Identification of the Intestinal Microbes Associated with Locomotion
Abstract
1. Introduction
2. Results
2.1. A Random Colonization of Human Gut Microbiome into Conventional Mice through FMT Had a Different Impact on Locomotion
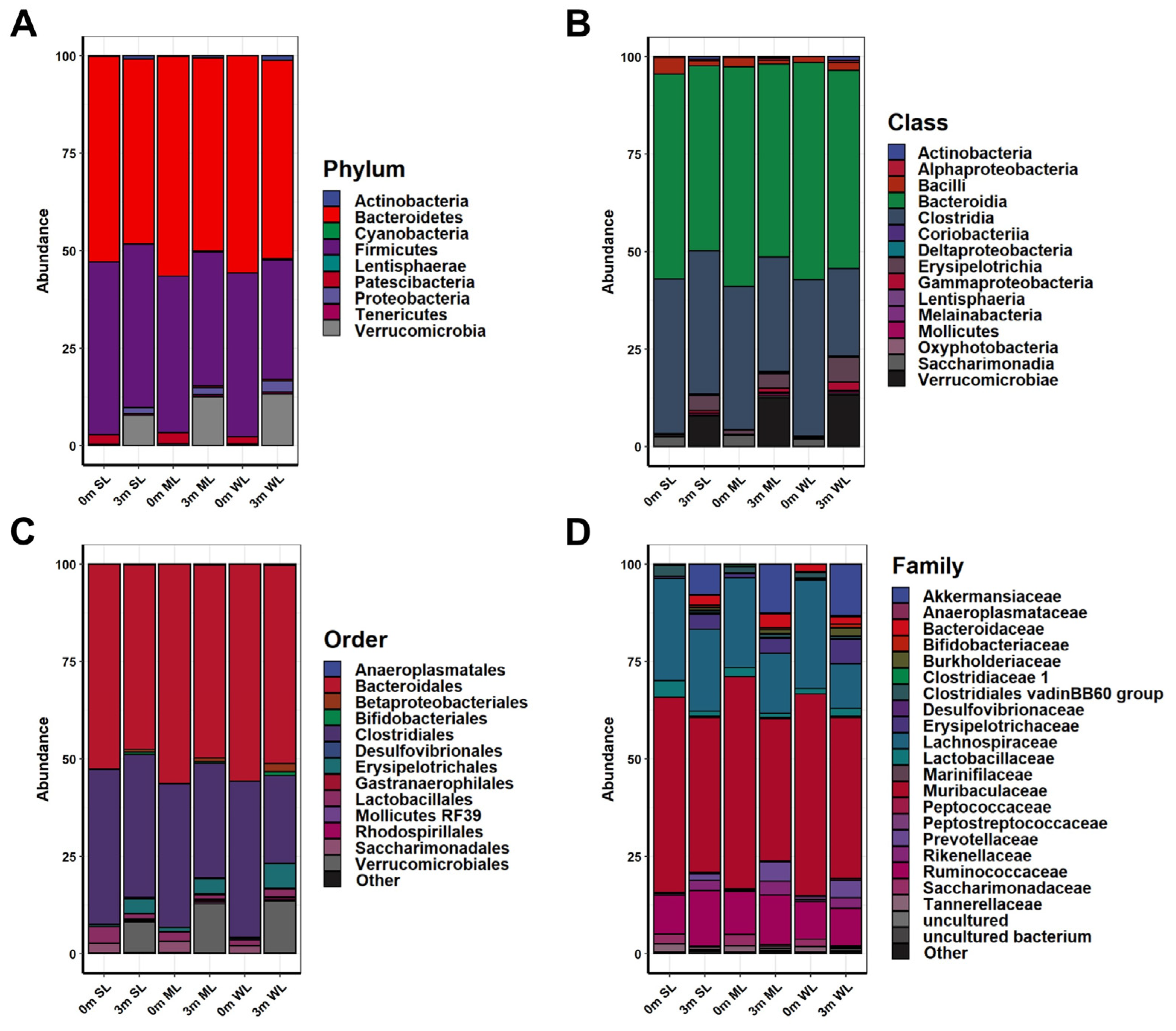
2.2. Different Types of Gut Microbiome Were Established in Each of the Experimental Mice after FMT with Human Feces
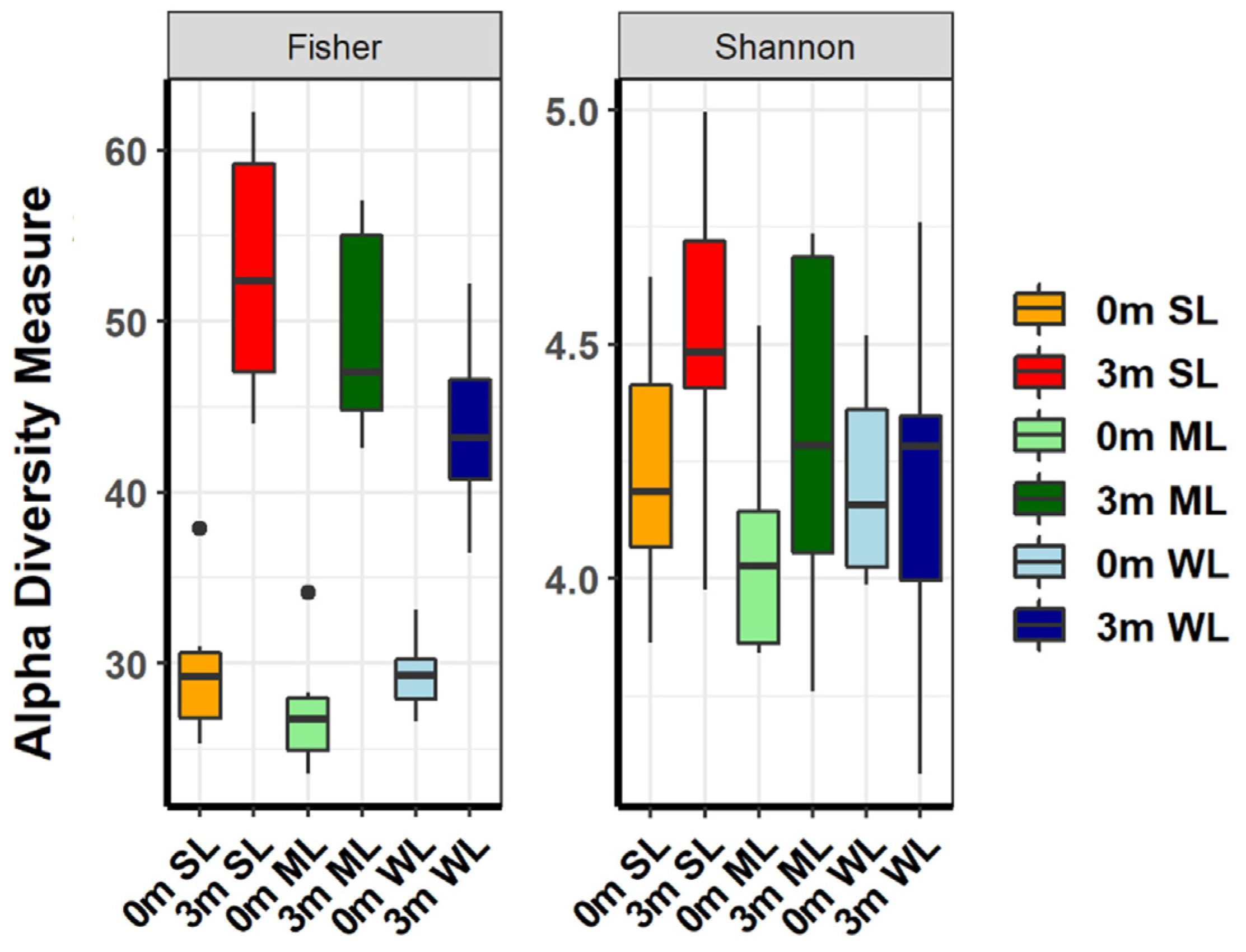
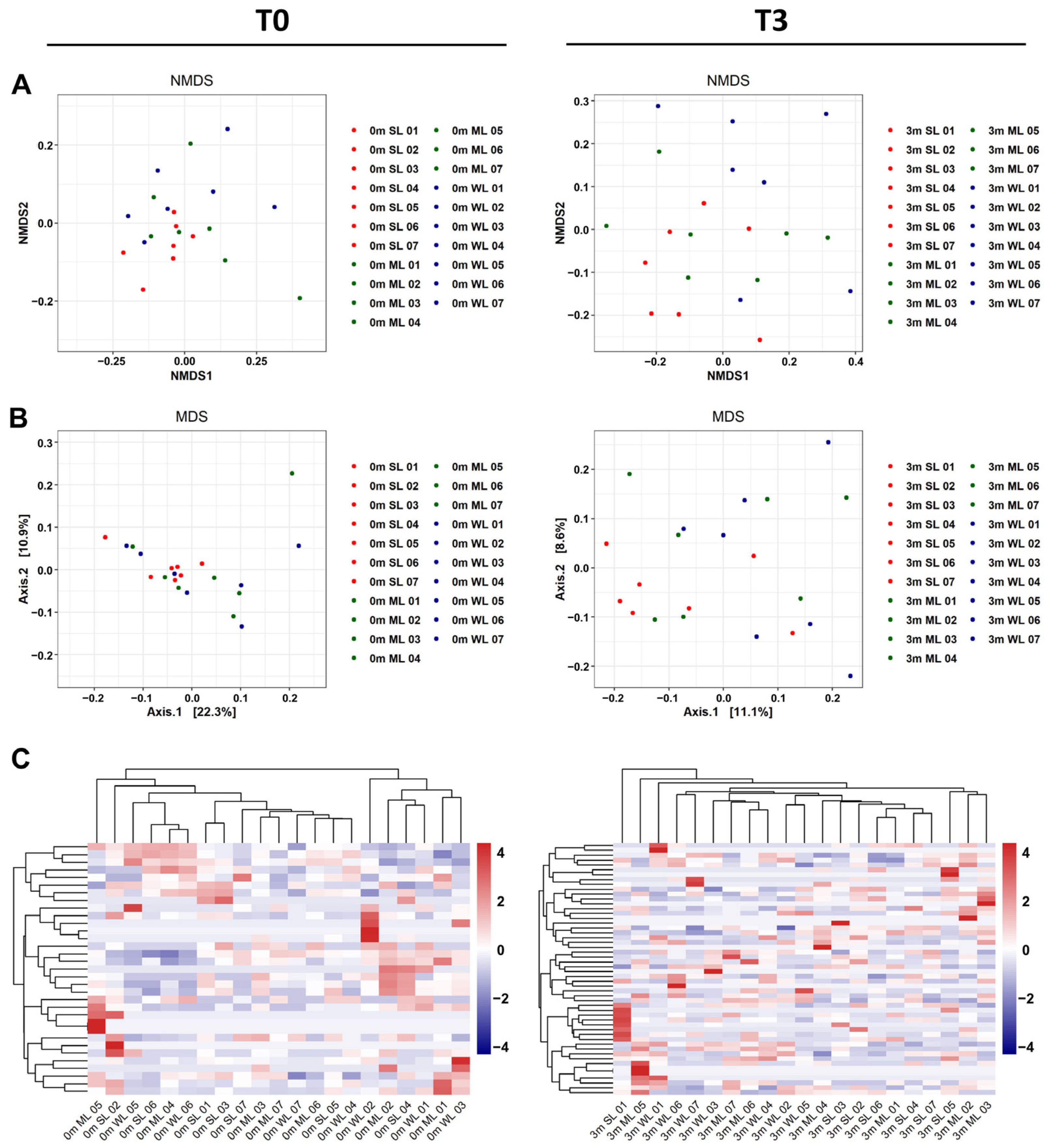
2.3. The Differences in Locomotion Were Correlated with the Shifts in the Composition of the Gut Microbiome
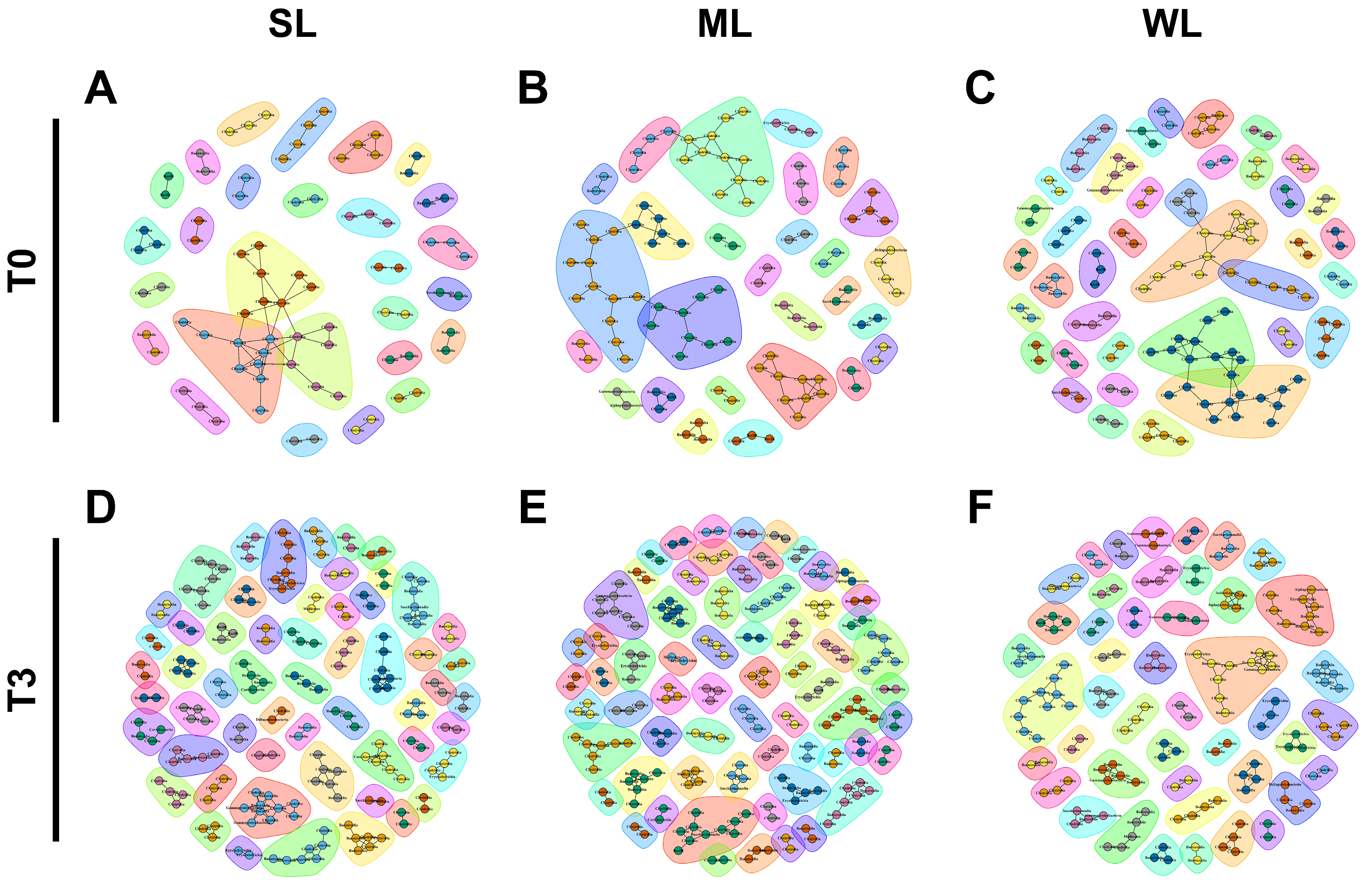
2.4. Different Microbial Communities Were Established in Each of the Three Groups of Mice following the FMT
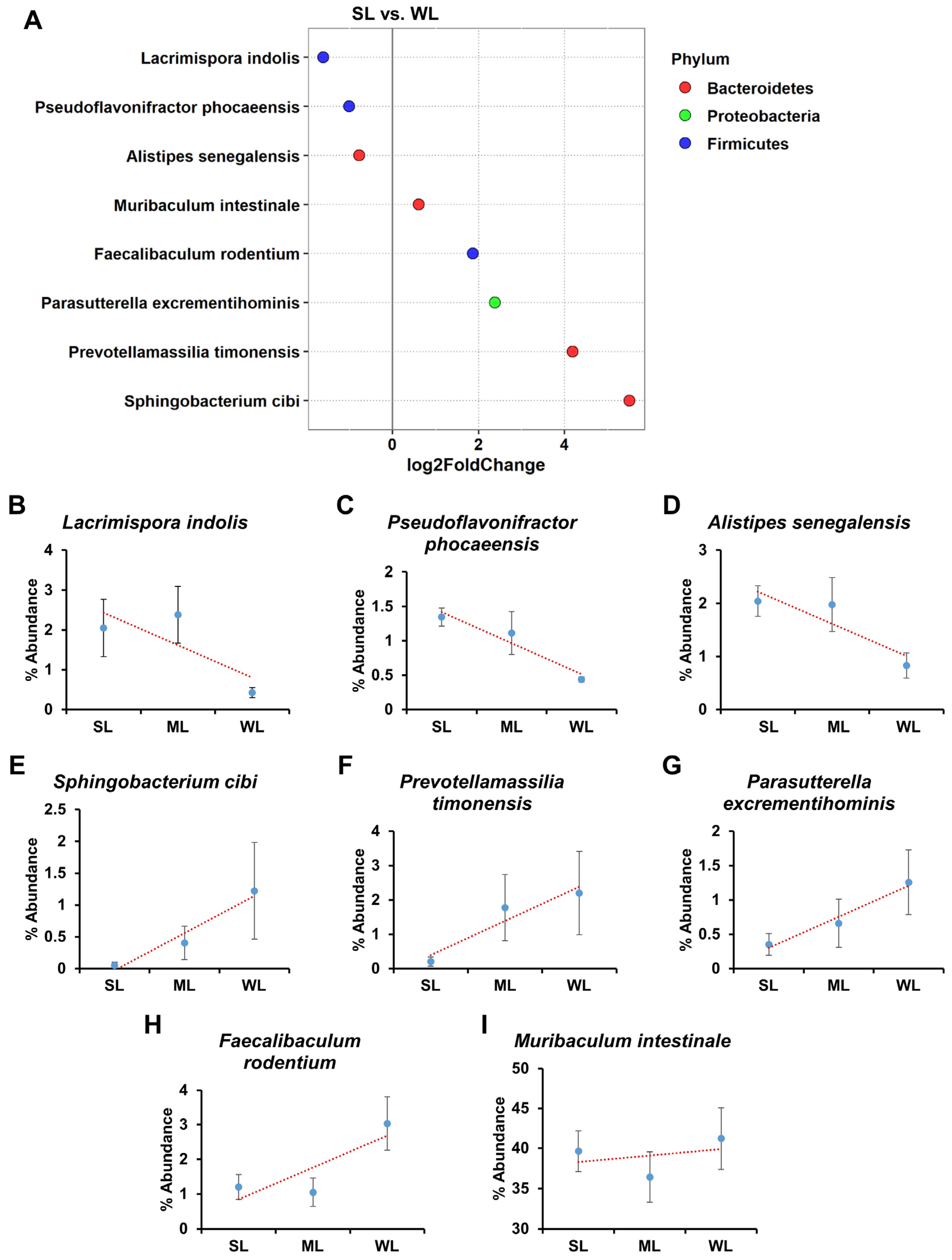
2.5. Intestinal Microbes That Affect Locomotion Were Identified at the Species Level
3. Discussion
4. Materials and Methods
4.1. Study Design and Animal Experiment
4.2. Wire Hanging Test
4.3. Analyses of Biochemical Parameters
4.4. Histological Analysis
4.5. DNA Extraction and 16S rRNA Gene Sequencing
4.6. Data and Statistical Analyses
4.7. The α-Diversity Analysis for Relative Abundance Evaluation of Material and Microbiome
4.8. The β-Analysis for Relative Abundance Evaluation of Material and Microbiome
4.9. Construction of Heatmap and Phylogenetic Tree
4.10. Co-Occurrence Network Construction
4.11. Differential Abundance
4.12. Quantification and Statistical Analysis
4.13. Ethics Approval
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Chung, H.J.; Nguyen, T.T.B.; Kim, H.J.; Hong, S.T. Gut Microbiota as a Missing Link Between Nutrients and Traits of Human. Front. Microbiol. 2018, 9, 1510. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469.e5. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef]
