Investigation of the Effects of Metallic Nanoparticles on Fertility Outcomes and Endocrine Modification of the Hypothalamic-Pituitary-Gonadal Axis
Abstract
1. Introduction
2. Results
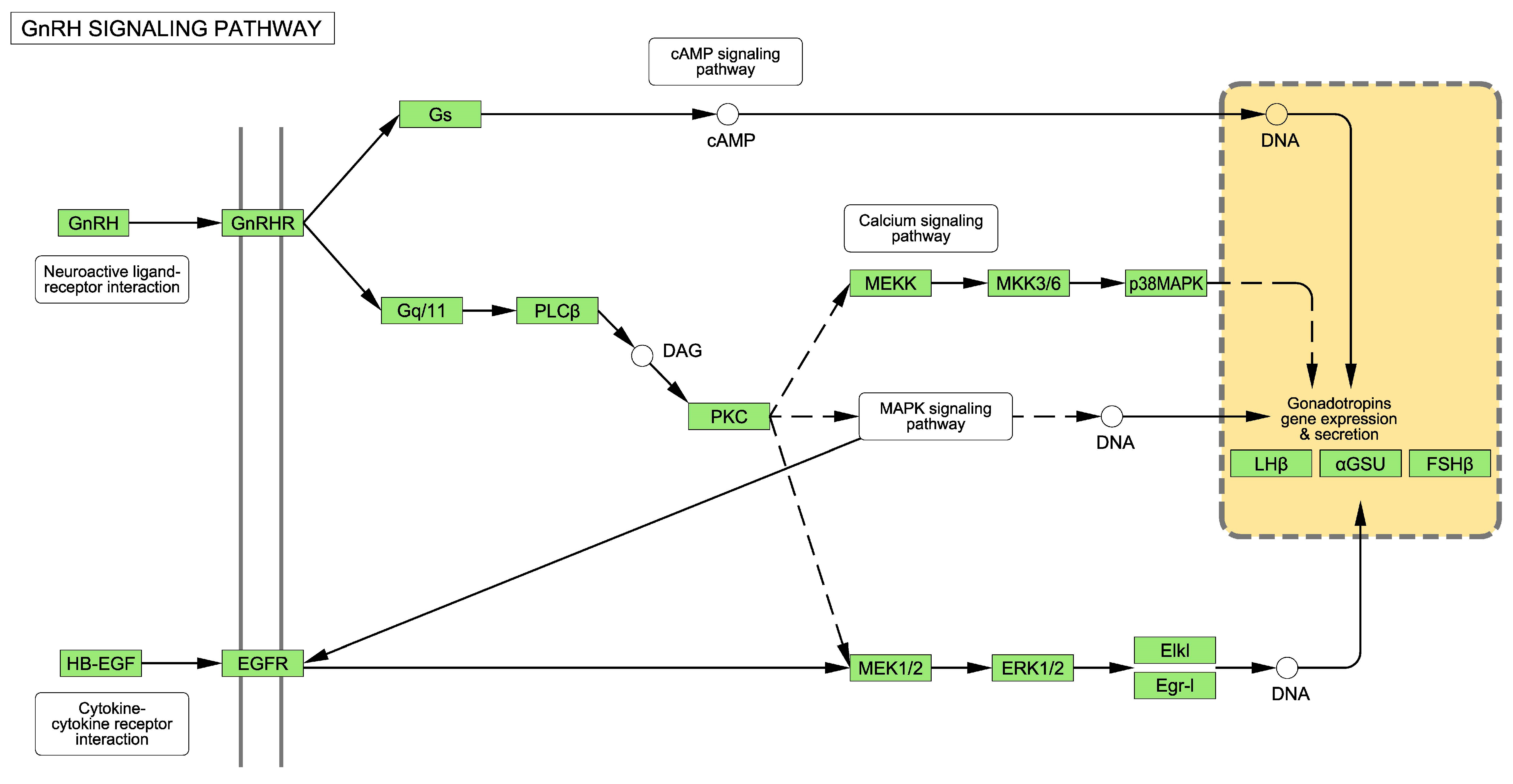
2.1. The GnRH Receptor Pathway
2.2. Effectof ZnO-NPs on Hypothalamic-Pituitary-Gonadal Axis in T98G Human Glioblastoma Cells
2.3. Effect of TiO2-NPs on Hypothalamic-Pituitary-Gonadal Axis in T98G Human Glioblastoma Cells Exposed to
2.4. Effect of Cd/Se-QD-NPs on Hypothalamic-Pituitary-Gonadal Axis in T98G Human Glioblastoma Cells
2.5. Effect of Ag-NPs on Hypothalamic-Pituitary-Gonadal Axis in T98G Human Glioblastoma Cells
