. USA 2008, 105, 13650–13655. [Google Scholar] [CrossRef]
- Eicher, J.D.; Chami, N.; Kacprowski, T.; Nomura, A.; Chen, M.-H.; Yanek, L.R.; Tajuddin, S.M.; Schick, U.M.; Slater, A.J.; Pankratz, N.; et al. Platelet-Related Variants Identified by Exomechip Meta-analysis in 157,293 Individuals. Am. J. Hum. Genet. 2016, 99, 40–55. [Google Scholar] [CrossRef]
- Eicher, J.D.; Xue, L.; Ben-Shlomo, Y.; Beswick, A.D.; Johnson, A.D. Replication and hematological characterization of human platelet reactivity genetic associations in men from the Caerphilly Prospective Study (CaPS). J. Thromb. Thrombolysis 2016, 41, 343–350. [Google Scholar] [CrossRef]
- Johnson, A.D.; Yanek, L.R.; Chen, M.-H.; Faraday, N.; Larson, M.; Tofler, G.; Lin, S.J.; Kraja, A.T.; Province, M.A.; Yang, Q.; et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat. Genet. 2010, 42, 608–613. [Google Scholar] [CrossRef]
- Kim, H.; Gillis, L.C.; Jarvis, J.D.; Yang, S.; Huang, K.; Der, S.; Barber, D.L. Tyrosine kinase chromosomal translocations mediate distinct and overlapping gene regulation events. BMC Cancer 2011, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Li, F.; Si, T.; Renzhi, P.; Yu, M.; Chen, D.; Ye, P.; Lu, Y. High Expression of CD300A Predicts Poor Survival in Acute Myeloid Leukemia. Acta Haematol. 2023, 146, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Do, J.H.; Bae, S.; Bae, D.-H.; Ahn, W.S. Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer 2010, 10, 576. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Xu, Y.-P.; Wang, L.-J.; Kong, Y. miR-940 potentially promotes proliferation and metastasis of endometrial carcinoma through regulation of MRVI1. Biosci. Rep. 2019, 39, BSR20190077. [Google Scholar] [CrossRef]
- Ji, H.; Li, K.; Jiang, W.; Li, J.; Zhang, J.-A.; Zhu, X. MRVI1 and NTRK3 Are Potential Tumor Suppressor Genes Commonly Inactivated by DNA Methylation in Cervical Cancer. Front. Oncol. 2021, 11, 802068. [Google Scholar] [CrossRef] [PubMed]
- Kusumawidjaja, G.; Kayed, H.; Giese, N.; Bauer, A.; Erkan, M.; Giese, T.; Hoheisel, J.D.; Friess, H.; Kleeff, J. Basic transcription factor 3 (BTF3) regulates transcription of tumor-associated genes in pancreatic cancer cells. Cancer Biol. Ther. 2007, 6, 367–376. [Google Scholar] [CrossRef]
- Jin, X.; Sun, Z.-Q.; Zhou, G.-L.; Li, G.-J.; Deng, S.-F. The Higher Expression of CDCA2 Associated with Poor Prognosis in Glioma. Dis. Markers 2022, 2022, 2184867. [Google Scholar] [CrossRef]
- Lee, J.-E.; Oum, B.S.; Choi, H.Y.; Lee, S.U.; Lee, J.S. Evaluation of differentially expressed genes identified in kerato-conus. Mol. Vis. 2009, 15, 2480–2487. [Google Scholar]
- Yaroslavskiy, B.B.; Turkova, I.; Wang, Y.; Robinson, L.J.; Blair, H.C. Functional osteoclast attachment requires inositol-1,4,5-trisphosphate receptor-associated cGMP-dependent kinase substrate. Lab. Investig. 2010, 90, 1533–1542. [Google Scholar] [CrossRef]
- Ye, J.; Li, Y.; Kong, C.; Ren, Y.; Lu, H. Label-free proteomic analysis and functional analysis in patients with intrauterine adhesion. J. Proteom. 2023, 277, 104854. [Google Scholar] [CrossRef]