- Chen, X.; D’Souza, R.; Hong, S.T. The role of gut microbiota in the gut-brain axis: Current challenges and perspectives. Protein Cell 2013, 4, 403–414. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Sheflin, A.M.; Whitney, A.K.; Weir, T.L. Cancer-promoting effects of microbial dysbiosis. Curr. Oncol. Rep. 2014, 16, 406. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, Z.; Liu, B.; Hou, D.; Liang, Y.; Zhang, J.; Shi, P. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genom. 2013, 14, 669. [Google Scholar] [CrossRef]
- Zou, R.; Wang, Y.; Duan, M.; Guo, M.; Zhang, Q.; Zheng, H. Dysbiosis of Gut Fungal Microbiota in Children with Autism Spectrum Disorders. J. Autism Dev. Disord. 2021, 51, 267–275. [Google Scholar] [CrossRef]
- Liu, C.; Cheung, W.H.; Li, J.; Chow, S.K.; Yu, J.; Wong, S.H.; Ip, M.; Sung, J.J.Y.; Wong, R.M.Y. Understanding the gut microbiota and sarcopenia: A systematic review. J. Cachexia Sarcopenia Muscle 2021, 12, 1393–1407. [Google Scholar] [CrossRef] [PubMed]
- Won, S.M.; Oh, K.K.; Gupta, H.; Ganesan, R.; Sharma, S.P.; Jeong, J.J.; Yoon, S.J.; Jeong, M.K.; Min, B.H.; Hyun, J.Y.; et al. The Link between Gut Microbiota and Hepatic Encephalopathy. Int. J. Mol. Sci. 2022, 23, 8999. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.L.; Gold, M.J.; Hartmann, M.; Willing, B.P.; Thorson, L.; Wlodarska, M.; Gill, N.; Blanchet, M.R.; Mohn, W.W.; McNagny, K.M.; et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012, 13, 440–447. [Google Scholar] [CrossRef]
- Han, H.; Li, Y.; Fang, J.; Liu, G.; Yin, J.; Li, T.; Yin, Y. Gut Microbiota and Type 1 Diabetes. Int. J. Mol. Sci. 2018, 19, 995. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.H.; Fåk, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Bäckhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef] [PubMed]