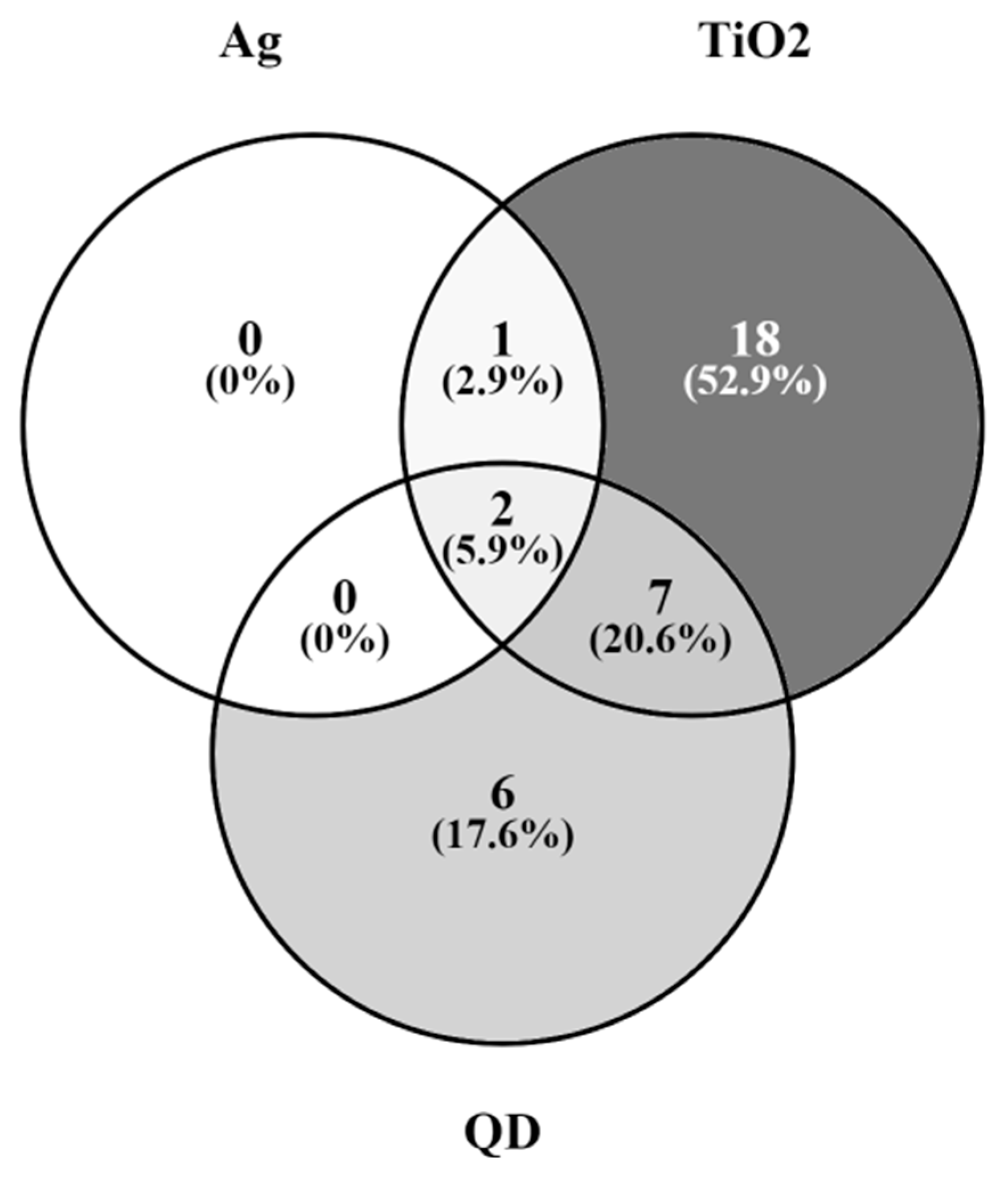
2.6. Genes of the GnRH Hormone Receptor Pathway Altered after Exposure to Several NPs
3. Discussion
3.1. Alterations to the GnRH Pathway
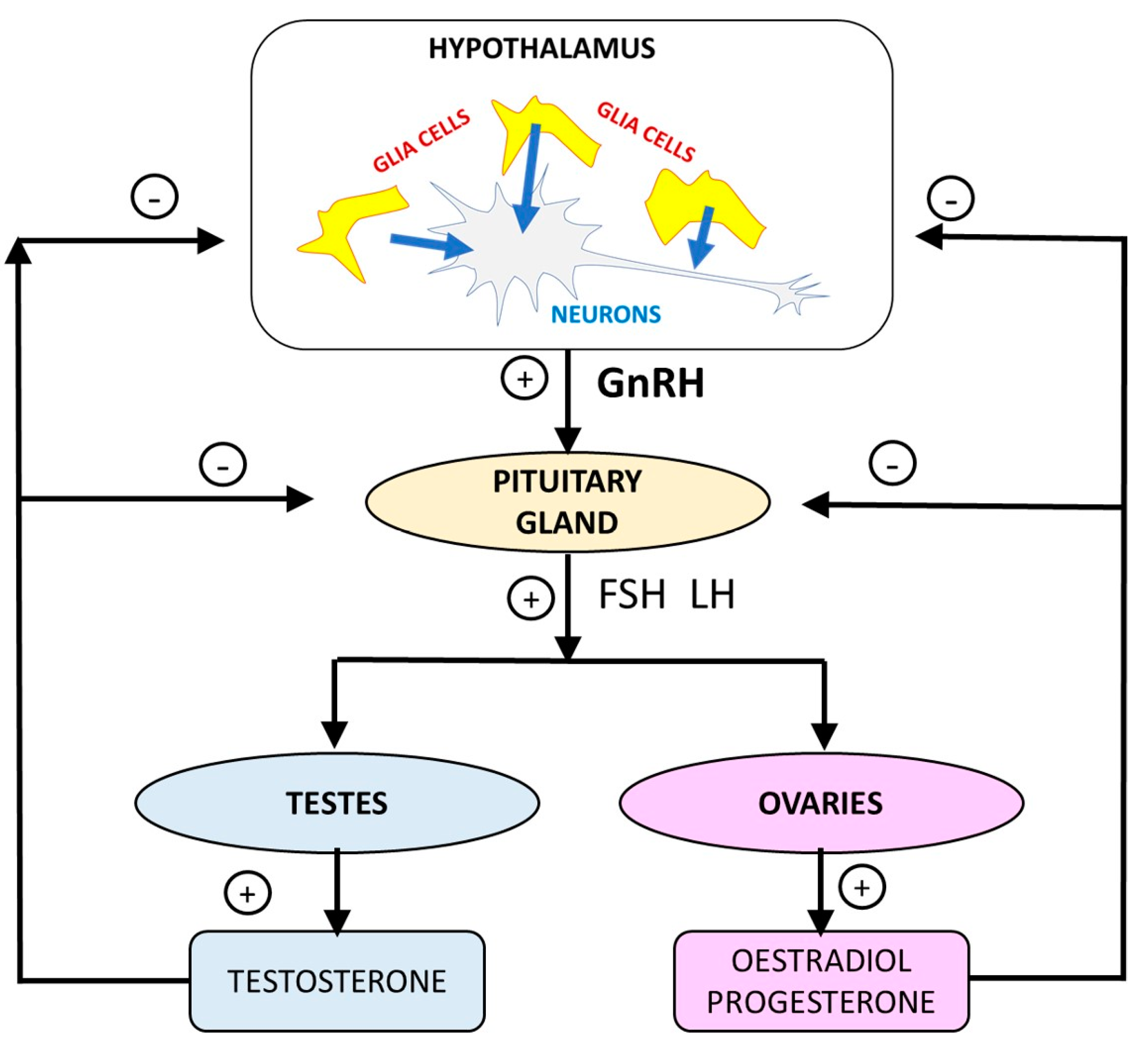
3.2. Alterations to the HPGA
3.3. Regulation of the Hypothalamic-Pituitary-Gonadal Axis by Glial Cells
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campos, E.V.R.; Pereira, A.E.S.; de Oliveira, J.L.; Carvalho, L.B.; Guilger-Casagrande, M.; de Lima, R.; Fraceto, L.F. How can nanotechnology help to combat COVID-19? Opportunities and urgent need. J. Nanobiotechnol. 2020, 18, 125. [Google Scholar] [CrossRef] [PubMed]
