cGMP Signaling in Photoreceptor Degeneration
Abstract
1. Introduction
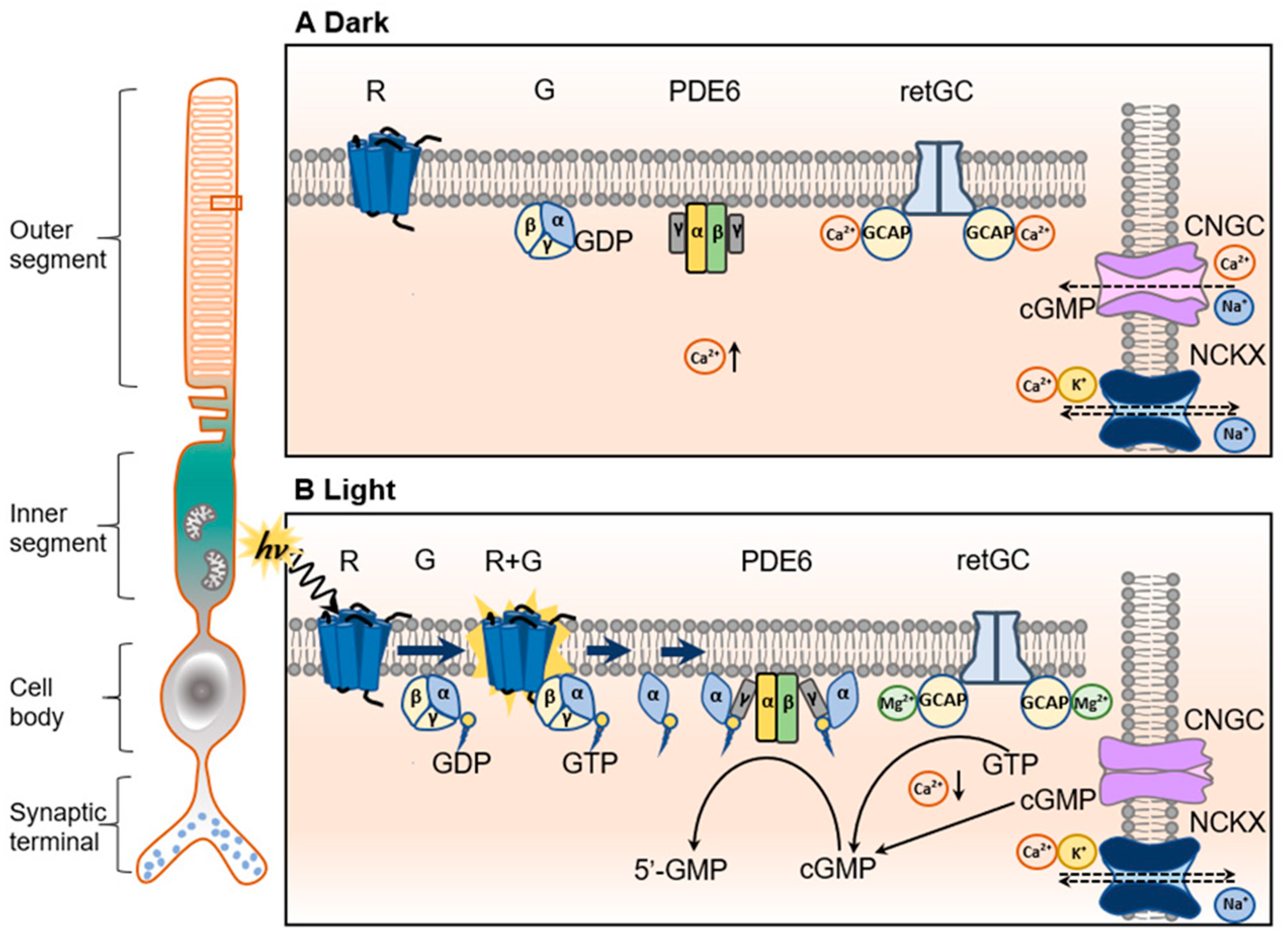
2. Regulation of Cellular cGMP Level in Photoreceptors
2.1. The Ca2+/Mg2+-GCAP/RetGC Complex
2.2. PDE6
2.3. CNG Channel
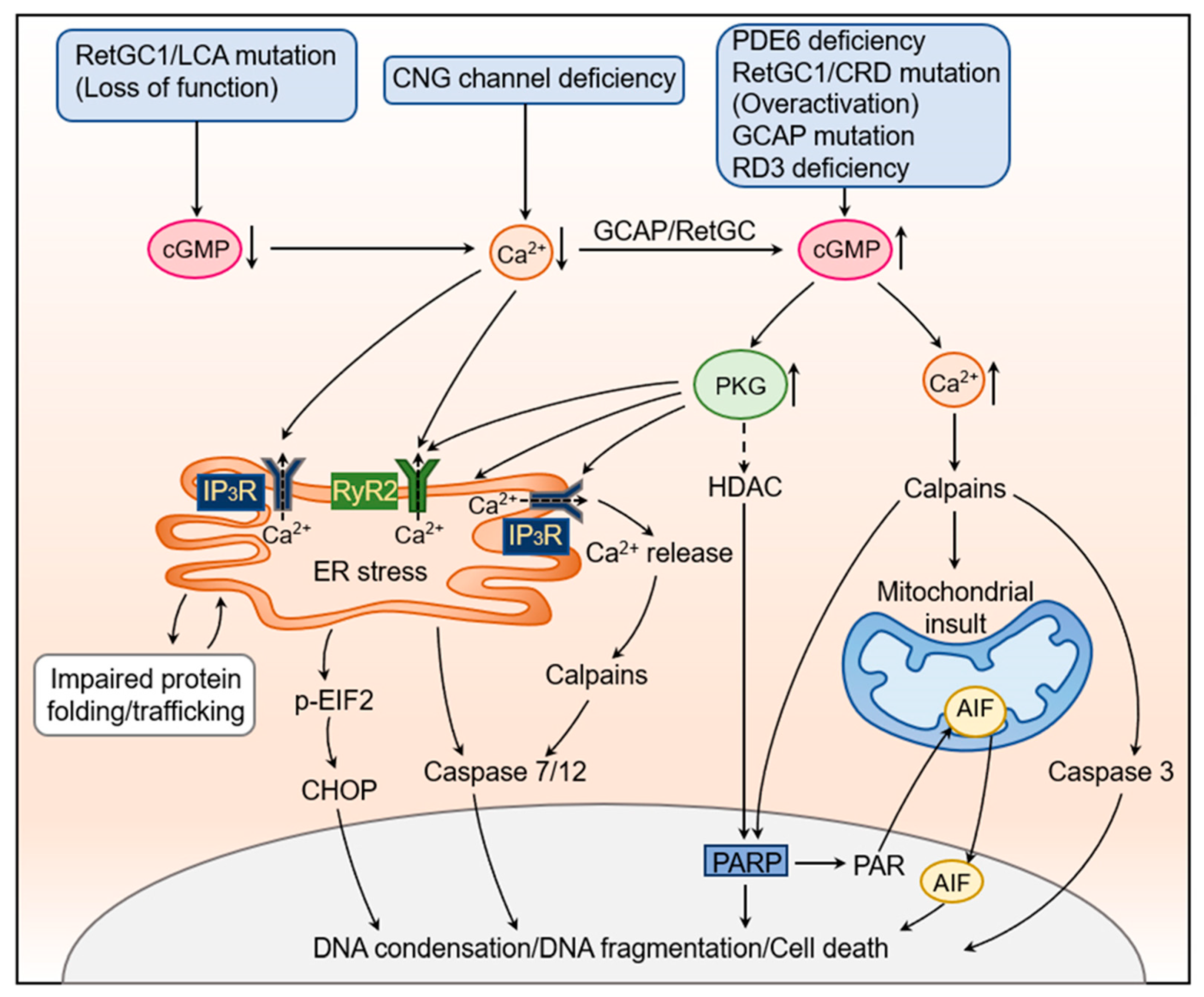
3. Dysregulation of cGMP Signaling in Photoreceptor Degeneration
3.1. Dysregulation of RetGC1, RD3, and GCAP1
3.2. Deficiency of PDE6
3.3. Deficiency of CNG Channel
3.4. Deficiency of Other Photoreceptor Specific Proteins
4. Experimental Evidence Supporting the Contribution of Elevated cGMP Signaling to Photoreceptor Degeneration
4.1. Depletion of cGMP Reduces Photoreceptor Degeneration
4.2. Inhibition of CNG Channel to Decrease Ca2+ Influx Reduces Photoreceptor Degeneration
4.3. Inhibition of PKG Reduces Photoreceptor Degeneration
5. The Cellular and Molecular Mechanisms Underlying cGMP Signaling-Induced Photoreceptor Degeneration
5.1. Elevated PKG Signaling
5.1.1. Excessive PKG Signaling Induces ER Stress
5.1.2. Excessive PKG Signaling Impairs ER Ca2+ Homeostasis
5.1.3. Other Potential Targets of PKG in Photoreceptor Degeneration
5.2. Impaired Cellular Ca2+ Homeostasis
5.2.1. Ca2+ Overload and Calpain Activation
5.2.2. Ca2+ Depletion
5.3. Other Factors
5.3.1. HDAC and PARP
5.3.2. AIF
6. Summary and Perspectives
Funding
Conflicts of Interest
References
- Kawamura, S.; Tachibanaki, S. Rod and cone photoreceptors: Molecular basis of the difference in their physiology. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2008, 150, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Korenbrot, J.I. Speed, sensitivity, and stability of the light response in rod and cone photoreceptors: Facts and models. Prog. Retin. Eye Res. 2012, 31, 442–466. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.E.; Arshavsky, V.Y. Beyond Counting Photons: Trials and Trends in Vertebrate Visual Transduction. Neuron 2005, 48, 387–401. [Google Scholar] [CrossRef]
- Ebrey, T.; Koutalos, Y. Vertebrate photoreceptors. Prog. Retin. Eye Res. 2001, 20, 49–94. [Google Scholar] [CrossRef] [PubMed]
- Molday, R.S.; Moritz, O.L. Photoreceptors at a glance. J. Cell Sci. 2015, 128, 4039–4045. [Google Scholar] [CrossRef]
- Barret, D.C.A.; Kaupp, U.B.; Marino, J. The structure of cyclic nucleotide-gated channels in rod and cone photoreceptors. Trends Neurosci. 2022, 45, 763–776. [Google Scholar] [CrossRef]
- Leskov, I.B.; Klenchin, V.A.; Handy, J.W.; Whitlock, G.G.; Govardovskii, V.I.; Bownds, M.D.; Lamb, T.D.; Pugh, E.N., Jr.; Arshavsky, V.Y. The gain of rod phototransduction: Reconciliation of biochemical and electrophysiological measurements. Neuron 2000, 27, 525–537. [Google Scholar] [CrossRef]
- Arshavsky, V.Y.; Lamb, T.D.; Pugh, E.N., Jr. G Proteins and Phototransduction. Annu. Rev. Physiol. 2002, 64, 153–187. [Google Scholar] [CrossRef]
- Stryer, L. Cyclic GMP cascade of vision. Annu. Rev. Neurosci. 1986, 9, 87–119. [Google Scholar] [CrossRef]
- Kaupp, U.B.; Seifert, R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 2002, 82, 769–824. [Google Scholar] [CrossRef]
- Deng, W.T.; Kolandaivelu, S.; Dinculescu, A.; Li, J.; Zhu, P.; Chiodo, V.A.; Ramamurthy, V.; Hauswirth, W.W. Cone Phosphodiesterase-6gamma’ Subunit Augments Cone PDE6 Holoenzyme Assembly and Stability in a Mouse Model Lacking Both Rod and Cone PDE6 Catalytic Subunits. Front. Mol. Neurosci. 2018, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- Lagman, D.; Franzen, I.E.; Eggert, J.; Larhammar, D.; Abalo, X.M. Evolution and expression of the phosphodiesterase 6 genes unveils vertebrate novelty to control photosensitivity. BMC Evol. Biol. 2016, 16, 124. [Google Scholar] [CrossRef] [PubMed]
- Tolone, A.; Belhadj, S.; Rentsch, A.; Schwede, F.; Paquet-Durand, F. The cGMP Pathway and Inherited Photoreceptor Degeneration: Targets, Compounds, and Biomarkers. Genes 2019, 10, 453. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Marigo, V.; Ekstrom, P. RD Genes Associated with High Photoreceptor cGMP-Levels (Mini-Review). Adv. Exp. Med. Biol. 2019, 1185, 245–249. [Google Scholar]
- Francis, S.H.; Blount, M.A.; Corbin, J.D. Mammalian Cyclic Nucleotide Phosphodiesterases: Molecular Mechanisms and Physiological Functions. Physiol. Rev. 2011, 91, 651–690. [Google Scholar] [CrossRef]
- Dizhoor, A.M.; Lowe, D.G.; Olshevskaya, E.V.; Laura, R.P.; Hurley, J.B. The human photoreceptor membrane guanylyl cyclase, RetGC, is present in outer segments and is regulated by calcium and a soluble activator. Neuron 1994, 12, 1345–1352. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Roseman, G.; Peshenko, I.; Manchala, G.; Cudia, D.; Dizhoor, A.; Millhauser, G.; Ames, J.B. Retinal guanylyl cyclase activating protein 1 forms a functional dimer. PLoS ONE 2018, 13, e0193947. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.-B.; Fülle, H.-J.; Garbers, D.L. Chromosomal Localization and Genomic Organization of Genes Encoding Guanylyl Cyclase Receptors Expressed in Olfactory Sensory Neurons and Retina. Genomics 1996, 31, 367–372. [Google Scholar] [CrossRef]
- Baehr, W.; Karan, S.; Maeda, T.; Luo, D.-G.; Li, S.; Bronson, J.D.; Watt, C.B.; Yau, K.-W.; Frederick, J.M.; Palczewski, K. The Function of Guanylate Cyclase 1 and Guanylate Cyclase 2 in Rod and Cone Photoreceptors. J. Biol. Chem. 2007, 282, 8837–8847. [Google Scholar] [CrossRef]
- Peshenko, I.V.; Olshevskaya, E.V.; Savchenko, A.B.; Karan, S.; Palczewski, K.; Baehr, W.; Dizhoor, A.M. Enzy-matic properties and regulation of the native isozymes of retinal membrane guanylyl cyclase (RetGC) from mouse photoreceptors. Biochemistry 2011, 50, 5590–5600. [Google Scholar] [CrossRef]