- Padhi, P.; Worth, C.; Zenitsky, G.; Jin, H.; Sambamurti, K.; Anantharam, V.; Kanthasamy, A.; Kanthasamy, A.G. Mechanistic Insights Into Gut Microbiome Dysbiosis-Mediated Neuroimmune Dysregulation and Protein Misfolding and Clearance in the Pathogenesis of Chronic Neurodegenerative Disorders. Front. Neurosci. 2022, 16, 836605. [Google Scholar] [CrossRef]
- Liu, J.; Lkhagva, E.; Chung, H.J.; Kim, H.J.; Hong, S.T. The Pharmabiotic Approach to Treat Hyperammonemia. Nutrients 2018, 10, 140. [Google Scholar] [CrossRef]
- Nguyen, T.T.B.; Chung, H.J.; Kim, H.J.; Hong, S.T. Establishment of an ideal gut microbiota to boost healthy growth of neonates. Crit. Rev. Microbiol. 2019, 45, 118–129. [Google Scholar] [CrossRef]
- Nguyen, T.T.B.; Jin, Y.Y.; Chung, H.J.; Hong, S.T. Pharmabiotics as an Emerging Medication for Metabolic Syndrome and Its Related Diseases. Molecules 2017, 22, 1795. [Google Scholar] [CrossRef]
- McFarland, L.V.; Dublin, S. Meta-analysis of probiotics for the treatment of irritable bowel syndrome. World J. Gastroenterol. 2008, 14, 2650–2661. [Google Scholar] [CrossRef]
- Chenoll, E.; Casinos, B.; Bataller, E.; Astals, P.; Echevarría, J.; Iglesias, J.R.; Balbarie, P.; Ramón, D.; Genovés, S. Novel probiotic Bifidobacterium bifidum CECT 7366 strain active against the pathogenic bacterium Helicobacter pylori. Appl. Environ. Microbiol. 2011, 77, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal permeability—A new target for disease prevention and therapy. BMC Gastroenterol. 2014, 14, 189. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Gährs, C.; Vidal-Gadea, A. Locomotion. In Encyclopedia of Animal Cognition and Behavior; Vonk, J., Shackelford, T.K., Eds.; Springer: Cham, Switzerland, 2022. [Google Scholar]