- Estevan, C.; Vilanova, E.; Sogorb, M.A. Case study: Risk associated to wearing silver or graphene nanoparticle-coated facemasks for protection against COVID-19. Arch. Toxicol. 2022, 96, 105–119. [Google Scholar] [CrossRef] [PubMed]
- EUON (European Union Observatory for Nanomaterials) Nanomaterials in the EU Market. Available online: https://euon.echa.europa.eu/search-for-nanomaterials (accessed on 23 May 2023).
- Bleeker, E.A.J.; Swart, E.; Braakhuis, H.; Fernández-Cruz, M.L.; Friedrichs, S.; Gosens, I.; Herzberg, F.; Jensen, K.A.; von der Kammer, F.; Kettelarij, J.A.B.; et al. Towards harmonisation of testing of nanomaterials for EU regulatory requirements on chemical safety—A proposal for further actions. Regul. Toxicol. Pharmacol. 2023, 139, 105360. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.B.; Mørck, T.A.; Andersen, D.N.; Hougaard, K.S. A Critical Review of Studies on the Reproductive and Developmental Toxicity of Nanomaterials; European Chemicals Agency: Helsinki, Finland, 2020; ISBN 978-92-9481-423-4. Available online: https://euon.echa.europa.eu/documents/2435000/3268573/critical_review_of_studies_on_reproductive_and_developmental_toxicity_of_nanomaterials_en.pdf/c83f78ef-7136-ef4b-268c-c5d9b7bf1fea?t=1586154196963 (accessed on 23 May 2023).