- Chen, W.; Oberwinkler, H.; Werner, F.; Gaßner, B.; Nakagawa, H.; Feil, R.; Hofmann, F.; Schlossmann, J.; Dietrich, A.; Gudermann, T.; et al. Atrial Natriuretic Peptide–Mediated Inhibition of Microcirculatory Endothelial Ca2+ and Permeability Response to Histamine Involves cGMP-Dependent Protein Kinase I and TRPC6 Channels. Arter. Thromb. Vasc. Biol. 2013, 33, 2121–2129. [Google Scholar] [CrossRef] [PubMed]
- Fung, K.; Ramírez, J.; Warren, H.R.; Aung, N.; Lee, A.M.; Tzanis, E.; Petersen, S.E.; Munroe, P.B. Genome-wide association study identifies loci for arterial stiffness index in 127,121 UK Biobank participants. Sci. Rep. 2019, 9, 9143. [Google Scholar] [CrossRef] [PubMed]
- Gormley, P.; International Headache Genetics Consortium; Anttila, V.; Winsvold, B.S.; Palta, P.; Esko, T.; Pers, T.H.; Farh, K.-H.; Cuenca-Leon, E.; Muona, M.; et al. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat. Genet. 2016, 48, 856–866. [Google Scholar] [CrossRef]
- Daghlas, I.; Sargurupremraj, M.; Danning, R.; Gormley, P.; Malik, R.; Amouyel, P.; Metso, T.; Pezzini, A.; Kurth, T.; Debette, S.; et al. Migraine, Stroke, and Cervical Arterial Dissection: Shared Genetics for a Triad of Brain Disorders With Vascular Involvement. Neurol. Genet. 2022, 8, e653. [Google Scholar] [CrossRef] [PubMed]
- Rudzik, R.; Dziedziejko, V.; Rać, M.E.; Sawczuk, M.; Maciejewska-Skrendo, A.; Safranow, K.; Pawlik, A. Polymorphisms in GP6, PEAR1A, MRVI1, PIK3CG, JMJD1C, and SHH Genes in Patients with Unstable Angina. Int. J. Environ. Res. Public Health 2020, 17, 7506. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.A.; Mathur, R.; Vonk, J.M.; Szwajda, A.; Brumpton, B.; Granell, R.; Brew, B.K.; Ullemar, V.; Lu, Y.; Jiang, Y.; et al. Genetic Architectures of Childhood- and Adult-Onset Asthma Are Partly Distinct. Am. J. Hum. Genet. 2019, 104, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Wang, Y.; Zhou, S.; Xia, X.; Han, R.; Fei, G.; Zeng, D.; Wang, R. Identification of three hub genes related to the prognosis of idiopathic pulmonary fibrosis using bioinformatics analysis. Int. J. Med. Sci. 2022, 19, 1417–1429. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, C.; Fan, R.; Liu, X.; Zhang, W. A prognostic model based on clusters of molecules related to epithelial–mesenchymal transition for idiopathic pulmonary fibrosis. Front. Genet. 2022, 13, 1109903. [Google Scholar] [CrossRef]
- Behrens, T.W.; Kearns, G.M.; Rivard, J.J.; Bernstein, H.D.; Yewdell, J.W.; Staudt, L.M. Carboxyl-terminal Targeting and Novel Post-translational Processing of JAW1, a Lymphoid Protein of the Endoplasmic Reticulum. J. Biol. Chem. 1996, 271, 23528–23534. [Google Scholar] [CrossRef]
- Kozono, T.; Tadahira, K.; Okumura, W.; Itai, N.; Tamura-Nakano, M.; Dohi, T.; Tonozuka, T.; Nishikawa, A. Jaw1/LRMP has a role in maintaining nuclear shape via interaction with SUN proteins. J. Biochem. 2018, 164, 303–311. [Google Scholar] [CrossRef]
- Horn, H.F.; Kim, D.I.; Wright, G.D.; Wong, E.S.M.; Stewart, C.L.; Burke, B.; Roux, K.J. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J. Cell Biol. 2013, 202, 1023–1039. [Google Scholar] [CrossRef] [PubMed]