- Lowe, D.G.; Dizhoor, A.M.; Liu, K.; Gu, Q.; Spencer, M.; Laura, R.; Lu, L.; Hurley, J.B. Cloning and expression of a second photoreceptor-specific membrane retina guanylyl cyclase (RetGC), RetGC-2. Proc. Natl. Acad. Sci. USA 1995, 92, 5535–5539. [Google Scholar] [CrossRef] [PubMed]
- Peshenko, I.V.; Olshevskaya, E.V.; Dizhoor, A.M. Dimerization Domain of Retinal Membrane Guanylyl Cyclase 1 (RetGC1) Is an Essential Part of Guanylyl Cyclase-activating Protein (GCAP) Binding Interface. J. Biol. Chem. 2015, 290, 19584–19596. [Google Scholar] [CrossRef] [PubMed]
- Peshenko, I.V.; Olshevskaya, E.V.; Dizhoor, A.M. Evaluating the Role of Retinal Membrane Guanylyl Cyclase 1 (RetGC1) Domains in Binding Guanylyl Cyclase-activating Proteins (GCAPs). J. Biol. Chem. 2015, 290, 6913–6924. [Google Scholar] [CrossRef]
- Dizhoor, A.M.; Peshenko, I.V. Regulation of retinal membrane guanylyl cyclase (RetGC) by negative calcium feedback and RD3 protein. Pflug. Arch. 2021, 473, 1393–1410. [Google Scholar] [CrossRef]
- Liu, Y.; Ruoho, A.E.; Rao, V.D.; Hurley, J.H. Catalytic mechanism of the adenylyl and guanylyl cyclases: Modeling and mutational analysis. Proc. Natl. Acad. Sci. USA 1997, 94, 13414–13419. [Google Scholar] [CrossRef] [PubMed]
- Olshevskaya, E.V.; Peshenko, I.V.; Savchenko, A.B.; Dizhoor, A.M. Retinal Guanylyl Cyclase Isozyme 1 Is the Preferential In Vivo Target for Constitutively Active GCAP1 Mutants Causing Congenital Degeneration of Photoreceptors. J. Neurosci. 2012, 32, 7208–7217. [Google Scholar] [CrossRef]
- Makino, C.L.; Peshenko, I.V.; Wen, X.H.; Olshevskaya, E.V.; Barrett, R.; Dizhoor, A.M. A role for GCAP2 in regulating the photoresponse. Guanylyl cyclase activation and rod electrophysiology in GUCA1B knock-out mice. J. Biol. Chem. 2008, 283, 29135–29143. [Google Scholar] [CrossRef]
- López-Begines, S.; Plana-Bonamaisó, A.; Méndez, A. Molecular determinants of Guanylate Cyclase Activating Protein subcellular distribution in photoreceptor cells of the retina. Sci. Rep. 2018, 8, 2903. [Google Scholar] [CrossRef]
- Dizhoor, A. Regulation of cGMP synthesis in photoreceptors: Role in signal transduction and congenital diseases of the retina. Cell. Signal. 2000, 12, 711–719. [Google Scholar] [CrossRef]
- Ames, J.B.; Dizhoor, A.M.; Ikura, M.; Palczewski, K.; Stryer, L. Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases. J. Biol. Chem. 1999, 274, 19329–19337. [Google Scholar] [CrossRef]
- Makino, C.L.; Wen, X.-H.; Olshevskaya, E.V.; Peshenko, I.V.; Savchenko, A.B.; Dizhoor, A.M. Enzymatic Relay Mechanism Stimulates Cyclic GMP Synthesis in Rod Photoresponse: Biochemical and Physiological Study in Guanylyl Cyclase Activating Protein 1 Knockout Mice. PLoS ONE 2012, 7, e47637. [Google Scholar] [CrossRef]
- Ames, J.B. Structural Insights into Retinal Guanylate Cyclase Activator Proteins (GCAPs). Int. J. Mol. Sci. 2021, 22, 8731. [Google Scholar] [CrossRef] [PubMed]
- Peshenko, I.V.; Yu, Q.; Lim, S.; Cudia, D.; Dizhoor, A.M.; Ames, J.B. Retinal degeneration 3 (RD3) protein, a retinal guanylyl cyclase regulator, forms a monomeric and elongated four-helix bundle. J. Biol. Chem. 2019, 294, 2318–2328. [Google Scholar] [CrossRef]
- Dizhoor, A.M.; Olshevskaya, E.V.; Peshenko, I.V. Retinal degeneration-3 protein promotes photoreceptor survival by suppressing activation of guanylyl cyclase rather than accelerating GMP recycling. J. Biol. Chem. 2021, 296, 100362. [Google Scholar] [CrossRef] [PubMed]
- Azadi, S.; Molday, L.L.; Molday, R.S. RD3, the protein associated with Leber congenital amaurosis type 12, is required for guanylate cyclase trafficking in photoreceptor cells. Proc. Natl. Acad. Sci. USA 2010, 107, 21158–21163. [Google Scholar] [CrossRef]
- Conti, M.; Beavo, J. Biochemistry and Physiology of Cyclic Nucleotide Phosphodiesterases: Essential Components in Cyclic Nucleotide Signaling. Annu. Rev. Biochem. 2007, 76, 481–511. [Google Scholar] [CrossRef]
- Cote, R.H.; Gupta, R.; Irwin, M.J.; Wang, X. Photoreceptor Phosphodiesterase (PDE6): Structure, Regulatory Mechanisms, and Implications for Treatment of Retinal Diseases. Adv. Exp. Med. Biol. 2022, 1371, 33–59. [Google Scholar]
- Cote, R.H. Characteristics of Photoreceptor PDE (PDE6): Similarities and differences to PDE5. Int. J. Impot. Res. 2004, 16 (Suppl. S1), S28–S33. [Google Scholar] [CrossRef]
- Cote, R.H. Photoreceptor phosphodiesterase (PDE6): Activation and inactivation mechanisms during visual transduction in rods and cones. Pflug. Arch. 2021, 473, 1377–1391. [Google Scholar] [CrossRef] [PubMed]
- Barren, B.; Gakhar, L.; Muradov, H.; Boyd, K.K.; Ramaswamy, S.; Artemyev, N.O. Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the gamma-subunit. Embo J. 2009, 28, 3613–3622. [Google Scholar] [CrossRef] [PubMed]
- Biel, M.; Michalakis, S. Cyclic nucleotide-gated channels. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 111–136. [Google Scholar]
- Zheng, J.; Trudeau, M.C.; Zagotta, W.N. Rod Cyclic Nucleotide-Gated Channels Have a Stoichiometry of Three CNGA1 Subunits and One CNGB1 Subunit. Neuron 2002, 36, 891–896. [Google Scholar] [CrossRef]
- Weitz, D.; Ficek, N.; Kremmer, E.; Bauer, P.J.; Kaupp, U. Subunit Stoichiometry of the CNG Channel of Rod Photoreceptors. Neuron 2002, 36, 881–889. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, Z.; Li, H.; Yang, J. Structure of the human cone photoreceptor cyclic nucleotide-gated channel. Nat. Struct. Mol. Biol. 2022, 29, 40–46. [Google Scholar] [CrossRef]
