Expression of Phospholipase D Family Member 6 in Bovine Testes and Its Molecular Characteristics
Abstract
:1. Introduction
2. Results
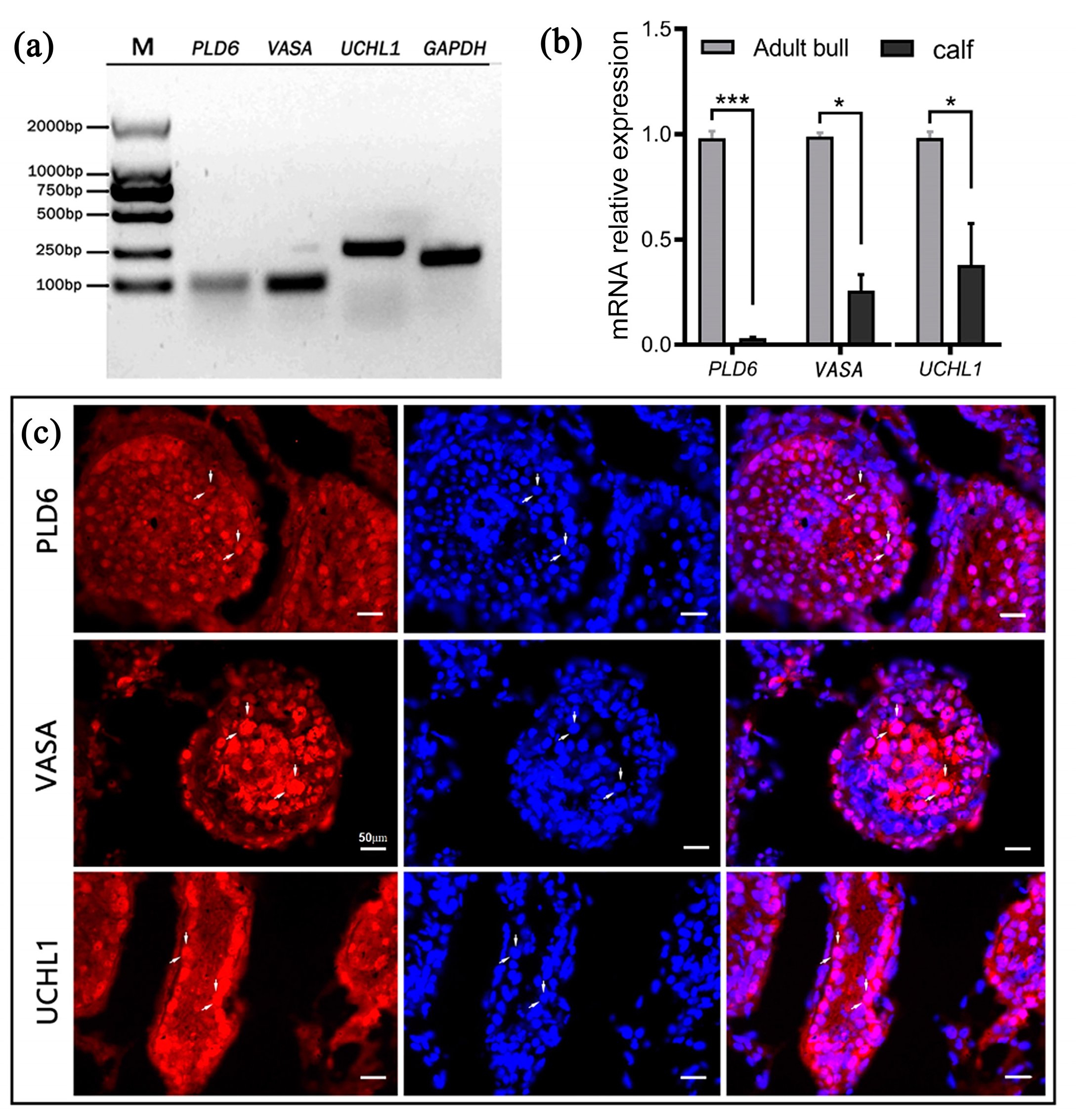
2.1. Expression of PLD6 in Bovine Testes
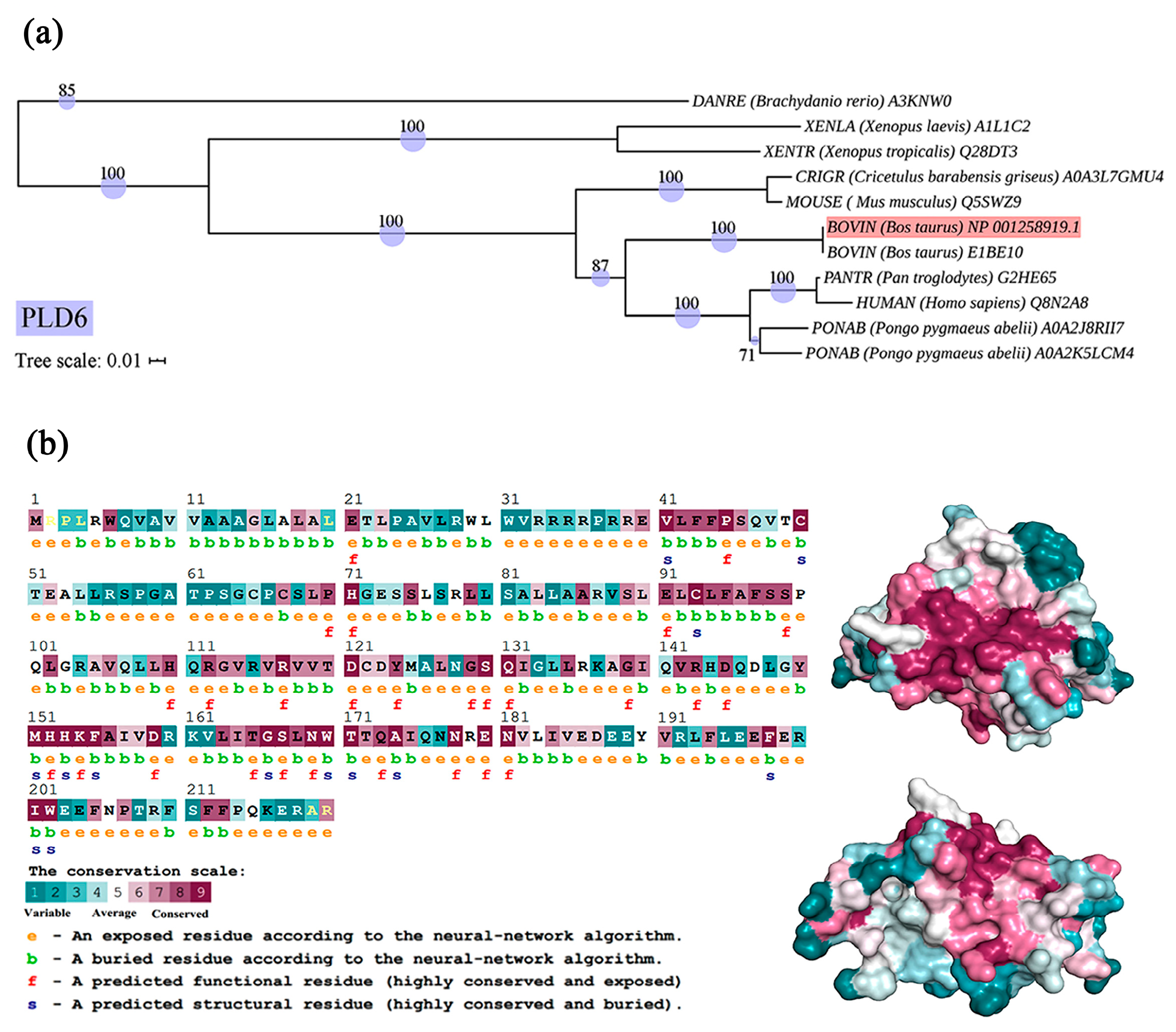
2.2. Phylogenetic and Structural Features of Bovine PLD6
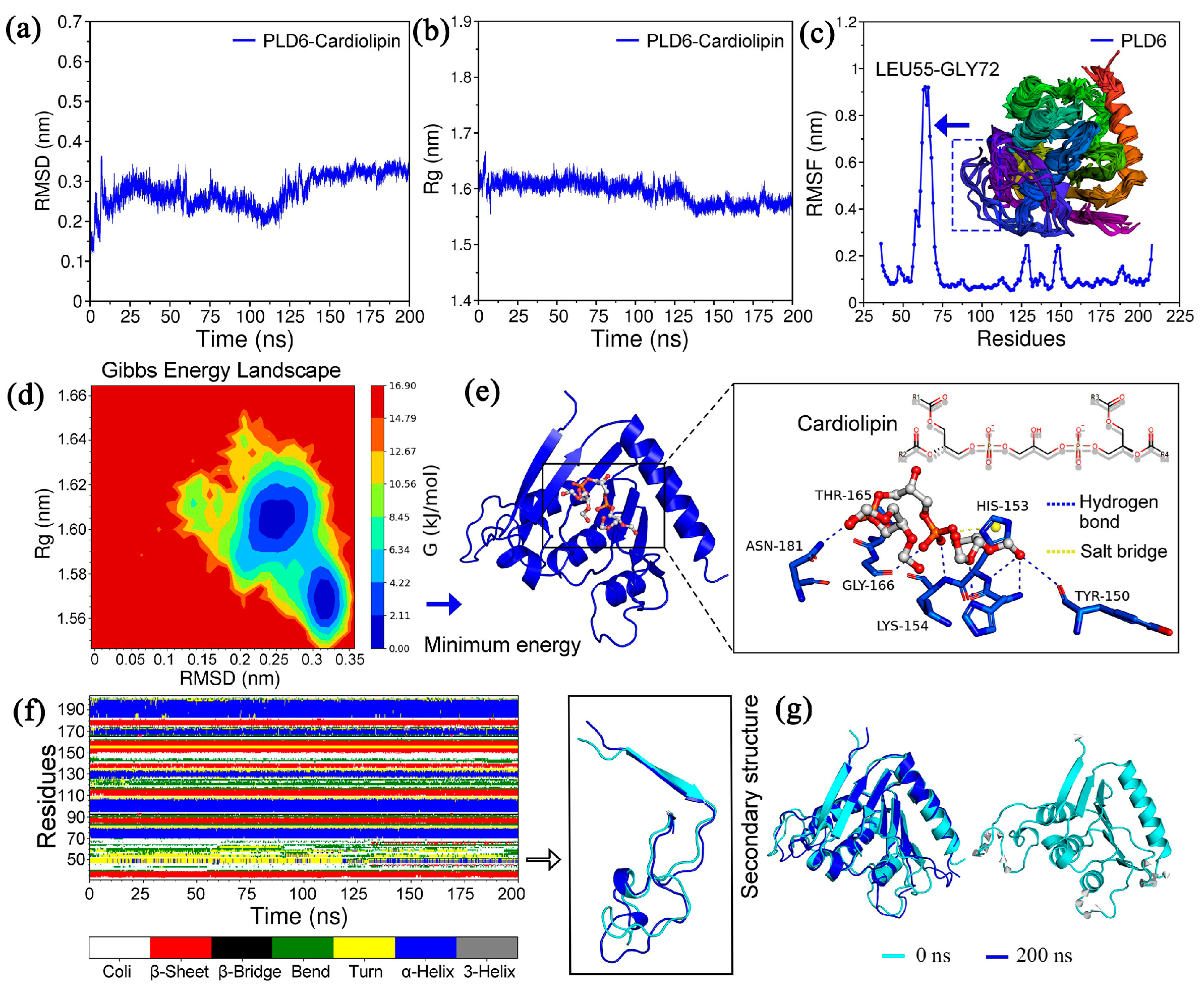
2.3. Molecular Dynamics Simulation and Binding Mode Analysis
2.4. Protein-Protein Interaction (PPI) Network
2.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
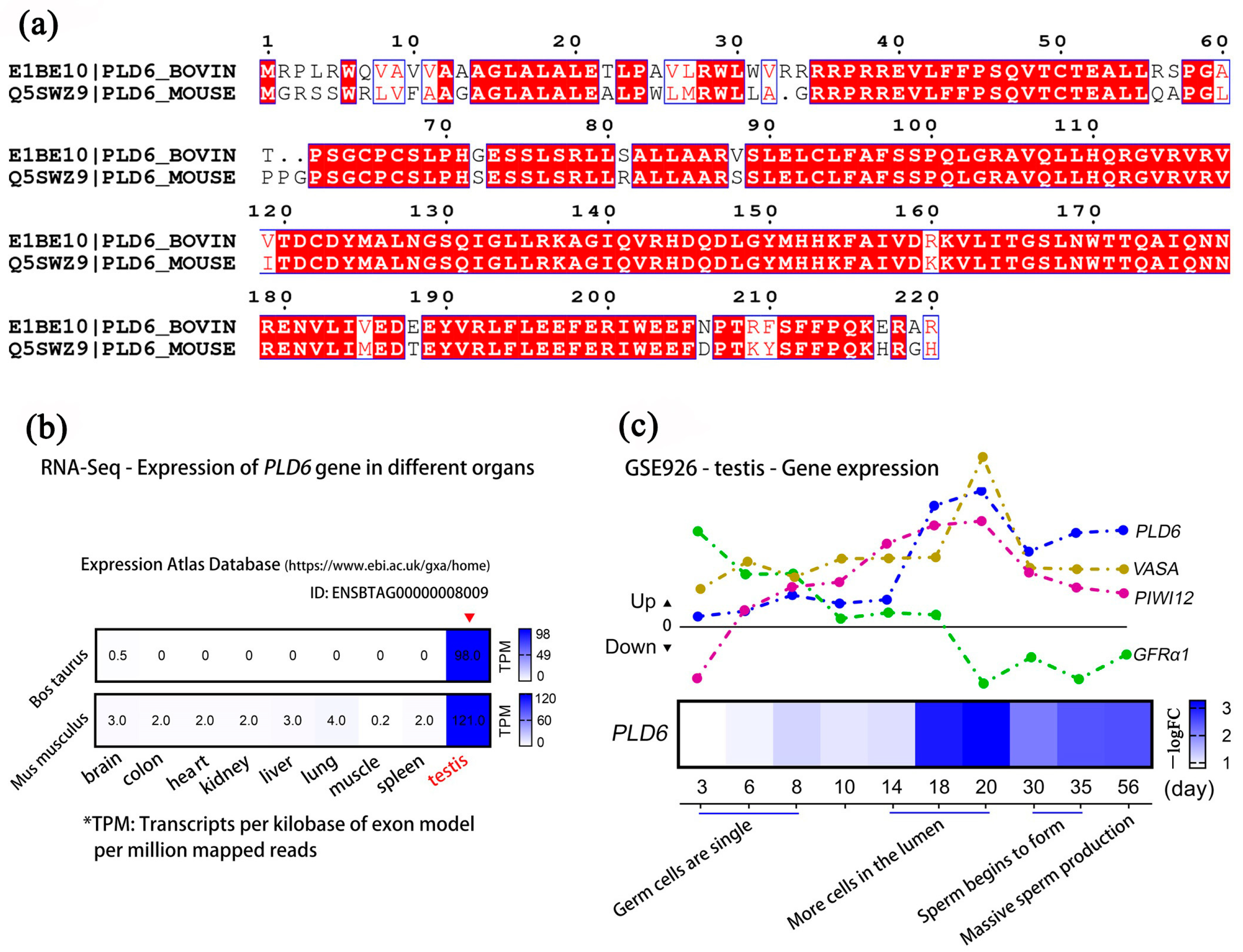
2.6. Multi-Sequence Alignment and Expression Analysis of PLD6 by GEO Mining
3. Discussion
4. Materials and Methods
4.1. Animal Tissues
4.2. RT-PCR and qRT-PCR
4.3. Immunostaining
4.4. Bioinformatics
4.4.1. Phylogenetic Analysis
4.4.2. Protein Structure Analysis
4.4.3. Molecular Docking
4.4.4. Molecular Dynamics Simulation
4.4.5. Protein-Protein Interaction (PPI) Network
4.4.6. GO and KEGG Analysis
4.4.7. Multiple Sequence Alignment and GEO Database Mining
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cazzolli, R.; Shemon, A.N.; Fang, M.Q.; Hughes, W.E. Phospholipid signalling through phospholipase D and phosphatidic acid. IUBMB Life 2006, 58, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, F.; Zhang, L.; Lei, P.; Zheng, Y.; Zeng, W. Stage-specific embryonic antigen 4 is a membrane marker for enrichment of porcine spermatogonial stem cells. Andrology 2020, 8, 1923–1934. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chi, X.; Wang, Y.; Heng, B.C.; Wei, Y.; Zhang, X.; Zhao, H.; Yin, Y.; Deng, X. Mitochondria transfer enhances proliferation, migration, and osteogenic differentiation of bone marrow mesenchymal stem cell and promotes bone defect healing. Stem Cell Res. Ther. 2020, 11, 245. