bra-miR167a Targets ARF8 and Negatively Regulates Arabidopsis thaliana Immunity against Plasmodiophora brassicae
Abstract
1. Introduction
2. Results
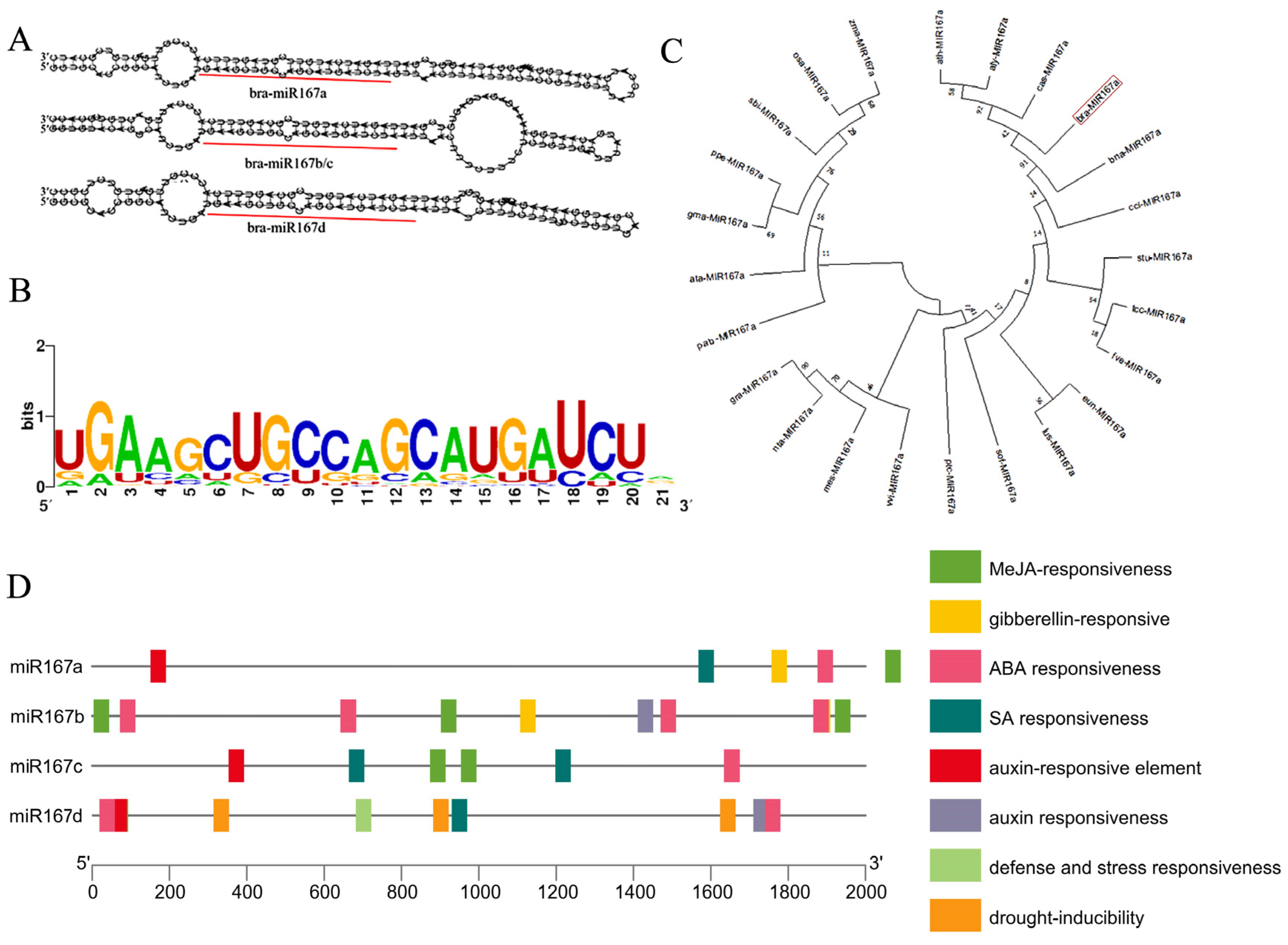
2.1. Bioinformatic Analysis of miR167s Reveales the Involvement of Cis-Acting Regulatory Elements
2.2. bra-miR167a Negatively Regulates the Expression of Targeted ARF and Auxin
2.3. STTM167 Negatively and OE-miR167a Positively Regulate the Expression of ARF
2.4. bra-miR167a Affects Lateral Root Development and Resistance to Clubroot Disease
2.5. bra-miR167a Regulate the Expression of Disease-Defense-Related Genes of Transgenic Arabidopsis under the Infection of Clubroot
2.6. Auxin Signaling Pathway Is Effected in OE-miR167a s under Infection with Clubroot
3. Discussion
4. Materials and Methods
4.1. Sources of Plant Materials and Transformation Vectors
4.2. Prediction of bra-miR167 Target Gene and Bioinformatic Analyses
4.3. Real-Time Quantitative PCR Assay
4.4. Transformation and Identification of Arabidopsis with STTM167a and OE-miR167a
4.5. Preparation of Clubroot Spore Solution and Investigation of Disease Incidence
4.6. Paraffin Sectioning
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jia, H.; Wei, X.; Yang, Y.; Yuan, Y.; Wei, F.; Zhao, Y.; Yang, S.; Yao, Q.; Wang, Z.; Tian, B.; et al. Root RNA-seq analysis reveals a distinct transcriptome landscape between clubroot-susceptible and clubroot-resistant Chinese cabbage lines after Plasmodiophora brassicae infection. Plant Soil 2017, 421, 93–105. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Prinsen, E.; Rolfe, S.A.; Scholes, J.D. Metabolism and Plant Hormone Action During Clubroot Disease. J. Plant Growth Regul. 2009, 28, 229–244. [Google Scholar] [CrossRef]
- Djavaheri, M.; Ma, L.; Klessig, D.F.; Mithofer, A.; Gropp, G.; Borhan, H. Mimicking the Host Regulation of Salicylic Acid: A Virulence Strategy by the Clubroot Pathogen Plasmodiophora brassicae. Mol. Plant Microbe Interact. 2019, 32, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wen, X.U.; Yuan, Y.; Yao, Q.; Zhao, Y.; Wang, Z.; Jiang, W.; Zhang, X. Genome-wide Investigation of micro RNAs and Their Targets in Brassica rapa ssp. pekinensis Root with Plasmodiophora brassicae Infection. Hortic. Plant J. 2016, 2, 209–216. [Google Scholar] [CrossRef]
- Javed, M.A.; Schwelm, A.; Zamani-Noor, N.; Salih, R.; Silvestre Vañó, M.; Wu, J.; González García, M.; Heick, T.M.; Luo, C.; Prakash, P.; et al. The clubroot pathogen Plasmodiophora brassicae: A profile update. Mol. Plant. Pathol. 2023, 24, 89–106. [Google Scholar] [CrossRef]
- Kageyama, K.; Asano, T. Life Cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 203–211. [Google Scholar] [CrossRef]