- Ding, X.Q.; Matveev, A.; Singh, A.; Komori, N.; Matsumoto, H. Biochemical characterization of cone cyclic nucle-otide-gated (CNG) channel using the infrared fluorescence detection system. Adv. Exp. Med. Biol. 2012, 723, 769–775. [Google Scholar] [PubMed]
- Kesters, D.; Brams, M.; Nys, M.; Wijckmans, E.; Spurny, R.; Voets, T.; Tytgat, J.; Kusch, J.; Ulens, C. Structure of the SthK Carboxy-Terminal Region Reveals a Gating Mechanism for Cyclic Nucleotide-Modulated Ion Channels. PLoS ONE 2015, 10, e0116369. [Google Scholar] [CrossRef] [PubMed]
- Korenbrot, J.I.; Rebrik, T.I. Tuning Outer Segment Ca2+Homeostasis to Phototransduction in Rods and Cones. Adv. Exp. Med. Biol. 2002, 514, 179–203. [Google Scholar] [CrossRef]
- Michalakis, S.; Becirovic, E.; Biel, M. Retinal Cyclic Nucleotide-Gated Channels: From Pathophysiology to Therapy. Int. J. Mol. Sci. 2018, 19, 749. [Google Scholar] [CrossRef] [PubMed]
- Paquet-Durand, F.; Hauck, S.M.; van Veen, T.; Ueffing, M.; Ekström, P. PKG activity causes photoreceptor cell death in two retinitis pigmentosa models. J. Neurochem. 2009, 108, 796–810. [Google Scholar] [CrossRef]
- Kelsell, R.E.; Gregory-Evans, K.; Payne, A.; Perrault, I.; Kaplan, J.; Yang, R.-B.; Garbers, D.L.; Bird, A.C.; Moore, A.T.; Hunt, D.M. Mutations in the Retinal Guanylate Cyclase (RETGC-1) Gene in Dominant Cone-Rod Dystrophy. Hum. Mol. Genet. 1998, 7, 1179–1184. [Google Scholar] [CrossRef]
- Tucker, C.L.; Woodcock, S.C.; Kelsell, R.E.; Ramamurthy, V.; Hunt, D.M.; Hurley, J.B. Biochemical analysis of a dimerization domain mutation in RetGC-1 associated with dominant cone–rod dystrophy. Proc. Natl. Acad. Sci. USA 1999, 96, 9039–9044. [Google Scholar] [CrossRef]
- Duda, T.; Koch, K.W. Retinal diseases linked with photoreceptor guanylate cyclase. Mol. Cell Biochem. 2002, 230, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Kitiratschky, V.B.; Wilke, R.; Renner, A.B.; Kellner, U.; Vadala, M.; Birch, D.G.; Wissinger, B.; Zrenner, E.; Kohl, S. Mutation analysis identifies GUCY2D as the major gene responsible for autosomal dominant progressive cone degeneration. Investig. Ophthalmol. Vis. Sci. 2008, 49, 5015–5023. [Google Scholar] [CrossRef] [PubMed]
- Sharon, D.; Wimberg, H.; Kinarty, Y.; Koch, K.-W. Genotype-functional-phenotype correlations in photoreceptor guanylate cyclase (GC-E) encoded by GUCY2D. Prog. Retin. Eye Res. 2018, 63, 69–91. [Google Scholar] [CrossRef] [PubMed]
- Wimberg, H.; Lev, D.; Yosovich, K.; Namburi, P.; Banin, E.; Sharon, D.; Koch, K.-W. Photoreceptor Guanylate Cyclase (GUCY2D) Mutations Cause Retinal Dystrophies by Severe Malfunction of Ca2+-Dependent Cyclic GMP Synthesis. Front. Mol. Neurosci. 2018, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Rozet, J.-M.; Perrault, I.; Gerber, S.; Hanein, S.; Barbet, F.; Ducroq, D.; Souied, E.; Munnich, A.; Kaplan, J. Complete abolition of the retinal-specific guanylyl cyclase (retGC-1) catalytic ability consistently leads to leber congenital amaurosis (LCA). Investig. Ophthalmol. Vis. Sci. 2001, 42, 1190–1192. [Google Scholar]
- Tucker, C.; Ramamurthy, V.; Pina, A.-L.; Loyer, M.; Dharmaraj, S.; Li, Y.; Maumenee, I.H.; Hurley, J.B.; Koenekoop, R.K. Functional analyses of mutant recessive GUCY2D alleles identified in Leber congenital amaurosis patients: Protein domain comparisons and dominant negative effects. Mol. Vis. 2004, 10, 297–303. [Google Scholar]
- Yi, Z.; Sun, W.; Xiao, X.; Li, S.; Jia, X.; Li, X.; Yu, B.; Wang, P.; Zhang, Q. Novel variants in GUCY2D causing retinopathy and the genotype-phenotype correlation. Exp. Eye Res. 2021, 208, 108637. [Google Scholar] [CrossRef]
- Liu, X.; Fujinami, K.; Kuniyoshi, K.; Kondo, M.; Ueno, S.; Hayashi, T.; Mochizuki, K.; Kameya, S.; Yang, L.; Fujinami-Yokokawa, Y.; et al. Clinical and Genetic Characteristics of 15 Affected Patients From 12 Japanese Families with GUCY2D-Associated Retinal Disorder. Transl. Vis. Sci. Technol. 2020, 9, 2. [Google Scholar] [CrossRef]
- Boye, S.E. Leber Congenital Amaurosis Caused by Mutations in GUCY2D. Cold Spring Harb. Perspect. Med. 2014, 5, a017350. [Google Scholar] [CrossRef]
- Jacobson, S.G.; Cideciyan, A.V.; Peshenko, I.V.; Sumaroka, A.; Olshevskaya, E.V.; Cao, L.; Schwartz, S.B.; Roman, A.J.; Olivares, M.B.; Sadigh, S.; et al. Determining consequences of retinal membrane guanylyl cyclase (RetGC1) deficiency in human Leber congenital amaurosis en route to therapy: Residual cone-photoreceptor vision correlates with biochemical properties of the mutants. Hum. Mol. Genet. 2013, 22, 168–183. [Google Scholar] [CrossRef]
- Dizhoor, A.M.; Olshevskaya, E.V.; Peshenko, I.V. The R838S Mutation in Retinal Guanylyl Cyclase 1 (RetGC1) Alters Calcium Sensitivity of cGMP Synthesis in the Retina and Causes Blindness in Transgenic Mice. J. Biol. Chem. 2016, 291, 24504–24516. [Google Scholar] [CrossRef]
- Sato, S.; Peshenko, I.V.; Olshevskaya, E.V.; Kefalov, V.J.; Dizhoor, A.M. GUCY2D Cone-Rod Dystrophy-6 Is a “Phototransduction Disease” Triggered by Abnormal Calcium Feedback on Retinal Membrane Guanylyl Cyclase 1. J. Neurosci. 2018, 38, 2990–3000. [Google Scholar] [CrossRef] [PubMed]