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liang, P.; Huang, Y.; Li, M.; Zhang, X.; Ding, C.; Feng, J.; Zhang, Z.; Zhang, X.; Gao, Y.; et al. Glycerol kinase-like proteins cooperate with Pld6 in regulating sperm mitochondrial sheath formation and male fertility. Cell Discov. 2017, 3, 17030. [Google Scholar] [CrossRef] [Green Version]
- Ipsaro, J.J.; Haase, A.D.; Knott, S.R.; Joshua-Tor, L.; Hannon, G.J. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 2012, 491, 279–283. [Google Scholar] [CrossRef] [Green Version]
- Kabayama, Y.; Toh, H.; Katanaya, A.; Sakurai, T.; Chuma, S.; Kuramochi-Miyagawa, S.; Saga, Y.; Nakano, T.; Sasaki, H. Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes. Nucleic Acids Res. 2017, 45, 5387–5398. [Google Scholar] [CrossRef] [Green Version]
- Nishimasu, H.; Ishizu, H.; Saito, K.; Fukuhara, S.; Kamatani, M.K.; Bonnefond, L.; Matsumoto, N.; Nishizawa, T.; Nakanaga, K.; Aoki, J.; et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 2012, 491, 284–287. [Google Scholar] [CrossRef]
- Huang, H.; Gao, Q.; Peng, X.; Choi, S.Y.; Sarma, K.; Ren, H.; Morris, A.J.; Frohman, M.A. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev. Cell 2011, 20, 376–387. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Chuma, S.; Yamamoto, Y.; Kuramochi-Miyagawa, S.; Totoki, Y.; Toyoda, A.; Hoki, Y.; Fujiyama, A.; Shibata, T.; Sado, T.; et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell 2011, 20, 364–375. [Google Scholar] [CrossRef] [Green Version]