- Arslan, N.P.; Keles, O.N.; Gonul-Baltaci, N. Effect of Titanium Dioxide and Silver Nanoparticles on Mitochondrial Dynamics in Mouse Testis Tissue. Biol. Trace Elem. Res. 2022, 200, 1650–1658. [Google Scholar] [CrossRef]
- Hong, X.; Shao, N.; Yin, L.; Li, C.; Tao, G.; Sun, Y.; Qian, K.; Yang, J.; Xiao, P.; Yu, X.; et al. Exposure to zinc oxide nanoparticles affects testicular structure, reproductive development and spermatogenesis in parental and offspring male rats. Ann. Transl. Med. 2022, 10, 751. [Google Scholar] [CrossRef] [PubMed]
- Szudrowicz, H.; Kamaszewski, M.; Adamski, A.; Skrobisz, M.; Frankowska-ukawska, J.; Wójcik, M.; Bochenek, J.; Kawalski, K.; Martynow, J.; Bujarski, P.; et al. The Effects of Seven-Day Exposure to Silver Nanoparticles on Fertility and Homeostasis of Zebrafish (Danio rerio). Int. J. Mol. Sci. 2022, 23, 11239. [Google Scholar] [CrossRef] [PubMed]
- Mawed, S.A.; Marini, C.; Alagawany, M.; Farag, M.R.; Reda, R.M.; El-Saadony, M.T.; Elhady, W.M.; Magi, G.E.; Di Cerbo, A.; El-Nagar, W.G. Zinc Oxide Nanoparticles (ZnO-NPs) Suppress Fertility by Activating Autophagy, Apoptosis, and Oxidative Stress in the Developing Oocytes of Female Zebrafish. Antioxidants 2022, 11, 1567. [Google Scholar] [CrossRef]
- Smedlund, K.B.; Hill, J.W. The role of non-neuronal cells in hypogonadotropic hypogonadism. Mol. Cell. Endocrinol. 2020, 518, 110996. [Google Scholar] [CrossRef] [PubMed]
- Warheit, D.B.; Oberdörster, G.; Kane, A.B.; Brown, S.C.; Klaper, R.D.; Hurt, R.H. Chapter 29: Nanoparticle Toxicology. In Casarett & Doull’s Toxicology: The Basic Science of Poisons, 9th ed.; Klaassen, C.D., Ed.; McGraw-Hill Education: Singapore, 2019; Available online: https://accesspharmacy.mhmedical.com/content.aspx?bookid=2462§ionid=202678282pp (accessed on 25 June 2023).
- Duittoz, A.H.; Tillet, Y.; Geller, S. The great migration: How glial cells could regulate GnRH neuron development and shape adult reproductive life. J. Chem. Neuroanat. 2022, 25, 102149. [Google Scholar] [CrossRef]
- Fields, R.D.; Araque, A.; Johansen-Berg, H.; Lim, S.S.; Lynch, G.; Nave, K.A.; Nedergaard, M.; Perez, R.; Sejnowski, T.; Wake, H. Glial biology in learning and cognition. Neuroscientist 2014, 20, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Barres, B.A.; Freeman, M.R.; Stevens, B. Glia; Cold Spring Harbor: New York, NY, USA, 2015; ISBN 978-1-62182-027-7. [Google Scholar]
- Prevot, V.; Sharif, A. The polygamous GnRH neuron: Astrocytic and tanycytic communication with a neuroendocrine neuronal population. J. Neuroendocrinol. 2022, 34, e13104. [Google Scholar] [CrossRef]
- Butruille, L.; Batailler, M.; Cateau, M.L.; Sharif, A.; Leysen, V.; Prévot, V.; Vaudin, P.; Pillon, D.; Migaud, M. Selective Depletion of Adult GFAP-Expressing Tanycytes Leads to Hypogonadotropic Hypogonadism in Males. Front. Endocrinol. 2022, 13, 869019. [Google Scholar] [CrossRef]
- Fuster, E.; Candela, H.; Estévez, J.; Vilanova, E.; Sogorb, M.A. Titanium Dioxide, but Not Zinc Oxide, Nanoparticles Cause Severe Transcriptomic Alterations in T98G Human Glioblastoma Cells. Int. J. Mol. Sci. 2021, 22, 2084. [Google Scholar] [CrossRef] [PubMed]
- Fuster, E.; Candela, H.; Estévez, J.; Arias, A.J.; Vilanova, E.; Sogorb, M.A. Effects of silver nanoparticles on T98G human glioblastoma cells. Toxicol. Appl. Pharmacol. 2020, 404, 115178. [Google Scholar] [CrossRef] [PubMed]
- Fuster, E.; Candela, H.; Estévez, J.; Vilanova, E.; Sogorb, M.A. A Transcriptomic Analysis of T98G Human Glioblastoma Cells after Exposure to Cadmium-Selenium Quantum Dots Mainly Reveals Alterations in Neuroinflammation Processes and Hypothalamus Regulation. Int. J. Mol. Sci. 2022, 23, 2267. [Google Scholar] [CrossRef]
- Protein Analysis through Evolutionary Relationships (PANTHER) Database. Available online: http://www.pantherdb.org/ (accessed on 10 July 2023).