- Peshenko, I.V.; Olshevskaya, E.V.; Dizhoor, A.M. GUCY2D mutations in retinal guanylyl cyclase 1 provide bio-chemical reasons for dominant cone-rod dystrophy but not for stationary night blindness. J. Biol. Chem. 2020, 295, 18301–18315. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, S.G.; Cideciyan, A.V.; Ho, A.C.; Roman, A.J.; Wu, V.; Garafalo, A.V.; Sumaroka, A.; Krishnan, A.K.; Swider, M.; Mascio, A.A.; et al. Night vision restored in days after decades of congenital blindness. iScience 2022, 25, 105274. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, S.G.; Cideciyan, A.V.; Ho, A.C.; Peshenko, I.V.; Garafalo, A.V.; Roman, A.J.; Sumaroka, A.; Wu, V.; Krishnan, A.K.; Sheplock, R.; et al. Safety and improved efficacy signals following gene therapy in childhood blindness caused by GUCY2D mutations. iScience 2021, 24, 102409. [Google Scholar] [CrossRef]
- Ugur Iseri, S.A.; Durlu, Y.K.; Tolun, A. A novel recessive GUCY2D mutation causing cone-rod dystrophy and not Leber’s congenital amaurosis. Eur. J. Hum. Genet. 2010, 18, 1121–1126. [Google Scholar] [CrossRef]
- Stunkel, M.L.; Brodie, S.; Cideciyan, A.V.; Pfeifer, W.L.; Kennedy, E.L.; Stone, E.M.; Jacobson, S.G.; Drack, A.V. Expanded Retinal Disease Spectrum Associated with Autosomal Recessive Mutations in GUCY2D. Am. J. Ophthalmol. 2018, 190, 58–68. [Google Scholar] [CrossRef]
- Friedman, J.S.; Chang, B.; Kannabiran, C.; Chakarova, C.; Singh, H.P.; Jalali, S.; Hawes, N.L.; Branham, K.; Othman, M.; Filippova, E.; et al. Premature Truncation of a Novel Protein, RD3, Exhibiting Subnuclear Localization Is Associated with Retinal Degeneration. Am. J. Hum. Genet. 2006, 79, 1059–1070. [Google Scholar] [CrossRef]
- Chang, B.; Heckenlively, J.R.; Hawes, N.L.; Roderick, T.H. New Mouse Primary Retinal Degeneration (rd-3). Genomics 1993, 16, 45–49. [Google Scholar] [CrossRef]
- Dell’orco, D.; Cortivo, G.D. Normal GCAPs partly compensate for altered cGMP signaling in retinal dystrophies associated with mutations in GUCA1A. Sci. Rep. 2019, 9, 20105. [Google Scholar] [CrossRef]
- Biasi, A.; Marino, V.; Cortivo, G.D.; Maltese, P.E.; Modarelli, A.M.; Bertelli, M.; Colombo, L.; Dell’orco, D. A Novel GUCA1A Variant Associated with Cone Dystrophy Alters cGMP Signaling in Photoreceptors by Strongly Interacting with and Hyperactivating Retinal Guanylate Cyclase. Int. J. Mol. Sci. 2021, 22, 10809. [Google Scholar] [CrossRef]
- Manes, G.; Mamouni, S.; Hérald, E.; Richard, A.-C.; Sénéchal, A.; Aouad, K.; Bocquet, B.; Meunier, I.; Hamel, C.P. Cone dystrophy or macular dystrophy associated with novel autosomal dominant GUCA1A mutations. Mol. Vis. 2017, 23, 198–209. [Google Scholar]
- Vocke, F.; Weisschuh, N.; Marino, V.; Malfatti, S.; Jacobson, S.G.; Reiff, C.M.; Dell’Orco, D.; Koch, K.W. Dys-function of cGMP signalling in photoreceptors by a macular dystrophy-related mutation in the calcium sensor GCAP1. Hum. Mol. Genet. 2017, 26, 133–144. [Google Scholar]
- Michaelides, M.; Wilkie, S.E.; Jenkins, S.; Holder, G.E.; Hunt, D.M.; Moore, A.T.; Webster, A.R. Mutation in the Gene GUCA1A, Encoding Guanylate Cyclase-Activating Protein 1, Causes Cone, Cone-Rod, and Macular Dystrophy. Ophthalmology 2005, 112, 1442–1447. [Google Scholar] [CrossRef]
- Payne, A.; Downes, S.M.; Bessant, D.A.; Taylor, R.; Holder, G.E.; Warren, M.; Bird, A.C.; Bhattacharya, S.S. A mutation in guanylate cyclase activator 1A (GUCA1A) in an autosomal dominant cone dystrophy pedigree mapping to a new locus on chromosome 6p21.1. Hum. Mol. Genet. 1998, 7, 273–277. [Google Scholar] [CrossRef]
- Jiang, L.; Katz, B.J.; Yang, Z.; Zhao, Y.; Faulkner, N.; Hu, J.; Baird, J.; Baehr, W.; Creel, D.J.; Zhang, K. Autosomal dominant cone dystrophy caused by a novel mutation in the GCAP1 gene (GUCA1A). Mol. Vis. 2005, 11, 143–151. [Google Scholar]
- Olshevskaya, E.V.; Calvert, P.D.; Woodruff, M.L.; Peshenko, I.V.; Savchenko, A.B.; Makino, C.L.; Ho, Y.-S.; Fain, G.L.; Dizhoor, A.M. The Y99C Mutation in Guanylyl Cyclase-Activating Protein 1 Increases Intracellular Ca2+ and Causes Photoreceptor Degeneration in Transgenic Mice. J. Neurosci. 2004, 24, 6078–6085. [Google Scholar] [CrossRef]
- Peshenko, I.V.; Cideciyan, A.V.; Sumaroka, A.; Olshevskaya, E.V.; Scholten, A.; Abbas, S.; Koch, K.-W.; Jacobson, S.G.; Dizhoor, A.M. A G86R mutation in the calcium-sensor protein GCAP1 alters regulation of retinal guanylyl cyclase and causes dominant cone-rod degeneration. J. Biol. Chem. 2019, 294, 3476–3488. [Google Scholar] [CrossRef]
- Huang, S.H.; Pittler, S.J.; Huang, X.; Oliveira, L.; Berson, E.L.; Dryja, T.P. Autosomal recessive retinitis pigmentosa caused by mutations in the α subunit of rod cGMP phosphodiesterase. Nat. Genet. 1995, 11, 468–471. [Google Scholar] [CrossRef]
- McLaughlin, M.E.; Ehrhart, T.L.; Berson, E.L.; Dryja, T.P. Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 1995, 92, 3249–3253. [Google Scholar] [CrossRef]
- Danciger, M.; Blaney, J.; Gao, Y.; Zhao, D.; Heckenlively, J.R.; Jacobson, S.G.; Farber, D.B. Mutations in the PDE6B Gene in Autosomal Recessive Retinitis Pigmentosa. Genomics 1995, 30, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dryja, T.P.; Rucinski, D.E.; Chen, S.H.; Berson, E.L. Frequency of mutations in the gene encoding the alpha subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1859–1865. [Google Scholar]
- Gopalakrishna, K.N.; Boyd, K.; Artemyev, N.O. Mechanisms of mutant PDE6 proteins underlying retinal diseases. Cell Signal. 2017, 37, 74–80. [Google Scholar] [CrossRef]