- Kozono, T.; Jogano, C.; Okumura, W.; Sato, H.; Matsui, H.; Takagi, T.; Okumura, N.; Takao, T.; Tonozuka, T.; Nishikawa, A. Cleavage of the Jaw1 C-terminal region enhances its augmentative effect on the Ca2+ release via IP3 receptors. J. Cell Sci. 2023, 136, jcs260439. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-Y.; Wang, J.; Zhao, Y.; Liu, J.; Yang, X.; Yue, X.; Wang, H.; Zhou, F.; Inclan-Rico, J.M.; Ponessa, J.J.; et al. Tumor suppressor p53 regulates intestinal type 2 immunity. Nat. Commun. 2021, 12, 3371. [Google Scholar] [CrossRef] [PubMed]
- Prüschenk, S.; Schlossmann, J. Function of IRAG2 Is Modulated by NO/cGMP in Murine Platelets. Int. J. Mol. Sci. 2022, 23, 6695. [Google Scholar] [CrossRef] [PubMed]
- Starr, D.A. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 2009, 122, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Ke, H.; Gao, F.; Ren, J.; Wang, M.; Huo, L.; Gong, W.; Feng, W. Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators. Structure 2016, 24, 80–91. [Google Scholar] [CrossRef]
- Sosa, B.A.; Rothballer, A.; Kutay, U.; Schwartz, T.U. LINC Complexes Form by Binding of Three KASH Peptides to Domain Interfaces of Trimeric SUN Proteins. Cell 2012, 149, 1035–1047. [Google Scholar] [CrossRef]
- Wang, W.; Shi, Z.; Jiao, S.; Chen, C.; Wang, H.; Liu, G.; Wang, Q.; Zhao, Y.; Greene, M.I.; Zhou, Z. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res. 2012, 22, 1440–1452. [Google Scholar] [CrossRef]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef]
- Kozono, T.; Sato, H.; Okumura, W.; Jogano, C.; Tamura-Nakano, M.; Kawamura, Y.I.; Rohrer, J.; Tonozuka, T.; Nishikawa, A. The N-terminal region of Jaw1 has a role to inhibit the formation of organized smooth endoplasmic reticulum as an intrinsically disordered region. Sci. Rep. 2021, 11, 753. [Google Scholar] [CrossRef]
- Snapp, E.; Hegde, R.; Francolini, M.; Lombardo, F.; Colombo, S.; Pedrazzini, E.; Borgese, N.; Lippincott-Schwartz, J. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 2003, 163, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Parrish, M.L.; Sengstag, C.; Rine, J.D.; Wright, R.L. Identification of the sequences in HMG-CoA reductase required for karmellae assembly. Mol. Biol. Cell 1995, 6, 1535–1547. [Google Scholar] [CrossRef] [PubMed]
- Fasana, E.; Fossati, M.; Ruggiano, A.; Brambillasca, S.; Hoogenraad, C.C.; Navone, F.; Francolini, M.; Borgese, N. A VAPB mutant linked to amyotrophic lateral sclerosis generates a novel form of organized smooth endoplasmic reticulum. FASEB J. 2010, 24, 1419–1430. [Google Scholar] [CrossRef]
- Mao, A.H.; Crick, S.L.; Vitalis, A.; Chicoine, C.L.; Pappu, R.V. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 8183–8188. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.M.; Kriwacki, R.W.; Pappu, R.V. Versatility from Protein Disorder. Science 2012, 337, 1460–1461. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Bigman, L.; Friedler, A. A role of disordered domains in regulating protein oligomerization and stability. Chem. Commun. 2014, 50, 10797–10800. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef]
- Hieda, M.; Matsumoto, T.; Isobe, M.; Kurono, S.; Yuka, K.; Kametaka, S.; Wang, J.-Y.; Chi, Y.-H.; Kameda, K.; Kimura, H.; et al. The SUN2-nesprin-2 LINC complex and KIF20A function in the Golgi dispersal. Sci. Rep. 2021, 11, 5358. [Google Scholar] [CrossRef] [PubMed]