- Clauss, M.; Gérard, P.; Mosca, A.; Leclerc, M. Interplay between Exercise and Gut Microbiome in the Context of Human Health and Performance. Front. Nutr. 2021, 8, 637010. [Google Scholar] [CrossRef]
- Monda, V.; Villano, I.; Messina, A.; Valenzano, A.; Esposito, T.; Moscatelli, F.; Viggiano, A.; Cibelli, G.; Chieffi, S.; Monda, M.; et al. Exercise Modifies the Gut Microbiota with Positive Health Effects. Oxid. Med. Cell Longev. 2017, 2017, 3831972. [Google Scholar] [CrossRef]
- Matsumoto, M.; Inoue, R.; Tsukahara, T.; Ushida, K.; Chiji, H.; Matsubara, N.; Hara, H. Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci. Biotechnol. Biochem. 2008, 72, 572–576. [Google Scholar] [CrossRef]
- Zheng, Q.; Cui, G.; Chen, J.; Gao, H.; Wei, Y.; Uede, T.; Chen, Z.; Diao, H. Regular Exercise Enhances the Immune Response against Microbial Antigens through Up-Regulation of Toll-like Receptor Signaling Pathways. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2015, 37, 735–746. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, Z.; Liu, T.; Zhang, W. Piezo-like Gene Regulates Locomotion in Drosophila Larvae. Cell Rep. 2019, 26, 1369–1377.e4. [Google Scholar] [CrossRef]
- Samson, M.M.; Crowe, A.; de Vreede, P.L.; Dessens, J.A.; Duursma, S.A.; Verhaar, H.J. Differences in gait parameters at a preferred walking speed in healthy subjects due to age, height and body weight. Aging 2001, 13, 16–21. [Google Scholar] [CrossRef]
- Kopp, R.; Legler, J.; Legradi, J. Alterations in locomotor activity of feeding zebrafish larvae as a consequence of exposure to different environmental factors. Environ. Sci. Pollut. Res. Int. 2018, 25, 4085–4093. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.; Ahn, J.S.; Islam, M.M.; Lkhagva, E.; Chung, H.J.; Hong, S.T. Comparative Analysis of Original and Replaced Gut Microbiomes within Same Individuals Identified the Intestinal Microbes Associated with Weight Gaining. Microorganisms 2022, 10, 1062. [Google Scholar] [CrossRef] [PubMed]