- Kohl, S.; Coppieters, F.; Meire, F.; Schaich, S.; Roosing, S.; Brennenstuhl, C.; Bolz, S.; van Genderen, M.M.; Riemslag, F.C.; Lukowski, R.; et al. A Nonsense Mutation in PDE6H Causes Autosomal-Recessive Incomplete Achromatopsia. Am. J. Hum. Genet. 2012, 91, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Grau, T.; Artemyev, N.O.; Rosenberg, T.; Dollfus, H.; Haugen, O.H.; Sener, E.C.; Jurklies, B.; Andreasson, S.; Kernstock, C.; Larsen, M.; et al. Decreased catalytic activity and altered activation properties of PDE6C mutants associated with autosomal recessive achromatopsia. Hum. Mol. Genet. 2011, 20, 719–730. [Google Scholar] [CrossRef]
- Thiadens, A.A.; Hollander, A.I.D.; Roosing, S.; Nabuurs, S.B.; Zekveld-Vroon, R.C.; Collin, R.W.; De Baere, E.; Koenekoop, R.K.; van Schooneveld, M.J.; Strom, T.M.; et al. Homozygosity Mapping Reveals PDE6C Mutations in Patients with Early-Onset Cone Photoreceptor Disorders. Am. J. Hum. Genet. 2009, 85, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.; Grau, T.; Dangel, S.; Hurd, R.; Jurklies, B.; Sener, E.C.; Andreasson, S.; Dollfus, H.; Baumann, B.; Bolz, S.; et al. A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc. Natl. Acad. Sci. USA 2009, 106, 19581–19586. [Google Scholar] [CrossRef]
- Farber, D.; Lolley, R.N. Enzymic basis for cyclic GMP accumulation in degenerative photoreceptor cells of mouse retina. J. Cycl. Nucleotide Res. 1976, 2, 139–148. [Google Scholar]
- Farber, D.B. From mice to men: The cyclic GMP phosphodiesterase gene in vision and disease. The Proctor Lecture. Investig. Ophthalmol. Vis. Sci. 1995, 36, 263–275. [Google Scholar]
- Farber, D.B.; Lolley, R.N. Cyclic Guanosine Monophosphate: Elevation in Degenerating Photoreceptor Cells of the C3H Mouse Retina. Science 1974, 186, 449–451. [Google Scholar] [CrossRef]
- Portera-Cailliau, C.; Sung, C.H.; Nathans, J.; Adler, R. Apoptotic photoreceptor cell death in mouse models of retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 1994, 91, 974–978. [Google Scholar] [CrossRef]
- Tsang, S.H.; Gouras, P.; Yamashita, C.K.; Kjeldbye, H.; Fisher, J.; Farber, D.B.; Goff, S.P. Retinal degeneration in mice lacking the gamma subunit of the rod cGMP phosphodiesterase. Science 1996, 272, 1026–1029. [Google Scholar] [CrossRef]
- Chang, B.; Hawes, N.; Hurd, R.; Davisson, M.; Nusinowitz, S.; Heckenlively, J. Retinal degeneration mutants in the mouse. Vis. Res. 2002, 42, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T.; Ikeda, H.O.; Nakano, N.; Muraoka, Y.; Tsuruyama, T.; Okamoto-Furuta, K.; Kohda, H.; Yoshimura, N. Changes in morphology and visual function over time in mouse models of retinal degeneration: An SD-OCT, histology, and electroretinography study. Jpn. J. Ophthalmol. 2016, 60, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Barhoum, R.; Martínez-Navarrete, G.; Corrochano, S.; Germain, F.; Fernandez-Sanchez, L.; de la Rosa, E.; de la Villa, P.; Cuenca, N. Functional and structural modifications during retinal degeneration in the rd10 mouse. Neuroscience 2008, 155, 698–713. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Reingruber, J.; Woodruff, M.L.; Majumder, A.; Camarena, A.; Artemyev, N.; Fain, G.; Chen, J. The PDE6 mutation in the rd10 retinal degeneration mouse model causes protein mislocalization and instability and promotes cell death through increased ion influx. J. Biol. Chem. 2018, 293, 15332–15346. [Google Scholar] [CrossRef] [PubMed]
- Trifunović, D.; Dengler, K.; Michalakis, S.; Zrenner, E.; Wissinger, B.; Paquet-Durand, F. cGMP-dependent cone photoreceptor degeneration in the cpfl1 mouse retina. J. Comp. Neurol. 2010, 518, 3604–3617. [Google Scholar] [CrossRef]
- Schaeferhoff, K.; Michalakis, S.; Tanimoto, N.; Fischer, M.D.; Becirovic, E.; Beck, S.C.; Huber, G.; Rieger, N.; Riess, O.; Wis-singer, B.; et al. Induction of STAT3-related genes in fast degenerating cone photo-receptors of cpfl1 mice. Cell Mol. Life Sci. 2010, 67, 3173–3186. [Google Scholar] [CrossRef]
- Fischer, M.D.; Tanimoto, N.; Beck, S.C.; Huber, G.; Schaeferhoff, K.; Michalakis, S.; Riess, O.; Wissinger, B.; Biel, M.; Bonin, M.; et al. Structural and Functional Phenotyping in the Cone-Specific Photoreceptor Function Loss 1 (cpfl1) Mouse Mutant—A Model of Cone Dystrophies. Adv. Exp. Med. Biol. 2010, 664, 593–599. [Google Scholar] [CrossRef]
- Kandaswamy, S.; Zobel, L.; John, B.; Santhiya, S.T.; Bogedein, J.; Przemeck, G.K.H.; Gailus-Durner, V.; Fuchs, H.; Biel, M.; de Angelis, M.H.; et al. Mutations within the cGMP-binding domain of CNGA1 causing autosomal recessive retinitis pigmentosa in human and animal model. Cell Death Discov. 2022, 8, 387. [Google Scholar] [CrossRef]
- Jin, X.; Qu, L.-H.; Hou, B.-K.; Xu, H.-W.; Meng, X.-H.; Pang, C.-P.; Yin, Z.-Q. Novel compound heterozygous mutation in the CNGA1 gene underlie autosomal recessive retinitis pigmentosa in a Chinese family. Biosci. Rep. 2016, 36, e00289. [Google Scholar] [CrossRef] [PubMed]
- Radojevic, B.; Jones, K.; Klein, M.; Mauro-Herrera, M.; Kingsley, R.; Birch, D.G.; Bennett, L.D. Variable expressivity in patients with autosomal recessive retinitis pigmentosa associated with the gene CNGB1. Ophthalmic Genet. 2021, 42, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, S.; Akahori, M.; Sergeev, Y.; Yoshitake, K.; Ikeo, K.; Furuno, M.; Hayashi, T.; Kondo, M.; Ueno, S.; Tsunoda, K.; et al. Whole Exome Analysis Identifies Frequent CNGA1 Mutations in Japanese Population with Autosomal Recessive Retinitis Pigmentosa. PLoS ONE 2014, 9, e108721. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Gotoh, N.; Kang, I.; Shimada, T.; Usui, T.; Terao, C. A case of retinitis pigmentosa homozygous for a rare CNGA1 causal variant. Sci. Rep. 2021, 11, 4681. [Google Scholar] [CrossRef]