- Subash, S.K.; Kumar, P.G. Self-renewal and differentiation of spermatogonial stem cells. Front. Biosci. 2021, 26, 163–205. [Google Scholar] [CrossRef]
- Law, N.C.; Oatley, M.J.; Oatley, J.M. Developmental kinetics and transcriptome dynamics of stem cell specification in the spermatogenic lineage. Nat. Commun. 2019, 10, 2787. [Google Scholar] [CrossRef] [Green Version]
- de Rooij, D.G.; Russell, L.D. All you wanted to know about spermatogonia but were afraid to ask. J. Androl. 2000, 21, 776–798. [Google Scholar] [CrossRef] [PubMed]
- Kanatsu-Shinohara, M.; Chen, G.; Morimoto, H.; Shinohara, T. CD2 is a surface marker for mouse and rat spermatogonial stem cells. J. Reprod. Dev. 2020, 66, 341–349. [Google Scholar] [CrossRef]
- Mutoji, K.; Singh, A.; Nguyen, T.; Gildersleeve, H.; Kaucher, A.V.; Oatley, M.J.; Oatley, J.M.; Velte, E.K.; Geyer, C.B.; Cheng, K.; et al. TSPAN8 Expression Distinguishes Spermatogonial Stem Cells in the Prepubertal Mouse Testis. Biol. Reprod. 2016, 95, 117. [Google Scholar] [CrossRef]
- Cai, H.; Jiang, Y.; Zhang, S.; Cai, N.N.; Zhu, W.Q.; Yang, R.; Tang, B.; Li, Z.Y.; Zhang, X.M. Culture bovine prospermatogonia with 2i medium. Andrologia 2021, 53, e14056. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Nie, J.; Zhu, X.; Lu, Y.; Liang, X.; Xu, H.; Yang, X.; Zhang, Y.; Lu, K.; Lu, S. In vitro differentiation of spermatogonial stem cells using testicular cells from Guangxi Bama mini-pig. J. Vet. Sci. 2018, 19, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Guo, Y.; Zheng, B.; Shao, B.; Jiang, M.; Wang, G.; Zhou, T.; Wang, L.; Zhou, Z.; Guo, X.; et al. Establishment of a proteome profile and identification of molecular markers for mouse spermatogonial stem cells. J. Cell. Mol. Med. 2015, 19, 521–534. [Google Scholar] [CrossRef]