- Jumpertz, R.; Le, D.S.; Turnbaugh, P.J.; Trinidad, C.; Bogardus, C.; Gordon, J.I.; Krakoff, J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011, 94, 58–65. [Google Scholar] [CrossRef]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Nichols, R.G.; Davenport, E.R. The relationship between the gut microbiome and host gene expression: A review. Hum. Genet. 2021, 140, 747–760. [Google Scholar] [CrossRef]
- Cortese, R.; Lu, L.; Yu, Y.; Ruden, D.; Claud, E.C. Epigenome-Microbiome crosstalk: A potential new paradigm influencing neonatal susceptibility to disease. Epigenetics 2016, 11, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.H.; Gadkari, M.; Zhou, Q.; Yu, S.; Gao, N.; Guan, Y.; Schady, D.; Roshan, T.N.; Chen, M.H.; Laritsky, E.; et al. Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol. 2015, 16, 211. [Google Scholar] [CrossRef]
- Pan, W.H.; Sommer, F.; Falk-Paulsen, M.; Ulas, T.; Best, P.; Fazio, A.; Kachroo, P.; Luzius, A.; Jentzsch, M.; Rehman, A.; et al. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med. 2018, 10, 27. [Google Scholar] [CrossRef]
- Davison, J.M.; Lickwar, C.R.; Song, L.; Breton, G.; Crawford, G.E.; Rawls, J.F. Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor Hepatocyte nuclear factor 4 alpha. Genome Res. 2017, 27, 1195–1206. [Google Scholar] [CrossRef]
- Collado, M.C.; Cernada, M.; Baüerl, C.; Vento, M.; Pérez-Martínez, G. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes 2012, 3, 352–365. [Google Scholar] [CrossRef]