- Dixon, G.R. The Occurrence and Economic Impact of Plasmodiophora brassicae and Clubroot Disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Yang, L.; Fang, Z.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y.; Ji, J.; Liu, Y.; Li, Z.; Han, F. Recent advances of disease and stress resistance breeding of cabbage in china. Acta Hortic. Sin. 2020, 47, 1678–1688. [Google Scholar]
- Yuan, Y.; Qin, L.; Su, H.; Yang, S.; Zhang, X. Transcriptome and Coexpression Network Analyses Reveal Hub Genes in Chinese Cabbage (Brassica rapa L. ssp. pekinensis) During Different Stages of Plasmodiophora brassicae Infection. Front. Plant Sci. 2021, 12, 650252. [Google Scholar] [CrossRef]
- Feng, L.; Xia, R.; Liu, Y. Comprehensive Characterization of miRNA and PHAS Loci in the Diploid Strawberry (Fragaria vesca) Genome. Hortic. Plant J. 2019, 5, 255–267. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys. Acta 2012, 1819, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Gu, X.; Liu, J.; He, Z. Roles of small RNAs in crop disease resistance. Stress Biol. 2021, 1, 6. [Google Scholar] [CrossRef]
- Li, T.; Gonzalez, N.; Inze, D.; Dubois, M. Emerging Connections between Small RNAs and Phytohormones. Trends Plant Sci. 2020, 25, 912–929. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Guilfoyle, T.; Hagen, G.; Ulmasov, T.; Murfett, J. How Does Auxin Turn On Genes? Plant Physiol. 1998, 118, 341–347. [Google Scholar] [CrossRef]
- Jiang, C.; Shen, Y.; Wang, Y.; Zhang, M.; Sun, C. Research Progress of Auxin Response Factor in Plants. Mol. Plant Breed. 2022, 1–8. [Google Scholar]
- Faivre-Rampant, O.; Cardle, L.; Marshall, D.; Viola, R.; Taylor, M.A. Changes in gene expression during meristem activation processes in Solanum tuberosum with a focus on the regulation of an auxin response factor gene. J. Exp. Bot. 2004, 55, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; Macpherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.H. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef]
- Wu, M.F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef]
- Ru, P.; Xu, L.; Ma, H.; Huang, H. Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res. 2006, 16, 457–465. [Google Scholar] [CrossRef]
- Kun, T.S. Functional Identificaiotn of Brassica Napus miR167 and Its Target under Cadmium Stress. Master Thesis, Nanjing Agricultural University, Nanjing, China, 2016. [Google Scholar]
- Wang, Y.; Li, K.; Chen, L.; Zou, Y.; Liu, H.; Tian, Y.; Li, D.; Wang, R.; Zhao, F.; Ferguson, B.J.; et al. MicroRNA167-Directed Regulation of the Auxin Response Factors GmARF8a and GmARF8b Is Required for Soybean Nodulation and Lateral Root Development. Plant Physiol. 2015, 168, 984–999. [Google Scholar] [CrossRef]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef]