- Zhang, P.; Qin, Y.; Zheng, Y.; Zeng, W. Phospholipase D Family Member 6 Is a Surface Marker for Enrichment of Undifferentiated Spermatogonia in Prepubertal Boars. Stem Cells Dev. 2018, 27, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Lehtiniemi, T.; Kotaja, N. Germ granule-mediated RNA regulation in male germ cells. Reproduction 2018, 155, R77–R91. [Google Scholar] [CrossRef]
- Zhang, G.W.; Wang, L.; Chen, H.; Guan, J.; Wu, Y.; Zhao, J.; Luo, Z.; Huang, W.; Zuo, F. Promoter hypermethylation of PIWI/piRNA pathway genes associated with diminished pachytene piRNA production in bovine hybrid male sterility. Epigenetics 2020, 15, 914–931. [Google Scholar] [CrossRef] [PubMed]
- Nakata, H. Morphology of mouse seminiferous tubules. Anat. Sci. Int. 2019, 94, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kulandaisamy, A.; Lathi, V.; ViswaPoorani, K.; Yugandhar, K.; Gromiha, M.M. Important amino acid residues involved in folding and binding of protein-protein complexes. Int. J. Biol. Macromol. 2017, 94, 438–444. [Google Scholar] [CrossRef]
- Ikeda, S.; Tanaka, K.; Ohtani, R.; Kanda, A.; Sotomaru, Y.; Kono, T.; Obata, Y. Disruption of piRNA machinery by deletion of ASZ1/GASZ results in the expression of aberrant chimeric transcripts in gonocytes. J. Reprod. Dev. 2022, 68, 125–136. [Google Scholar] [CrossRef]
- Hutt, K.J.; Lim, S.L.; Zhang, Q.H.; Gonzalez, M.; O’Connor, A.E.; Merriner, D.J.; Liew, S.H.; Al-Zubaidi, U.; Yuen, W.S.; Adhikari, D.; et al. HENMT1 is involved in the maintenance of normal female fertility in the mouse. Mol. Hum. Reprod. 2021, 27, gaab061. [Google Scholar] [CrossRef]