- Johnson, K.V. Gut microbiome composition and diversity are related to human personality traits. Hum. Microb. J. 2020, 15, 100069. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.; Panebianco, C.; Picchianti-Diamanti, A.; Laganà, B.; Cavalieri, D.; Potenza, A.; Pracella, R.; Binda, E.; Copetti, M.; Pazienza, V. Gut Microbiota Profiles Differ among Individuals Depending on Their Region of Origin: An Italian Pilot Study. Int. J. Environ. Res. Public Health 2019, 16, 4065. [Google Scholar] [CrossRef] [PubMed]
- Biddle, A.S.; Leschine, S.; Huntemann, M.; Han, J.; Chen, A.; Kyrpides, N.; Markowitz, V.; Palaniappan, K.; Ivanova, N.; Mikhailova, N.; et al. The complete genome sequence of Clostridium indolis DSM 755(T.). Stand. Genom. Sci. 2014, 9, 1089–1104. [Google Scholar] [CrossRef] [PubMed]
- Ricaboni, D.; Mailhe, M.; Benezech, A.; Andrieu, C.; Fournier, P.E.; Raoult, D. ‘Pseudoflavonifractor phocaeensis’ gen. nov., sp. nov., isolated from human left colon. New Microbes New Infect. 2016, 17, 15–17. [Google Scholar] [CrossRef]
- Mishra, A.K.; Gimenez, G.; Lagier, J.C.; Robert, C.; Raoult, D.; Fournier, P.E. Genome sequence and description of Alistipes senegalensis sp. nov. Stand. Genom. Sci. 2012, 6, 304–314. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Lai, W.A.; Hameed, A.; Liu, Y.C.; Hsu, Y.H.; Lin, S.Y.; Young, C.C. Sphingobacterium cibi sp. nov., isolated from the food-waste compost and emended descriptions of Sphingobacterium spiritivorum (Holmes et al. 1982) Yabuuchi et al. 1983 and Sphingobacteriumthermophilum Yabe et al. 2013. Int. J. Syst. Evol. Microbiol. 2016, 66, 5336–5344. [Google Scholar] [CrossRef]
- Ndongo, S.; Lagier, J.C.; Fournier, P.E.; Raoult, D.; Khelaifia, S. “Prevotellamassilia timonensis”, a new bacterial species isolated from the human gut. New Microbes New Infect. 2016, 13, 102–103. [Google Scholar] [CrossRef]