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Naor, Z. Signaling by G-protein-coupled receptor (GPCR): Studies on the GnRH receptor. Front. Neuroendocrinol. 2009, 30, 10–29. [Google Scholar] [CrossRef]
- Naor, Z.; Benard, O.; Seger, R. Activation of MAPK cascades by G-protein-coupled receptors: The case of gonadotropin-releasing hormone receptor. Trends Endocrinol. Metab. 2000, 11, 91–99. [Google Scholar] [CrossRef]
- Farshori, P.Q.; Shah, B.H.; Arora, K.K.; Martinez-Fuentes, A.; Catt, K.J. Activation and nuclear translocation of PKCdelta, Pyk2 and ERK1/2 by gonadotropin releasing hormone in HEK293 cells. J. Steroid Biochem. Mol. Biol. 2003, 85, 337–347. [Google Scholar] [CrossRef]
- Desroziers, E. Unusual suspects: Glial cells in fertility regulation and their suspected role in polycystic ovary syndrome. J. Neuroendocrinol. 2022, 34, e13136. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Segura, L.M.; Lorenz, B.; DonCarlos, L.L. The role of glia in the hypothalamus: Implications for gonadal steroid feedback and reproductive neuroendocrine output. Reproduction 2008, 135, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Ojeda, S.R.; Lomniczi, A.; Sandau, U.S. Glial-gonadotrophin hormone (GnRH) neurone interactions in the median eminence and the control of GnRH secretion. J. Neuroendocrinol. 2008, 20, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Sharif, A.; Baroncini, M.; Prevot, V. Role of glia in the regulation of gonadotropin-releasing hormone neuronal activity and secretion. Neuroendocrinology 2013, 98, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Oliveros, J.C. (2007–2015) Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 23 May 2023).
| PANTHER Pathway | Total Genes in the Pathway | DEGs after NP Exposure | Expected among DEGs | EF | p Value | FDR |
|---|---|---|---|---|---|---|
| TiO2-NPs | 237 | 28 | 11 | 2.5 | 4.25 × 10−5 | 2.27 × 10−3 |
| Cd/Se-QDs-NPs | 237 | 15 | 6 | 2.5 | 1.5 × 10−3 | 4.7 × 10−2 |
| Ag-NPs | 237 | 3 | 0.5 | 6 | 1.2 × 10−2 | 4.8 × 10−1 |
| Biological Process | Total Genes Biological Process | DEGs after TiO2-NP Exposure | Expected among DEGs | EF | p Value | FDR |
|---|---|---|---|---|---|---|
| Cellular response to FSH stimulus (GO:0071372) | 12 | 5 | 0.6 | 8.3 | 8.0 × 10−4 | 2.3 × 10−2 |
| Negative regulation of ERK1 and ERK2 cascade (GO:0070373) | 71 | 12 | 3.4 | 3.5 | 4.0 × 10−4 | 1.4 × 10−2 |
| Positive regulation of pathway-restricted SMAD protein phosphorylation (GO:0060394) | 50 | 9 | 2.4 | 3.7 | 1.4 × 10−3 | 3.6 × 10−2 |
| Cellular response to gonadotropin stimulus (GO:0071371) | 20 | 6 | 1 | 6.0 | 9.7 × 10−4 | 2.7× 10−2 |
| Cellular response to hormone stimulus (GO:0032870) | 509 | 50 | 25 | 2.0 | 7.9 × 10−6 | 5.0 × 10−4 |
| Response to steroid hormone (GO:0048545) | 289 | 34 | 14 | 2.4 | 1.0 × 10−5 | 6.3 × 10−4 |