- Hempfling, A.L.; Lim, S.L.; Adelson, D.L.; Evans, J.; O’Connor, A.E.; Qu, Z.P.; Kliesch, S.; Weidner, W.; O’Bryan, M.K.; Bergmann, M. Expression patterns of HENMT1 and PIWIL1 in human testis: Implications for transposon expression. Reproduction 2017, 154, 363–374. [Google Scholar] [CrossRef] [Green Version]
- Ding, D.; Liu, J.; Midic, U.; Wu, Y.; Dong, K.; Melnick, A.; Latham, K.E.; Chen, C. TDRD5 binds piRNA precursors and selectively enhances pachytene piRNA processing in mice. Nat. Commun. 2018, 9, 127. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef]
- Philip, F.; Ha, E.E.; Seeliger, M.A.; Frohman, M.A. Measuring Phospholipase D Enzymatic Activity Through Biochemical and Imaging Methods. Methods Enzymol. 2017, 583, 309–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riew, T.R.; Kim, S.; Jin, X.; Kim, H.L.; Hwang, W.C.; Kang, M.; Yang, E.S.; Lee, M.Y.; Min, D.S. Cellular and subcellular localization of endogenous phospholipase D6 in seminiferous tubules of mouse testes. Cell Tissue Res. 2021, 385, 191–205. [Google Scholar] [CrossRef]
- Ramalho-Santos, J.; Varum, S.; Amaral, S.; Mota, P.C.; Sousa, A.P.; Amaral, A. Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 2009, 15, 553–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, E.E.; Frohman, M.A. Regulation of mitochondrial morphology by lipids. Biofactors 2014, 40, 419–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [Green Version]