- Nagai, F.; Morotomi, M.; Sakon, H.; Tanaka, R. Parasutterella excrementihominis gen. nov., sp. nov., a member of the family Alcaligenaceae isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 7, 1793–1797. [Google Scholar] [CrossRef]
- Chang, D.H.; Rhee, M.S.; Ahn, S.; Bang, B.H.; Oh, J.E.; Lee, H.K.; Kim, B.C. Faecalibaculum rodentium gen. nov., sp. nov., isolated from the faeces of a laboratory mouse. Antonie Van Leeuwenhoek 2015, 108, 1309–1318. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Pukall, R.; Abt, B.; Foesel, B.U.; Meier-Kolthoff, J.P.; Kumar, N.; Bresciani, A.; Martínez, I.; Just, S.; Ziegler, C.; et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat. Microbiol. 2016, 1, 16131. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.S.; Koo, B.C.; Choi, Y.J.; Jung, W.W.; Kim, H.S.; Lee, S.J.; Chung, H.J.; Hong, S.T. Identification of the intestinal microbes associated with muscle strength. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Chung, H.J.; Lee, H.K.; Kim, H.J.; Baek, S.H.; Hong, S.T. Gene expression profiles and physiological data from mice fed resveratrol-enriched rice DJ526. Sci. Data 2016, 3, 160114. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.J.; Sharma, S.P.; Kim, H.J.; Baek, S.H.; Hong, S.T. The resveratrol-enriched rice DJ526 boosts motor coordination and physical strength. Sci. Rep. 2016, 6, 23958. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.J.; Lee, H.K.; Kwon, K.B.; Kim, H.J.; Hong, S.T. Transferrin as a thermosensitizer in radiofrequency hyperthermia for cancer treatment. Sci. Rep. 2018, 8, 13505. [Google Scholar] [CrossRef] [PubMed]
- Lkhagva, E.; Chung, H.J.; Hong, J.; Tang, W.H.W.; Lee, S.I.; Hong, S.T.; Lee, S. The regional diversity of gut microbiome along the GI tract of male C57BL/6 mice. BMC Microbiol. 2021, 21, 44. [Google Scholar] [CrossRef]