- Nassisi, M.; Smirnov, V.; Hernandez, C.S.; Mohand-Saïd, S.; Condroyer, C.; Antonio, A.; Kühlewein, L.; Kempf, M.; Kohl, S.; Wissinger, B.; et al. CNGB1-related rod-cone dystrophy: A mutation review and update. Hum. Mutat. 2021, 42, 641–666. [Google Scholar] [CrossRef]
- Kohl, S.; Baumann, B.; Broghammer, M.; Jägle, H.; Sieving, P.; Kellner, U.; Spegal, R.; Anastasi, M.; Zrenner, E.; Sharpe, L.T.; et al. Mutations in the CNGB3 gene encoding the beta-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum. Mol. Genet. 2000, 9, 2107–2116. [Google Scholar] [CrossRef]
- Wissinger, B.; Gamer, D.; Jägle, H.; Giorda, R.; Marx, T.; Mayer, S.; Tippmann, S.; Broghammer, M.; Jurklies, B.; Rosenberg, T.; et al. CNGA3 Mutations in Hereditary Cone Photoreceptor Disorders. Am. J. Hum. Genet. 2001, 69, 722–737. [Google Scholar] [CrossRef]
- Sidjanin, D.J.; Lowe, J.K.; McElwee, J.; Milne, B.S.; Phippen, T.M.; Sargan, D.R.; Aguirre, G.D.; Acland, G.M.; Ostrander, E. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum. Mol. Genet. 2002, 11, 1823–1833. [Google Scholar] [CrossRef]
- Kohl, S.; Varsanyi, B.; Antunes, G.A.; Baumann, B.; Hoyng, C.B.; Jägle, H.; Rosenberg, T.; Kellner, U.; Lorenz, B.; Salati, R.; et al. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur. J. Hum. Genet. 2005, 13, 302–308. [Google Scholar] [CrossRef]
- Nishiguchi, K.M.; Sandberg, M.A.; Gorji, N.; Berson, E.L.; Dryja, T.P. Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum. Mutat. 2005, 25, 248–258. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Xiao, Y.; Li, X.; Ruan, S.; Luo, X.; Wan, X.; Wang, F.; Sun, X. Retinal degeneration in mice lacking the cyclic nucleotide-gated channel subunit CNGA1. FASEB J. 2021, 35, e21859. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Morris, L.; Thapa, A.; Ma, H.; Michalakis, S.; Biel, M.; Baehr, W.; Peshenko, I.V.; Dizhoor, A.M.; Ding, X.Q. cGMP accumulation causes photoreceptor degeneration in CNG channel deficiency: Evidence of cGMP cytotoxicity independently of enhanced CNG channel function. J. Neurosci. 2013, 33, 14939–14948. [Google Scholar] [CrossRef]
- Thapa, A.; Morris, L.; Xu, J.; Ma, H.; Michalakis, S.; Biel, M.; Ding, X.-Q. Endoplasmic Reticulum Stress-associated Cone Photoreceptor Degeneration in Cyclic Nucleotide-gated Channel Deficiency. J. Biol. Chem. 2012, 287, 18018–18029. [Google Scholar] [CrossRef]
- Michalakis, S.; Xu, J.; Biel, M.; Ding, X.-Q. Detection of cGMP in the Degenerating Retina. Methods Mol. Biol. 2013, 1020, 235–245. [Google Scholar] [CrossRef]
- Michalakis, S.; Muhlfriedel, R.; Tanimoto, N.; Krishnamoorthy, V.; Koch, S.; Fischer, M.D.; Becirovic, E.; Bai, L.; Huber, G.; Beck, S.C.; et al. Restoration of cone vision in the CNGA3-/- mouse model of congenital complete lack of cone photoreceptor function. Mol. Ther. 2010, 18, 2057–2063. [Google Scholar] [CrossRef]
- Wang, T.; Tsang, S.H.; Chen, J. Two pathways of rod photoreceptor cell death induced by elevated cGMP. Hum. Mol. Genet. 2017, 26, 2299–2306. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Bernhard-Kurz, S.; Arango-Gonzalez, B.; Zrenner, E.; Ueffing, M. Cell Death in rd2/rds Retina: An Apoptotic Process? Investig. Ophthalmol. Vis. Sci. 2012, 53, 6891. [Google Scholar]
- Ding, X.Q.; Nour, M.; Ritter, L.M.; Goldberg, A.F.; Fliesler, S.J.; Naash, M.I. The R172W mutation in peripherin/rds causes a cone-rod dystrophy in transgenic mice. Hum. Mol. Genet. 2004, 13, 2075–2087. [Google Scholar] [CrossRef]
- Stricker, H.M.; Ding, X.Q.; Quiambao, A.; Fliesler, S.J.; Naash, M.I. The Cys214-->Ser mutation in peripherin/rds causes a loss-of-function phenotype in transgenic mice. Biochem. J. 2005, 388, 605–613. [Google Scholar] [CrossRef]
- Tosi, J.; Davis, R.J.; Wang, N.-K.; Naumann, M.; Lin, C.-S.; Tsang, S.H. shRNA knockdown of guanylate cyclase 2e or cyclic nucleotide gated channel alpha 1 increases photoreceptor survival in a cGMP phosphodiesterase mouse model of retinitis pigmentosa. J. Cell Mol. Med. 2011, 15, 1778–1787. [Google Scholar] [CrossRef]
- Ma, H.; Butler, M.R.; Thapa, A.; Belcher, J.; Yang, F.; Baehr, W.; Biel, M.; Michalakis, S.; Ding, X.-Q. cGMP/Protein Kinase G Signaling Suppresses Inositol 1,4,5-Trisphosphate Receptor Phosphorylation and Promotes Endoplasmic Reticulum Stress in Photoreceptors of Cyclic Nucleotide-gated Channel-deficient Mice. J. Biol. Chem. 2015, 290, 20880–20892. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.R.; Ma, H.; Yang, F.; Belcher, J.; Le, Y.-Z.; Mikoshiba, K.; Biel, M.; Michalakis, S.; Iuso, A.; Križaj, D.; et al. Endoplasmic reticulum (ER) Ca2+-channel activity contributes to ER stress and cone death in cyclic nucleotide-gated channel deficiency. J. Biol. Chem. 2017, 292, 11189–11205. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Lockard, R.; Titus, H.; Hiblar, J.; Weller, K.; Wafai, D.; Weleber, R.G.; Duvoisin, R.M.; Morgans, C.W.; Pennesi, M.E. Suppression of cGMP-Dependent Photoreceptor Cytotoxicity with Mycophenolate Is Neuroprotective in Murine Models of Retinitis Pigmentosa. Investig. Ophthalmol. Vis. Sci. 2020, 61, 25. [Google Scholar] [CrossRef]
- Peshenko, I.V.; Olshevskaya, E.V.; Dizhoor, A.M. Retinal degeneration-3 protein attenuates photoreceptor de-generation in transgenic mice expressing dominant mutation of human retinal guanylyl cyclase. J. Biol. Chem. 2021, 297, 101201. [Google Scholar] [CrossRef] [PubMed]