- Gimpel, P.; Lee, Y.L.; Sobota, R.M.; Calvi, A.; Koullourou, V.; Patel, R.; Mamchaoui, K.; Nédélec, F.; Shackleton, S.; Schmoranzer, J.; et al. Nesprin-1α-Dependent Microtubule Nucleation from the Nuclear Envelope via Akap450 Is Necessary for Nuclear Positioning in Muscle Cells. Curr. Biol. 2017, 27, 2999–3009.e9. [Google Scholar] [CrossRef]
- Roux, K.J.; Crisp, M.L.; Liu, Q.; Kim, D.; Kozlov, S.; Stewart, C.L.; Burke, B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA 2009, 106, 2194–2199. [Google Scholar] [CrossRef]
- Okumura, W.; Tadahira, K.; Kozono, T.; Tamura-Nakano, M.; Sato, H.; Matsui, H.; Dohi, T.; Rohrer, J.; Tonozuka, T.; Nishikawa, A. Jaw1/LRMP is associated with the maintenance of Golgi ribbon structure. J. Biochem. 2023, 173, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J. Inositol trisphosphate and calcium signalling. Nature 1993, 361, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Okumura, W.; Kozono, T.; Sato, H.; Matsui, H.; Takagi, T.; Tonozuka, T.; Nishikawa, A. Jaw1/LRMP increases Ca2+ influx upon GPCR stimulation with heterogeneous effect on the activity of each ITPR subtype. Sci. Rep. 2022, 12, 9476. [Google Scholar] [CrossRef] [PubMed]
- Prüschenk, S. Molekulare und Physiologische Funktion von IRAG2 in Thrombozyten und im Pankreas; Universität Regensburg: Regensburg, Germany, 2023. [Google Scholar] [CrossRef]
- Makhoul, S.; Walter, E.; Pagel, O.; Walter, U.; Sickmann, A.; Gambaryan, S.; Smolenski, A.; Zahedi, R.P.; Jurk, K. Effects of the NO/soluble guanylate cyclase/cGMP system on the functions of human platelets. Nitric Oxide 2018, 76, 71–80. [Google Scholar] [CrossRef]
- Butt, E.; Nolte, C.; Schulz, S.; Beltman, J.; Beavo, J.A.; Jastorff, B.; Walter, U. Analysis of the functional role of cGMP-dependent protein kinase in intact human platelets using a specific activator 8-para-chlorophenylthio-cGMP. Biochem. Pharmacol. 1992, 43, 2591–2600. [Google Scholar] [CrossRef]
- Luo, X.-C.; Chen, Z.-H.; Xue, J.-B.; Zhao, D.-X.; Lu, C.; Li, Y.-H.; Li, S.-M.; Du, Y.-W.; Liu, Q.; Wang, P.; et al. Infection by the parasitic helminth Trichinella spiralis activates a Tas2r-mediated signaling pathway in intestinal tuft cells. Proc. Natl. Acad. Sci. USA 2019, 116, 5564–5569. [Google Scholar] [CrossRef]
- DiFrancesco, D.; Tortora, P. Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature 1991, 351, 145–147. [Google Scholar] [CrossRef]
- Wainger, B.J.; DeGennaro, M.; Santoro, B.; Siegelbaum, S.A.; Tibbs, G.R. Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature 2001, 411, 805–810. [Google Scholar] [CrossRef]
- Wicks, N.L.; Wong, T.; Sun, J.; Madden, Z.; Young, E.C. Cytoplasmic cAMP-sensing domain of hyperpolarization-activated cation (HCN) channels uses two structurally distinct mechanisms to regulate voltage gating. Proc. Natl. Acad. Sci. USA 2011, 108, 609–614. [Google Scholar] [CrossRef]
- Hisatsune, C.; Yasumatsu, K.; Takahashi-Iwanaga, H.; Ogawa, N.; Kuroda, Y.; Yoshida, R.; Ninomiya, Y.; Mikoshiba, K. Abnormal Taste Perception in Mice Lacking the Type 3 Inositol 1,4,5-Trisphosphate Receptor. J. Biol. Chem. 2007, 282, 37225–37231. [Google Scholar] [CrossRef]
