Dynamic Properties of the DNA Damage Response Mre11/Rad50 Complex
Abstract
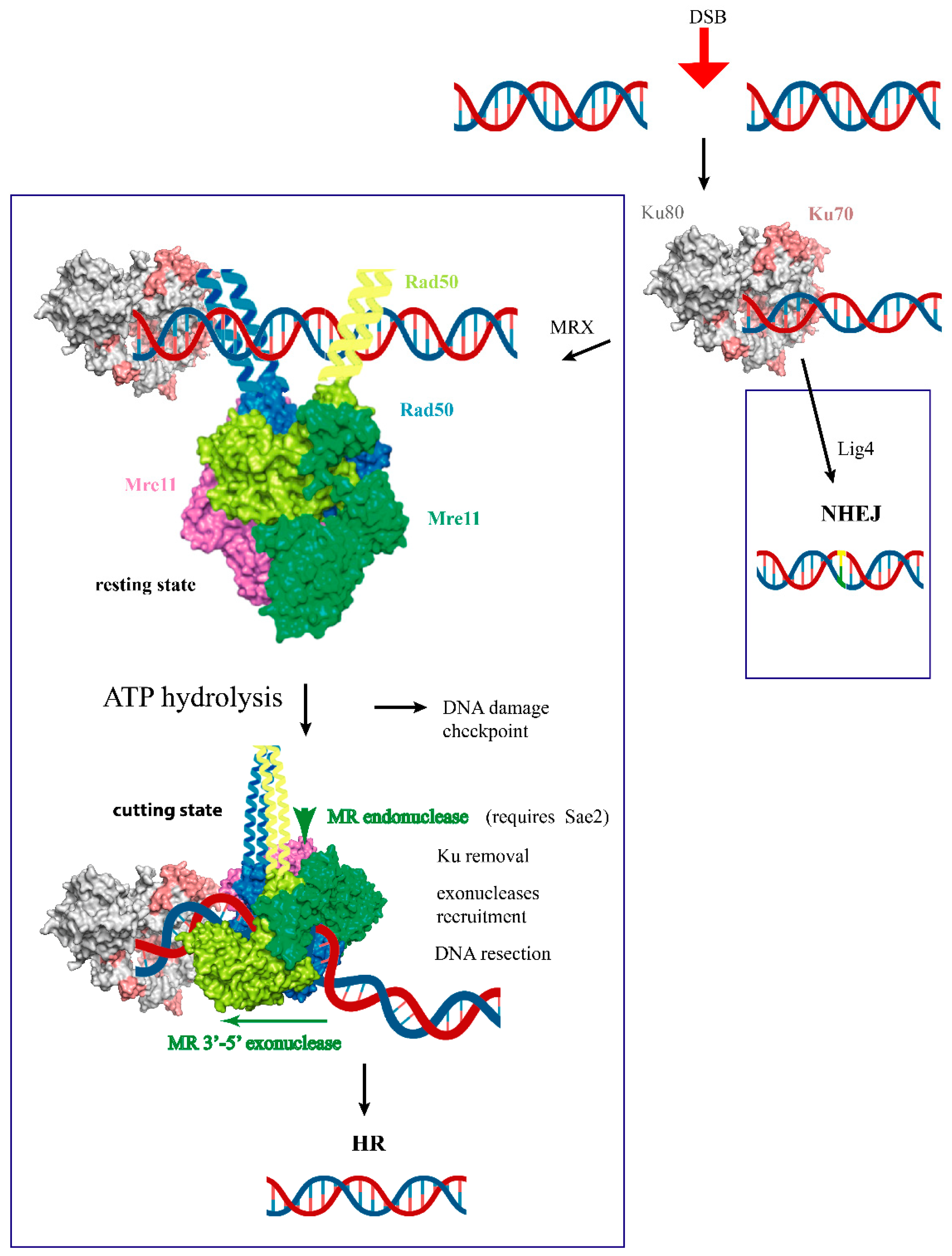
:1. Introduction
2. Rad50 Intrinsic Dynamic Properties