- Ahn, J.S.; Lkhagva, E.; Jung, S.; Kim, H.J.; Chung, H.J.; Hong, S.T. Fecal Microbiome Does Not Represent Whole Gut Microbiome. Cell. Microbiol. 2023, 2023, 6868417. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.N.; Stine, O.C.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: From raw reads to community analyses. F1000Res 2016, 5, 1492. [Google Scholar] [CrossRef]
- Lkhagva, E.; Chung, H.J.; Ahn, J.S.; Hong, S.T. Host Factors Affect the Gut Microbiome More Significantly than Diet Shift. Microorganisms 2021, 9, 2520. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, Y.; Huang, L. ImageGP: An easy-to-use data visualization web server for scientific researchers. Imeta 2022, 1, e5. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, J.-S.; Choi, Y.-J.; Kim, H.-B.; Chung, H.-J.; Hong, S.-T. Identification of the Intestinal Microbes Associated with Locomotion. Int. J. Mol. Sci. 2023, 24, 11392. https://doi.org/10.3390/ijms241411392
Ahn J-S, Choi Y-J, Kim H-B, Chung H-J, Hong S-T. Identification of the Intestinal Microbes Associated with Locomotion. International Journal of Molecular Sciences. 2023; 24(14):11392. https://doi.org/10.3390/ijms241411392
Chicago/Turabian StyleAhn, Ji-Seon, Yu-Jin Choi, Han-Byeol Kim, Hea-Jong Chung, and Seong-Tshool Hong. 2023. "Identification of the Intestinal Microbes Associated with Locomotion" International Journal of Molecular Sciences 24, no. 14: 11392. https://doi.org/10.3390/ijms241411392
APA StyleAhn, J.-S., Choi, Y.-J., Kim, H.-B., Chung, H.-J., & Hong, S.-T. (2023). Identification of the Intestinal Microbes Associated with Locomotion. International Journal of Molecular Sciences, 24(14), 11392. https://doi.org/10.3390/ijms241411392