- Aravin, A.A.; Chan, D.C. piRNAs meet mitochondria. Dev. Cell 2011, 20, 287–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Q.; Frohman, M.A. Roles for the lipid-signaling enzyme MitoPLD in mitochondrial dynamics, piRNA biogenesis, and spermatogenesis. BMB Rep. 2012, 45, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Helsel, A.R.; Yang, Q.; Oatley, M.J.; Lord, T.; Sablitzky, F.; Oatley, J.M. ID4 levels dictate the stem cell state in mouse spermatogonia. Development 2017, 144, 624–634. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Tang, B.; Wu, J.Y.; Zhao, X.X.; Wang, Z.Z.; An, X.L.; Lai, L.X.; Li, Z.Y.; Zhang, X.M. Enrichment and in vitro features of the putative gonocytes from cryopreserved testicular tissue of neonatal bulls. Andrology 2016, 4, 1150–1158. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.H.; Choi, Y.R.; Kim, B.J.; Jung, S.E.; Kim, S.M.; Jin, J.H.; Yun, M.H.; Kim, S.U.; Kim, Y.H.; Hwang, S.; et al. GDNF family receptor alpha 1 is a reliable marker of undifferentiated germ cells in bulls. Theriogenology 2019, 132, 172–181. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, W.Q.; Zhu, X.C.; Cai, N.N.; Yang, R.; Cai, H.; Zhang, X.M. Cryopreservation of calf testicular tissues with knockout serum replacement. Cryobiology 2020, 92, 255–257. [Google Scholar] [CrossRef]
- Geng, N.; Sun, G.; Liu, W.J.; Gao, B.C.; Sun, C.; Xu, C.; Hua, E.; Xu, L. Distribution, Phylogeny and Evolution of Clinical and Environmental Vibrio vulnificus Antibiotic-Resistant Genes. Evol. Bioinform. Online 2022, 18, 1041271744. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002. Chapter 2, Unit 2.3. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Weg, K.J.; Gohlke, H. TopEnzyme: A framework and database for structural coverage of the functional enzyme space. Bioinformatics 2023, 39, btad116. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yu, P.; Luo, J.; Jiang, Y. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm. Genome 2003, 14, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Rigsby, R.E.; Parker, A.B. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem. Mol. Biol. Educ. 2016, 44, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Sousa, D.S.A.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [Green Version]


| Importance Score | Number | Trace Residues |
|---|---|---|
| <25% | 34 | 44, 45, 69, 71, 72, 83, 86, 93, 95, 97, 106, 109, 112, 113, 114, 116, 121, 151, 152, 154, 155, 159, 166, 167, 169, 170, 171, 174, 178, 180, 181, 194, 198, 202 |
| <5% | 17 | 86, 93, 95, 112, 121, 152, 154, 159, 166, 167, 169, 170, 174, 178, 180, 198, 202 |
| Mnemonic Name | Species | Common Name | Accession Number |
|---|---|---|---|
| DANRE | Danio rerio | Zebrafish | A3KNW0 |
| XENLA | Xenopus laevis | African clawed frog | A1L1C2 |
| XENTR | Xenopus tropicalis | Western clawed frog | Q28DT3 |
| CRIGR | Cricetulus griseus | Chinese hamster | A0A3L7GMU4 |
| MOUSE | Mus musculus | Mouse | Q5SWZ9 |
| BOVINE | Bos taurus | Bovine | NP_001258919.1 |
| BOVINE | Bos taurus | Bovine | E1BE10 |
| PANTR | Pan troglodytes | Chimpanzee | G2HE65 |
| HUMAN | Homo sapiens | Human | Q8N2A8 |
| PONAB | Pongo abelii | Sumatran orangutan | A0A2J8RII7 |
| PONAB | Cercocebus atys | Sooty mangabey | A0A2K5LCM4 |
| Potential Biomarker | Proteins Involved (%) | KEGG Pathway ID | Description |
|---|---|---|---|
| PLD6 | 61.08 | map 00564 | Glycerophospholipid metabolism |
| 14.77 | map 00440 | Aminophosphonate metabolism | |
| 9.55 | map 00260 | Glycine, serine and threonine metabolism | |
| 7.03 | map 00565 | Ether lipid metabolism | |