| Ovulation cycle (GO:0042698) | 75 | 12 | 3.6 | 3.3 | 6.2 × 10−4 | 1.9 × 10−2 |
| Response to estradiol (GO:0032355) | 121 | 15 | 5.8 | 2.6 | 1.5 × 10−3 | 3.8 × 10−2 |
| MAPK cascade (GO:0000165) | 216 | 24 | 10 | 2.4 | 4.6 × 10−4 | 1.5 × 10−2 |
| Positive regulation of MAPK cascade (GO:0043410) | 483 | 48 | 23 | 2.1 | 8.1 × 10−6 | 5.1 × 10−4 |
| Biological Process | Total Genes Biological Process | DEGs after Cd/Se-QDs Exposure | Expected among DEGs | EF | p Value | FDR |
|---|---|---|---|---|---|---|
| Regulation of ERK1 and ERK2 cascade (GO:0070372) | 301 | 21 | 7.6 | 2.8 | 5.2 × 10−5 | 4.4 × 10−3 |
| Response to estradiol (GO:0032355) | 121 | 11 | 3.0 | 3.6 | 4.1 × 10−4 | 2.3 × 10−2 |
| Response to hormone (GO:0009725) | 789 | 39 | 20 | 2.0 | 1.2 × 10−4 | 8.8 × 10−3 |
| Regulation of MAPK cascade (GO:0043408) | 672 | 35 | 17 | 2.1 | 9.5 × 10−5 | 8.0 × 10−3 |
| Regulation of hormone secretion (GO:0046883) | 253 | 16 | 6.4 | 2.5 | 1.1 × 10−3 | 4.7 × 10−2 |
| Biological Process | Total Genes Biological Process | DEGs after Cd/Se-QDs Exposure | Expected among DEGs | EF | p Value | FDR |
|---|---|---|---|---|---|---|
| Negative regulation of ERK1 and ERK2 cascade (GO:0070373) | 71 | 3 | 0.14 | 21 | 4.3 × 10−4 | 4.3 × 10−2 |
| Positive regulation of ERK1 and ERK2 cascade (GO:0070374) | 215 | 5 | 0.43 | 12 | 7.2 × 10−5 | 1.2 × 10−2 |
| Negative regulation of MAPK cascade (GO:0043409) | 171 | 5 | 0.34 | 15 | 2.5 × 10−5 | 5.1 × 10−3 |
| Negative regulation of MAP kinase activity (GO:0043407) | 58 | 3 | 0.12 | 26 | 2.4 × 10−4 | 3.0 × 10−2 |
| MAPK cascade (GO:0000165) | 216 | 6 | 0.43 | 14 | 4.8 × 10−6 | 1.9 × 10−3 |
| Response to hormone (GO:0009725) | 789 | 8 | 1.6 | 5.1 | 1.5 × 10−4 | 2.0 × 10−2 |
| FC | ||||
|---|---|---|---|---|
| Gene | Name | TiO2 | Cd/Se-QDs | Ag |
| ENSG00000131791 | Protein kinase AMP-activated non-catalytic subunit beta 2 (PRKAB2) | 1.35 | - | - |
| ENSG00000185950 | Insulin receptor substrate 2 (IRS2) | 1.45 | 0.77 | - |
| ENSG00000164111 | Annexin A5 (ANXA5) | 1.30 | - | - |
| ENSG00000204388 | Heat shock protein family A (Hsp70) member 1B (HSPA1B) | 2.44 | - | - |
| ENSG00000107968 | Mitogen-activated protein kinase kinase kinase 8 (MAP3K8) | 0.56 | 0.59 | - |
| ENSG00000104899 | Anti-Mullerian hormone (AMH) | 0.50 | - | - |
| ENSG00000164742 | Adenylate cyclase 1 (ADCY1) | 1.88 | 1.61 | - |
| ENSG00000095015 | Mitogen-activated protein kinase kinase kinase 1 (MAP3K1) | 0.73 | - | - |
| ENSG00000204389 | Heat shock protein family A (Hsp70) member 1A (HSPA1A) | 2.52 | - | - |
| ENSG00000096433 | Inositol 1,4,5-trisphosphate receptor type 3 (ITPR3) | 1.43 | - | - |
| ENSG00000162734 | Proliferation and apoptosis adaptor protein 15 (PEA15) | 1.62 | - | - |
| ENSG00000106617 | Protein kinase AMP-activated non-catalytic subunit gamma 2 (PRKAG2) | 1.57 | - | - |