- Jia, X.; Ding, N.; Fan, W.; Yan, J.; Gu, Y.; Tang, X.; Li, R.; Tang, G. Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. 2015, 233, 11–21. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Yan, J.; Gou, F.; Mao, Y.; Tang, G.; Botella, J.R.; Zhu, J.K. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. USA 2017, 114, 5277–5282. [Google Scholar] [CrossRef]
- Damodharan, S.; Zhao, D.; Arazi, T. A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 2016, 86, 458–471. [Google Scholar] [CrossRef]
- Nizampatnam, N.R.; Schreier, S.J.; Damodaran, S.; Adhikari, S.; Subramanian, S. microRNA160 dictates stage-specific auxin and cytokinin sensitivities and directs soybean nodule development. Plant J. 2015, 84, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Wang, Y.; Teotia, S.; Wang, Z.; Shi, C.; Sun, H.; Gu, Y.; Zhang, Z.; Tang, G. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci. Rep. 2019, 9, 2832. [Google Scholar] [CrossRef]
- Chen, J.F.; Zhao, Z.X.; Li, Y.; Li, T.T.; Zhu, Y.; Yang, X.M.; Zhou, S.X.; Wang, H.; Zhao, J.Q.; Pu, M.; et al. Fine-tuning roles of Osa-miR159a in rice immunity against Magnaporthe oryzae and development. Rice 2021, 14, 26. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Huang, C.; Li, F.; Zhou, X. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 2014, 12, 638–649. [Google Scholar] [CrossRef]
- Liu, X.; Liu, S.; Wang, R.; Chen, X.; Zhou, T. Analyses of MiRNA Functions in Maize Using a Newly Developed ZMBJ-CMV-2bN81-STTM Vector. Front. Plant Sci. 2019, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Meng, J.; Cui, J.; Sun, G.; Luan, Y. Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans. Hortic. Res. 2018, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, S.; Xie, H. Advances in the regulation of plant development and stress response by miR167. FBL 2021, 26, 655–665. [Google Scholar]
- Caifu, C.; Ridzon, D.A.; Broomer, A.J.; Zhaohui, Z.; Lee, D.H.; Nguyen, J.T.; Maura, B.; Lan, X.N.; Mahuvakar, V.R.; Andersen, M.R. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 30, e179. [Google Scholar]
- Wu, D.; Cai, C.; Wei, G.; Xiang, Y. Genome-Wide Analysis of NBS-Encoding Disease Resistance Genes in Populus trichocarpa. China Sci. 2009, 45, 152–157. [Google Scholar]
- Wei, X.; Liao, R.; Zhang, X.; Zhao, Y.; Xie, Z.; Yang, S.; Su, H.; Wang, Z.; Zhang, L.; Tian, B.; et al. Integrative Transcriptome, miRNAs, Degradome, and Phytohormone Analysis of Brassica rapa L. in Response to Plasmodiophora brassicae. Int. J. Mol. Sci. 2023, 24, 2414. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an Auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Li, W.; Zhu, Y.; Liu, Z.; Huang, W.; Zhan, J. Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in Vitis vinifera. Plant Cell Rep. 2014, 33, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Matthieu, P.; Nikita, K.; Yi, Z.; Kathleen, G.; Whitnie, S.; Kumar, P.B.; Arvind, B.R.; Bennett, M.J.; Wolfgang, B.; et al. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 2020, 9, e54740. [Google Scholar] [CrossRef]