- Morris, L.; Ma, Z.; Thapa, A.; Ma, H.; Michalakis, S.; Biel, M.; Baehr, W.; Ding, X.Q. Exploration of the Mechanisms of Cone Photoreceptor Death in the Deficiency of Phosphodiesterase. Investig. Ophthalmol. Vis. Sci. 2013, 54, 5953. [Google Scholar]
- Paquet-Durand, F.; Beck, S.; Michalakis, S.; Goldmann, T.; Huber, G.; Mühlfriedel, R.; Trifunović, D.; Fischer, M.D.; Fahl, E.; Duetsch, G.; et al. A key role for cyclic nucleotide gated (CNG) channels in cGMP-related retinitis pigmentosa. Hum. Mol. Genet. 2011, 20, 941–947. [Google Scholar] [CrossRef]
- Hofmann, F.; Bernhard, D.; Lukowski, R.; Weinmeister, P. cGMP Regulated Protein Kinases (cGK). In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 137–162. [Google Scholar] [CrossRef]
- Hofmann, F. The Biology of Cyclic GMP-dependent Protein Kinases. J. Biol. Chem. 2005, 280, 1–4. [Google Scholar] [CrossRef]
- Vighi, E.; Trifunovic, D.; Veiga-Crespo, P.; Rentsch, A.; Hoffmann, D.; Sahaboglu, A.; Strasser, T.; Kulkarni, M.; Bertolotti, E.; van den Heuvel, A.; et al. Combination of cGMP analogue and drug delivery system provides functional protection in hereditary retinal degeneration. Proc. Natl. Acad. Sci. USA 2018, 115, E2997–E3006. [Google Scholar] [CrossRef]
- Koch, M.; Scheel, C.; Ma, H.; Yang, F.; Stadlmeier, M.; Glück, A.F.; Murenu, E.; Traube, F.R.; Carell, T.; Biel, M.; et al. The cGMP-Dependent Protein Kinase 2 Contributes to Cone Photoreceptor Degeneration in the Cnga3-Deficient Mouse Model of Achromatopsia. Int. J. Mol. Sci. 2020, 22, 52. [Google Scholar] [CrossRef]
- Szegezdi, E.; Logue, S.E.; Gorman, A.M.; Samali, A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006, 7, 880–885. [Google Scholar] [CrossRef]
- Kim, I.; Xu, W.; Reed, J.C. Cell death and endoplasmic reticulum stress: Disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 2008, 7, 1013–1030. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.; Stolz, J.; Kohl, S.; Chiang, W.-C.; Lin, J.H. Endoplasmic reticulum stress in human photoreceptor diseases. Brain Res. 2016, 1648, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Thapa, A.; Morris, L.M.; Michalakis, S.; Biel, M.; Frank, M.B.; Bebak, M.; Ding, X.-Q. Loss of cone cyclic nucleotide-gated channel leads to alterations in light response modulating system and cellular stress response pathways: A gene expression profiling study. Hum. Mol. Genet. 2013, 22, 3906–3919. [Google Scholar] [CrossRef]
- Nakagawa, T.; Yuan, J. Cross-Talk between Two Cysteine Protease Families: Activation of caspase-12 by calpain in apoptosis. J. Cell Biol. 2000, 150, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Zhu, H.; Morishima, N.; Li, E.; Xu, J.; Yankner, B.A.; Yuan, J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 2000, 403, 98–103. [Google Scholar] [CrossRef]
- Haidara, K.; Marion, M.; Gascon-Barré, M.; Denizeau, F.; Averill-Bates, D.A. Implication of caspases and subcellular compartments in tert-butylhydroperoxide induced apoptosis. Toxicol. Appl. Pharmacol. 2008, 229, 65–76. [Google Scholar] [CrossRef]
- Sanges, D.; Comitato, A.; Tammaro, R.; Marigo, V. Apoptosis in retinal degeneration involves cross-talk between apoptosis-inducing factor (AIF) and caspase-12 and is blocked by calpain inhibitors. Proc. Natl. Acad. Sci. USA 2006, 103, 17366–17371. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Zakar, M.; Shmuelivich, F.; Nahon, E.; Vardi, N. Retina expresses a novel variant of the ryanodine receptor. Eur. J. Neurosci. 2007, 26, 3113–3125. [Google Scholar] [CrossRef]
- Yang, F.; Ma, H.; Butler, M.R.; Ding, X. Potential contribution of ryanodine receptor 2 upregulation to cGMP/PKG signaling-induced cone degeneration in cyclic nucleotide-gated channel deficiency. FASEB J. 2020, 34, 6335–6350. [Google Scholar] [CrossRef]
- Ma, H.; Yang, F.; Butler, M.R.; Rapp, J.; Le, Y.-Z.; Ding, X.-Q. Ryanodine Receptor 2 Contributes to Impaired Protein Localization in Cyclic Nucleotide-Gated Channel Deficiency. Eneuro 2019, 6, 0119–19. [Google Scholar] [CrossRef]
- Yang, F.; Ma, H.; Butler, M.R.; Ding, X.Q. Preservation of endoplasmic reticulum (ER) Ca2+ stores by deletion of inositol-1,4,5-trisphosphate receptor type 1 promotes ER retrotranslocation, proteostasis, and protein outer segment localization in cyclic nucleotide-gated channel-deficient cone photoreceptors. FASEB J. 2021, 35, e21579. [Google Scholar]
- Roy, A.; Tolone, A.; Hilhorst, R.; Groten, J.; Tomar, T.; Paquet-Durand, F. Kinase activity profiling identifies putative downstream targets of cGMP/PKG signaling in inherited retinal neurodegeneration. Cell Death Discov. 2022, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rasmussen, M.; Ekstrom, P. cGMP-PKG dependent transcriptome in normal and degenerating retinas: Novel insights into the retinitis pigmentosa pathology. Exp. Eye Res. 2021, 212, 108752. [Google Scholar] [CrossRef] [PubMed]
- Vosler, P.S.; Sun, D.; Wang, S.; Gao, Y.; Kintner, D.B.; Signore, A.P.; Cao, G.; Chen, J. Calcium dysregulation induces apoptosis-inducing factor release: Cross-talk between PARP-1- and calpain- signaling pathways. Exp. Neurol. 2009, 218, 213–220. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, K.M.; Gnegy, M.E.; Park, Y.H.; Mukerjee, N.; Wang, K.K. Procaspase-3 and poly(ADP)ribose polymerase (PARP) are calpain substrates. Biochem. Biophys. Res. Commun. 1999, 263, 94–99. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Azadi, S.; Hauck, S.M.; Ueffing, M.; van Veen, T.; Ekström, P. Calpain is activated in degenerating photoreceptors in the rd1 mouse. J. Neurochem. 2006, 96, 802–814. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Sanges, D.; McCall, J.; Silva, J.; Van Veen, T.; Marigo, V.; Ekström, P. Photoreceptor rescue and toxicity induced by different calpain inhibitors. J. Neurochem. 2010, 115, 930–940. [Google Scholar] [CrossRef]