| ENSG00000168610 | Signal transducer and activator of transcription 3 (STAT3) | 0.73 | - | - |
| ENSG00000177606 | Jun proto-oncogene, AP-1 transcription factor subunit (JUN) | 1.34 | - | - |
| ENSG00000171223 | Jun B proto-oncogene | 0.42 | 0.45 | 0.48 |
| ENSG00000117394 | Solute carrier family 2 member 1 (SLC2A1) | 1.31 | - | - |
| ENSG00000167552 | Tubulin alpha 1a (TUBA1A) | 1.30 | 1.25 | - |
| ENSG00000117318 | Inhibitor of DNA binding 3 (ID3) | 0.67 | 0.48 | - |
| ENSG00000069011 | Paired like homeodomain 1 (PITX1) | 0.52 | - | - |
| ENSG00000122641 | Inhibin subunit beta A (INHBA) | 2.20 | - | - |
| ENSG00000162772 | Activating transcription factor 3 (ATF3) | 2.16 | 0.73 | - |
| ENSG00000169083 | Androgen receptor (AR) | 0.77 | - | - |
| ENSG00000171951 | Secretogranin II (SCG2) | 0.73 | - | - |
| ENSG00000146648 | Epidermal growth factor receptor (EGFR) | 1.61 | - | 1.26 |
| ENSG00000170345 | Fos proto-oncogene | 0.31 | 0.22 | - |
| ENSG00000100968 | Nuclear factor of activated T cells 4 (NFATC4) | 0.48 | - | - |
| ENSG00000122420 | Prostaglandin F receptor (PTGFR) | 0.77 | - | - |
| ENSG00000120738 | Early growth response 1 (EGR1) | 0.18 | 0.19 | 0.074 |
| ENSG00000123416 | Tubulin alpha 1b (TUBA1B) | - | 1.42 | - |
| ENSG00000134909 | Rho GTPase activating protein 32 (ARHGAP32) | - | 1.52 | - |
| ENSG00000149295 | Dopamine receptor D2 (DRD2) | - | 2.10 | - |
| ENSG00000166949 | SMAD family member 3 (SMAD3) | - | 1.39 | - |
| ENSG00000163083 | Inhibin subunit beta B (INHBB) | - | 1.82 | - |
| ENSG00000130522 | Jun D proto-oncogene | - | 1.64 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sogorb, M.A.; Candela, H.; Estévez, J.; Vilanova, E. Investigation of the Effects of Metallic Nanoparticles on Fertility Outcomes and Endocrine Modification of the Hypothalamic-Pituitary-Gonadal Axis. Int. J. Mol. Sci. 2023, 24, 11687. https://doi.org/10.3390/ijms241411687
Sogorb MA, Candela H, Estévez J, Vilanova E. Investigation of the Effects of Metallic Nanoparticles on Fertility Outcomes and Endocrine Modification of the Hypothalamic-Pituitary-Gonadal Axis. International Journal of Molecular Sciences. 2023; 24(14):11687. https://doi.org/10.3390/ijms241411687
Chicago/Turabian StyleSogorb, Miguel A., Héctor Candela, Jorge Estévez, and Eugenio Vilanova. 2023. "Investigation of the Effects of Metallic Nanoparticles on Fertility Outcomes and Endocrine Modification of the Hypothalamic-Pituitary-Gonadal Axis" International Journal of Molecular Sciences 24, no. 14: 11687. https://doi.org/10.3390/ijms241411687
APA StyleSogorb, M. A., Candela, H., Estévez, J., & Vilanova, E. (2023). Investigation of the Effects of Metallic Nanoparticles on Fertility Outcomes and Endocrine Modification of the Hypothalamic-Pituitary-Gonadal Axis. International Journal of Molecular Sciences, 24(14), 11687. https://doi.org/10.3390/ijms241411687