- Caruana, J.C.; Dhar, N.; Raina, R. Overexpression of Arabidopsis microRNA167 induces salicylic acid-dependent defense against Pseudomonas syringae through the regulation of its targets ARF6 and ARF8. Plant Direct 2020, 4, e00270. [Google Scholar] [CrossRef]
- Meng, Y.; Huang, F.; Shi, Q.; Cao, J.; Chen, D.; Zhang, J.; Ni, J.; Wu, P.; Chen, M. Genome-wide survey of rice microRNAs and microRNA–target pairs in the root of a novel auxin-resistant mutant. Planta 2009, 230, 883–898. [Google Scholar] [CrossRef]
- Gifford, M.L.; Dean, A.; Gutierrez, R.A.; Coruzzi, G.M.; Birnbaum, K.D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 2008, 105, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Bussell, J.D.; Pacurar, D.I.; Schwambach, J.; Pacurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef] [PubMed]
- Couzigou, J.-M.; Combier, J.-P. Plant microRNAs: Key regulators of root architecture and biotic interactions. New Phytol. 2016, 212, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.; Das, A.; Thakur, S.; Jalali, B. Recent Understanding on Structure, Function and Evolution of Plant Disease Resistance Genes. Proc. Indian Natl. Sci. Acad. 2015, 80, 83–93. [Google Scholar] [CrossRef]
- Bi, G.; Zhou, J.M. MAP Kinase Signaling Pathways: A Hub of Plant-Microbe Interactions. Cell Host Microbe 2017, 21, 270. [Google Scholar] [CrossRef]
- Sun, T.; Zhang, Y. MAP kinase cascades in plant development and immune signaling. EMBO Rep. 2022, 23, e53817. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Zhang, S.; Klessig, D.F. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001, 6, 520–527. [Google Scholar] [CrossRef]
- Okushima, Y.; Koizumi, N.; Kusano, T.; Sano, H. Secreted proteins of tobacco cultured BY2 cells: Identification of a new member of pathogenesis-related proteins. Plant Mol. Biol. 2000, 42, 479–488. [Google Scholar] [CrossRef]
- Han, Z.; Xiong, D.; Schneiter, R.; Tian, C. The function of plant PR1 and other members of the CAP protein superfamily in plant-pathogen interactions. Mol. Plant Pathol. 2023, 24, 651–668. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P. Pathogenesis-Related Proteins and Peptides as Promising Tools for Engineering Plants with Multiple Stress Tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic Acid Inhibits Pathogen Growth in Plants through Repression of the Auxin Signaling Pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.X.; Tasset, C.; Hanemian, M.; Barlet, X.; Hu, J.; Tremousaygue, D.; Deslandes, L.; Marco, Y. Biological control of bacterial wilt in Arabidopsis thaliana involves abscissic acid signalling. New Phytol. 