- Zhang, Y.; Hoon, M.A.; Chandrashekar, J.; Mueller, K.L.; Cook, B.; Wu, D.; Zuker, C.S.; Ryba, N.J. Coding of Sweet, Bitter, and Umami Tastes: Different Receptor Cells Sharing Similar Signaling Pathways. Cell 2003, 112, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Matthews, E.K.; Petersen, O.H.; Williams, J.A. Pancreatic acinar cells: Acetylcholine-induced membrane depolarization, calcium efflux and amylase release. J. Physiol. 1973, 234, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Petersen, O. Ca2+ signaling in pancreatic acinar cells: Physiology and pathophysiology. Braz. J. Med. Biol. Res. 2009, 42, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Gaisano, H.Y.; Dolai, S.; Takahashi, T. Physiologic Exocytosis in Pancreatic Acinar Cells and Pathologic Fusion Underlying Pancreatitis. Exocrine Pancreas Knowl. Base Pancreapedia 2020. [CrossRef]
- Ruggeri, Z.M. Platelets in atherothrombosis. Nat. Med. 2002, 8, 1227–1234. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef]
- Rosenwald, A.; Wright, G.; Chan, W.C.; Connors, J.M.; Campo, E.; Fisher, R.I.; Gascoyne, R.D.; Muller-Hermelink, H.K.; Smeland, E.B.; Giltnane, J.M.; et al. The Use of Molecular Profiling to Predict Survival after Chemotherapy for Diffuse Large-B-Cell Lymphoma. N. Engl. J. Med. 2002, 346, 1937–1947. [Google Scholar] [CrossRef]
- Lossos, I.S.; Czerwinski, D.K.; Alizadeh, A.A.; Wechser, M.A.; Tibshirani, R.; Botstein, D.; Levy, R. Prediction of Survival in Diffuse Large-B-Cell Lymphoma Based on the Expression of Six Genes. New Engl. J. Med. 2004, 350, 1828–1837. [Google Scholar] [CrossRef]
- Snyder, H.L.; Bačík, I.; Bennink, J.R.; Kearns, G.; Behrens, T.W.; Bächi, T.; Orlowski, M.; Yewdell, J.W. Two Novel Routes of Transporter Associated with Antigen Processing (TAP)-independent Major Histocompatibility Complex Class I Antigen Processing. J. Exp. Med. 1997, 186, 1087–1098. [Google Scholar] [CrossRef]
- Miller, T.P.; Lippman, S.M.; Spier, C.M.; Slymen, D.J.; Grogan, T.M. HLA-DR (Ia) immune phenotype predicts outcome for patients with diffuse large cell lymphoma. J. Clin. Investig. 1988, 82, 370–372. [Google Scholar] [CrossRef]
- Rimsza, L.M.; Roberts, R.A.; Miller, T.P.; Unger, J.M.; LeBlanc, M.; Braziel, R.M.; Weisenberger, D.D.; Chan, W.C.; Muller-Hermelink, H.K.; Jaffe, E.S.; et al. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: A follow-up study from the Leukemia and Lymphoma Molecular Profiling Project. Blood 2004, 103, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, P.; Antonov, A.; Anemona, L.; Vangapandou, C.; Montanaro, M.; Botticelli, A.; Mauriello, A.; Melino, G.; Catani, M.V. New immunological potential markers for triple negative breast cancer: IL18R1, CD53, TRIM, Jaw1, LTB, PTPRCAP. Discov. Oncol. 2021, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Chen, L.; Zhou, N.; Ni, H.; Zu, L.; He, J.; Yang, L.; Zhu, Y.; Sun, X.; Li, X.; et al. LRMP Associates With Immune Infiltrates and Acts as a Prognostic Biomarker in Lung Adenocarcinoma. Front. Mol. Biosci. 2021, 8, 711928. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Itoh, T.; Imamichi, S.; Kikuhara, S.; Fujimori, H.; Hirai, T.; Saito, S.; Sakurai, Y.; Tanaka, H.; Nakamura, H.; et al. Proteomic analysis of cellular response induced by boron neutron capture reaction in human squamous cell carcinoma SAS cells. Appl. Radiat. Isot. 2015, 106, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Gariboldi, M.; Manenti, G.; Canzian, F.; Falvella, F.S.; Radice, M.T.; Pierotti, M.A.; Della Porta, G.; Binelli, G.; Dragani, T.A. A major susceptibility locus to murine lung carcinogenesis maps on chromosome 6. Nat. Genet. 1993, 3, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Manenti, G.; Dragani, T.A. Pas1 haplotype-dependent genetic predisposition to lung tumorigenesis in rodents: A meta-analysis. Carcinog. 2005, 26, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Manenti, G.; Galbiati, F.; Giannì-Barrera, R.; Pettinicchio, A.; Acevedo, A.; Dragani, T.A. Haplotype sharing suggests that a genomic segment containing six genes accounts for the pulmonary adenoma susceptibility 1 (Pas1) locus activity in mice. Oncogene 2004, 23, 4495–4504. [Google Scholar] [CrossRef] [PubMed]
- Manenti, G.; Galbiati, F.; Pettinicchio, A.; Spinola, M.; Piconese, S.; Leoni, V.P.; Conti, B.; Ravagnani, F.; Incarbone, M.; Pastorino, U.; et al. A V141L polymorphism of the human LRMP gene is associated with survival of lung cancer patients. Carcinog. 2006, 27, 1386–1390. [Google Scholar] [CrossRef]
- Maria, D.A.; Manenti, G.; Galbiati, F.; Ribeiro, O.G.; Cabrera, W.H.K.; Barrera, R.G.; Pettinicchio, A.; De Franco, M.; Starobinas, N.; Siqueira, M.; et al. Pulmonary adenoma susceptibility 1 (Pas1) locus affects inflammatory response. Oncogene 2003, 22, 426–432. [Google Scholar] [CrossRef]

| Function by Interaction with Proteins | References | |
|---|---|---|
| IRAG1 | IP3R/PKGIβ: | |
| cGMP- and cCMP-dependent inhibition of Ca2+ signaling (smooth muscle and platelets) | [1,2,3,4,5,13,21,22,30,36] | |
| TRPM4: | ||
| NO/cGMP-dependent modulation of TRPM4 activity | [20] | |
| HCN4: | ||
| enhanced opening of the channels | [18,34] | |
| IRAG2 | IP3R: | |
| activation of Ca2+ signaling (platelets, pancreas, and MEF and HEK cells) | [9,63,64,83,84] | |
| HCN4: | ||
| reduced opening of the channels | [18,34] | |
| SUN proteins: | ||
| IRAG2 takes part in LINC complex as KASH protein: microtubule interaction and maintaining nuclear shape | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prüschenk, S.; Majer, M.; Schlossmann, J. Novel Functional Features of cGMP Substrate Proteins IRAG1 and IRAG2. Int. J. Mol. Sci. 2023, 24, 9837. https://doi.org/10.3390/ijms24129837
Prüschenk S, Majer M, Schlossmann J. Novel Functional Features of cGMP Substrate Proteins IRAG1 and IRAG2. International Journal of Molecular Sciences. 2023; 24(12):9837. https://doi.org/10.3390/ijms24129837
Chicago/Turabian StylePrüschenk, Sally, Michael Majer, and Jens Schlossmann. 2023. "Novel Functional Features of cGMP Substrate Proteins IRAG1 and IRAG2" International Journal of Molecular Sciences 24, no. 12: 9837. https://doi.org/10.3390/ijms24129837
APA StylePrüschenk, S., Majer, M., & Schlossmann, J. (2023). Novel Functional Features of cGMP Substrate Proteins IRAG1 and IRAG2. International Journal of Molecular Sciences, 24(12), 9837. https://doi.org/10.3390/ijms24129837