| 7.03 | map 04912 | GnRH signaling pathway | |
| 0.18 | map 00623 | 2,4-Dichlorobenzoate degradation | |
| 0.18 | map 00960 | Alkaloid biosynthesis II | |
| 0.18 | map 00650 | Butanoate metabolism |
| Gene | Primer Sequence (5′→3′) | Fragment Size (bp) | Gene ID | Accession Number |
|---|---|---|---|---|
| GAPDH | F: TGACCCCTTCATTGACCTTC | 250 | 281181 | NM_001034034 |
| R: TACTCAGCACCAGCATCACC | ||||
| VASA | F: TTGGGACTTGTGTAAGAGCTGT | 98 | 493725 | NM_001278643 |
| R: CTTGACTTGTTTGAGGC | ||||
| UCHL1 | F: CCTTCCCTGGGCAGGTGCGCGC | 280 | 514394 | NM_001046172 |
| R: GGCTGTAGAACGCAAGAA | ||||
| PLD6 | F: GTGGACAGGAAGGTGCTGAT | 108 | 526651 | NM_001271990 |
| R: TACTCCTCGTCCTCCACGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Zhang, B.; Zhu, W.; Zhu, C.; Chen, L.; Zhao, Y.; Wang, Y.; Zhang, Y.; Riaz, A.; Tang, B.; et al. Expression of Phospholipase D Family Member 6 in Bovine Testes and Its Molecular Characteristics. Int. J. Mol. Sci. 2023, 24, 12172. https://doi.org/10.3390/ijms241512172
Yang R, Zhang B, Zhu W, Zhu C, Chen L, Zhao Y, Wang Y, Zhang Y, Riaz A, Tang B, et al. Expression of Phospholipase D Family Member 6 in Bovine Testes and Its Molecular Characteristics. International Journal of Molecular Sciences. 2023; 24(15):12172. https://doi.org/10.3390/ijms241512172
Chicago/Turabian StyleYang, Rui, Boyang Zhang, Wenqian Zhu, Chunling Zhu, Lanxin Chen, Yansen Zhao, Yueqi Wang, Yan Zhang, Amjad Riaz, Bo Tang, and et al. 2023. "Expression of Phospholipase D Family Member 6 in Bovine Testes and Its Molecular Characteristics" International Journal of Molecular Sciences 24, no. 15: 12172. https://doi.org/10.3390/ijms241512172