2012, 194, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O.; Leyser, O. Dynamic integration of auxin transport and signalling. Curr. Biol. 2006, 16, R424–R433. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhao, Y.; Wei, X.; Yao, Q.; Jiang, W.; Wang, Z.; Yang, L.I.; Qian, X.U.; Yang, S.; Zhang, X. Pathotype Identification of Plasmodiophora brassicae Woron. Collected from Chinese Cabbage in Henan Province. J. Henan Agric. Sci. 2017, 46, 71–76. [Google Scholar]
- Cheng, X. Molecular Mechanism of Sly-miR319b Regulating TCPs in Response to Low Potassium Stress in Tomato. Master Thesis, Shenyang Agricultural University, Shenyang, China, 2020. [Google Scholar]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49. [Google Scholar] [CrossRef]
- Nour-Eldin, H.H.; Hansen, B.G.; Nørholm, M.H.; Jensen, J.K.; Halkier, B.A. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 2006, 34, e122. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Jubault, M.; Lariagon, C.; Taconnat, L.; Renou, J.P.; Gravot, A.; Delourme, R.; Manzanares-Dauleux, M.J. Partial resistance to clubroot in Arabidopsis is based on changes in the host primary metabolism and targeted cell division and expansion capacity. Funct. Integr. Genom. 2013, 13, 191–205. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, R.; Wei, X.; Zhao, Y.; Xie, Z.; Nath, U.K.; Yang, S.; Su, H.; Wang, Z.; Li, L.; Tian, B.; et al. bra-miR167a Targets ARF8 and Negatively Regulates Arabidopsis thaliana Immunity against Plasmodiophora brassicae. Int. J. Mol. Sci. 2023, 24, 11850. https://doi.org/10.3390/ijms241411850
Liao R, Wei X, Zhao Y, Xie Z, Nath UK, Yang S, Su H, Wang Z, Li L, Tian B, et al. bra-miR167a Targets ARF8 and Negatively Regulates Arabidopsis thaliana Immunity against Plasmodiophora brassicae. International Journal of Molecular Sciences. 2023; 24(14):11850. https://doi.org/10.3390/ijms241411850
Chicago/Turabian StyleLiao, Rujiao, Xiaochun Wei, Yanyan Zhao, Zhengqing Xie, Ujjal Kumar Nath, Shuangjuan Yang, Henan Su, Zhiyong Wang, Lin Li, Baoming Tian, and et al. 2023. "bra-miR167a Targets ARF8 and Negatively Regulates Arabidopsis thaliana Immunity against Plasmodiophora brassicae" International Journal of Molecular Sciences 24, no. 14: 11850. https://doi.org/10.3390/ijms241411850
APA StyleLiao, R., Wei, X., Zhao, Y., Xie, Z., Nath, U. K., Yang, S., Su, H., Wang, Z., Li, L., Tian, B., Wei, F., Yuan, Y., & Zhang, X. (2023). bra-miR167a Targets ARF8 and Negatively Regulates Arabidopsis thaliana Immunity against Plasmodiophora brassicae. International Journal of Molecular Sciences, 24(14), 11850. https://doi.org/10.3390/ijms241411850