- Power, M.J.; Rogerson, L.E.; Schubert, T.; Berens, P.; Euler, T.; Paquet-Durand, F. Systematic spatiotemporal mapping reveals divergent cell death pathways in three mouse models of hereditary retinal degeneration. J. Comp. Neurol. 2020, 528, 1113–1139. [Google Scholar] [CrossRef]
- Vu, J.T.; Wang, E.; Wu, J.; Sun, Y.J.; Velez, G.; Bassuk, A.G.; Lee, S.H.; Mahajan, V.B. Calpains as mechanistic drivers and therapeutic targets for ocular disease. Trends Mol. Med. 2022, 28, 644–661. [Google Scholar] [CrossRef]
- Comitato, A.; Sanges, D.; Rossi, A.; Humphries, M.M.; Marigo, V. Activation of Bax in Three Models of Retinitis Pigmentosa. Investig. Ophthalmol. Vis. Sci. 2014, 55, 3555–3561. [Google Scholar] [CrossRef]
- Oka, T.; Nakajima, T.; Tamada, Y.; Shearer, T.R.; Azuma, M. Contribution of calpains to photoreceptor cell death in N-methyl-N-nitrosourea-treated rats. Exp. Neurol. 2007, 204, 39–48. [Google Scholar] [CrossRef]
- Baehr, W.; Palczewski, K. Guanylate Cyclase-Activating Proteins and Retina Disease. Subcell. Biochem. 2007, 45, 71–91. [Google Scholar] [CrossRef]
- Plana-Bonamaisó, A.; López-Begines, S.; Andilla, J.; Fidalgo, M.J.; Loza-Alvarez, P.; Estanyol, J.M.; de la Villa, P.; Méndez, A. GCAP neuronal calcium sensor proteins mediate photoreceptor cell death in the rd3 mouse model of LCA12 congenital blindness by involving endoplasmic reticulum stress. Cell Death Dis. 2020, 11, 62. [Google Scholar] [CrossRef]
- Sancho-Pelluz, J.; Alavi, M.V.; Sahaboglu, A.; Kustermann, S.; Farinelli, P.; Azadi, S.; van Veen, T.; Romero, F.J.; Paquet-Durand, F.; Ekström, P. Excessive HDAC activation is critical for neurodegeneration in the rd1 mouse. Cell Death Dis. 2010, 1, e24. [Google Scholar] [CrossRef] [PubMed]
- Trifunović, D.; Arango-Gonzalez, B.; Comitato, A.; Barth, M.; Del Amo, E.M.; Kulkarni, M.; Sahaboglu, A.; Hauck, S.M.; Urtti, A.; Arsenijevic, Y.; et al. HDAC inhibition in the cpfl1 mouse protects de-generating cone photoreceptors in vivo. Hum. Mol. Genet. 2016, 25, 4462–4472. [Google Scholar] [PubMed]
- Ekstrom, P.A.; Ueffing, M.; Zrenner, E.; Paquet-Durand, F. Novel in situ activity assays for the quantitative molecular analysis of neurodegenerative processes in the retina. Curr. Med. Chem. 2014, 21, 3478–3493. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ge, P. Parthanatos in the pathogenesis of nervous system diseases. Neuroscience 2020, 449, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Paquet-Durand, F.; Silva, J.; Talukdar, T.; Johnson, L.E.; Azadi, S.; van Veen, T.; Ueffing, M.; Hauck, S.M.; Ekström, P.A.R. Excessive Activation of Poly(ADP-Ribose) Polymerase Contributes to Inherited Photoreceptor Degeneration in the Retinal Degeneration 1 Mouse. J. Neurosci. 2007, 27, 10311–10319. [Google Scholar] [CrossRef]
- Zhang, F.; Xie, R.; Munoz, F.M.; Lau, S.S.; Monks, T.J. PARP-1 hyperactivation and reciprocal elevations in intracellular Ca2+ during ROS-induced nonapoptotic cell death. Toxicol. Sci. 2014, 140, 118–134. [Google Scholar] [CrossRef]
- Geistrikh, I.; Visochek, L.; Klein, R.; Miller, L.; Mittelman, L.; Shainberg, A.; Cohen-Armon, M. Ca2+-induced PARP-1 activation and ANF expression are coupled events in cardiomyocytes. Biochem. J. 2011, 438, 337–347. [Google Scholar] [CrossRef]
- Wenzel, A.; Grimm, C.; Samardzija, M.; Remé, C.E. Molecular mechanisms of light-induced photoreceptor apoptosis and neuroprotection for retinal degeneration. Prog. Retin. Eye Res. 2005, 24, 275–306. [Google Scholar] [CrossRef] [PubMed]
- Candé, C.; Vahsen, N.; Garrido, C.; Kroemer, G. Apoptosis-inducing factor (AIF): Caspase-independent after all. Cell Death Differ. 2004, 11, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; An, R.; Umanah, G.K.; Park, H.; Nambiar, K.; Eacker, S.M.; Kim, B.; Bao, L.; Harraz, M.M.; Chang, C.; et al. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science 2016, 354, 6308. [Google Scholar] [CrossRef]
- Mizukoshi, S.; Nakazawa, M.; Sato, K.; Ozaki, T.; Metoki, T.; Ishiguro, S.-I. Activation of mitochondrial calpain and release of apoptosis-inducing factor from mitochondria in RCS rat retinal degeneration. Exp. Eye Res. 2010, 91, 353–361. [Google Scholar] [CrossRef]
- Comitato, A.; Schiroli, D.; Montanari, M.; Marigo, V. Calpain Activation Is the Major Cause of Cell Death in Photoreceptors Expressing a Rhodopsin Misfolding Mutation. Mol. Neurobiol. 2020, 57, 589–599. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Ma, H.; Yang, F.; Ding, X. cGMP Signaling in Photoreceptor Degeneration. Int. J. Mol. Sci. 2023, 24, 11200. https://doi.org/10.3390/ijms241311200
Li S, Ma H, Yang F, Ding X. cGMP Signaling in Photoreceptor Degeneration. International Journal of Molecular Sciences. 2023; 24(13):11200. https://doi.org/10.3390/ijms241311200
Chicago/Turabian StyleLi, Shujuan, Hongwei Ma, Fan Yang, and Xiqin Ding. 2023. "cGMP Signaling in Photoreceptor Degeneration" International Journal of Molecular Sciences 24, no. 13: 11200. https://doi.org/10.3390/ijms241311200
APA StyleLi, S., Ma, H., Yang, F., & Ding, X. (2023). cGMP Signaling in Photoreceptor Degeneration. International Journal of Molecular Sciences, 24(13), 11200. https://doi.org/10.3390/ijms241311200