2.1. A Dynamic Allosteric Pattern Is the Key to Rad50 Conformational Change
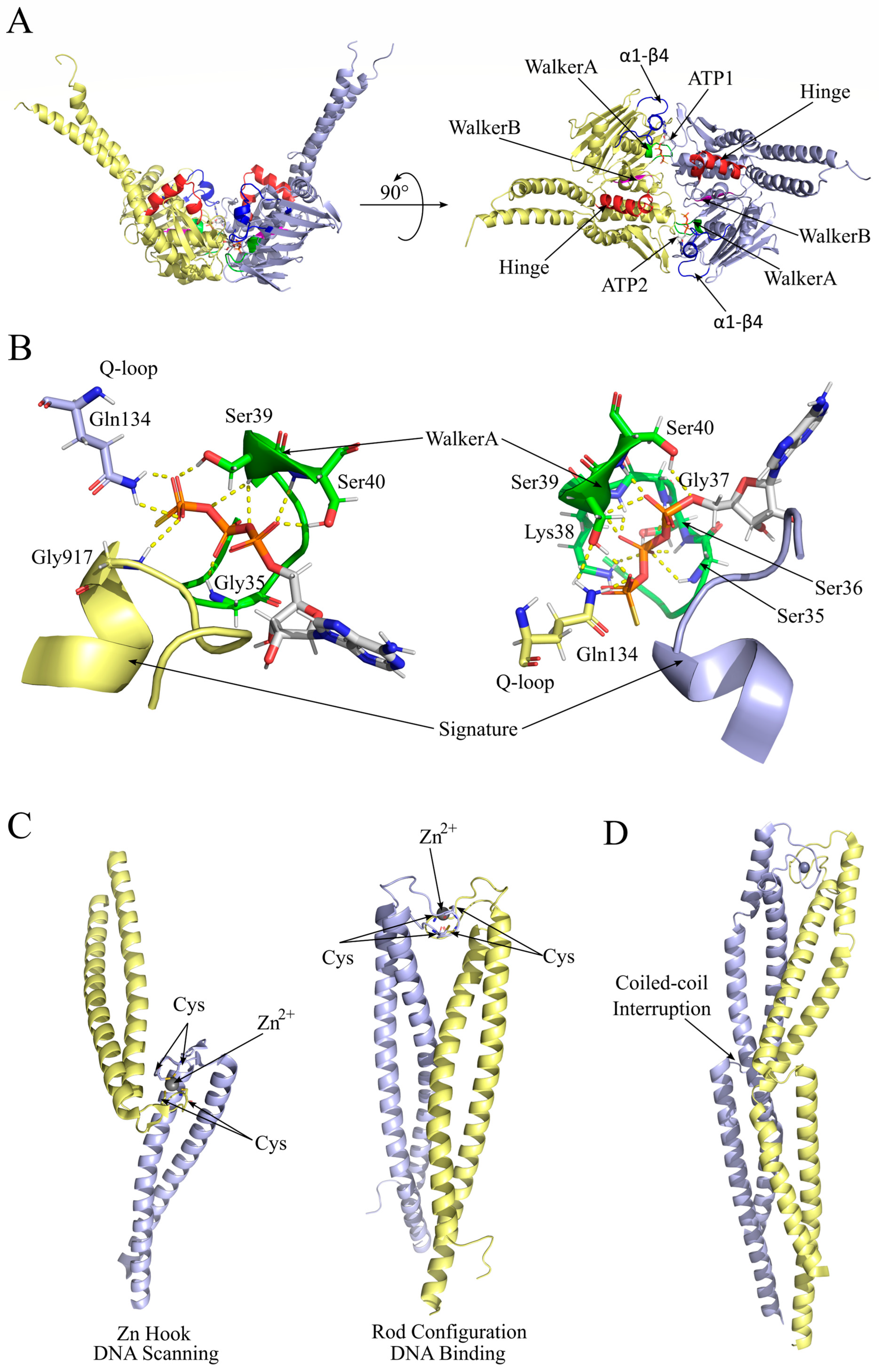
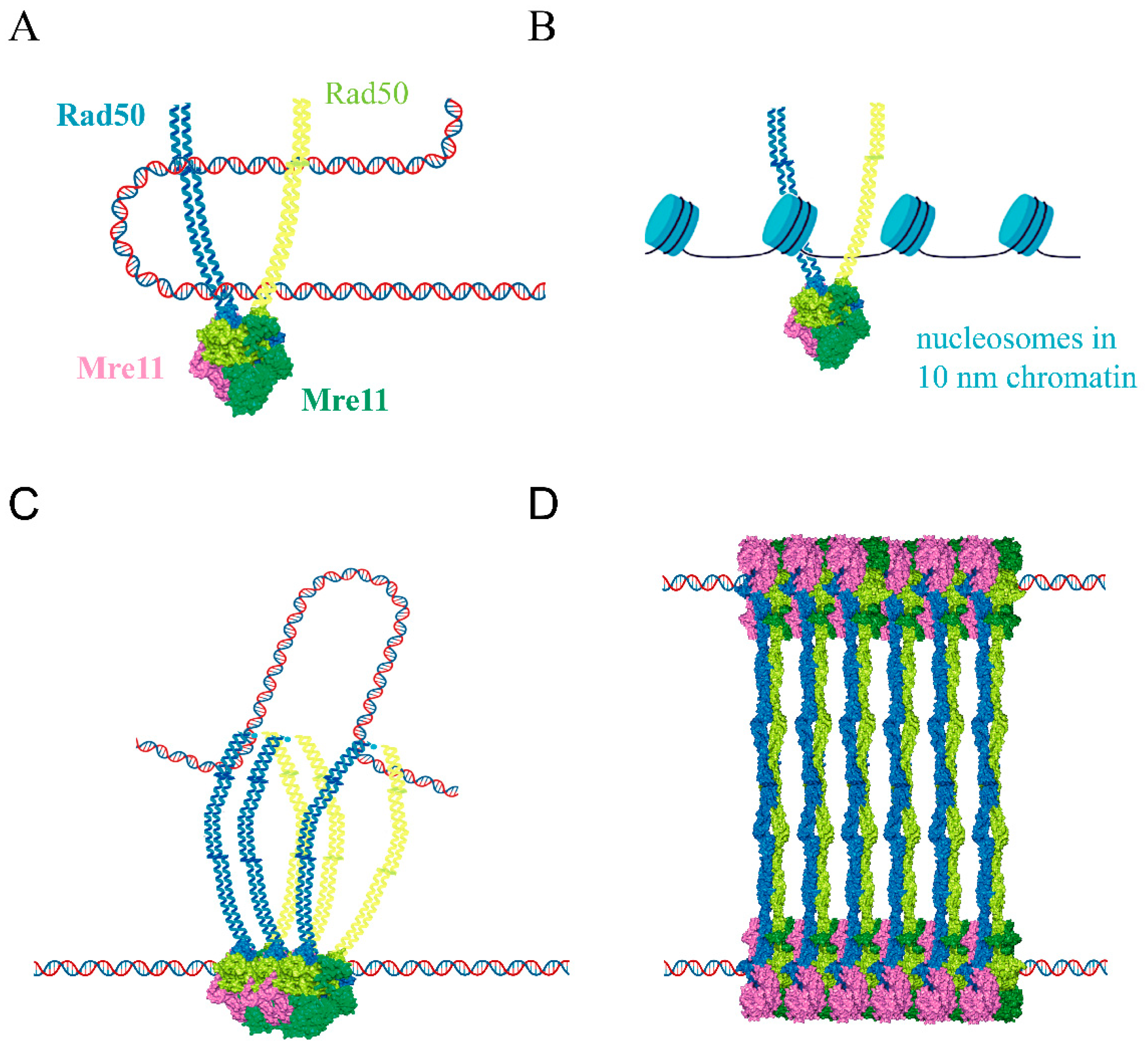
2.2. Rad50 Coiled-Coil Region Has Roles in DNA Tethering
3. Mre11 Intrinsic Dynamic Properties
4. Mre11/Rad50 Complex Adopts Different Conformations for Different Functions
4.1. The Resting State Is Competent for DNA Ends Tethering, DNA Scanning, and Multicomplex Assembly
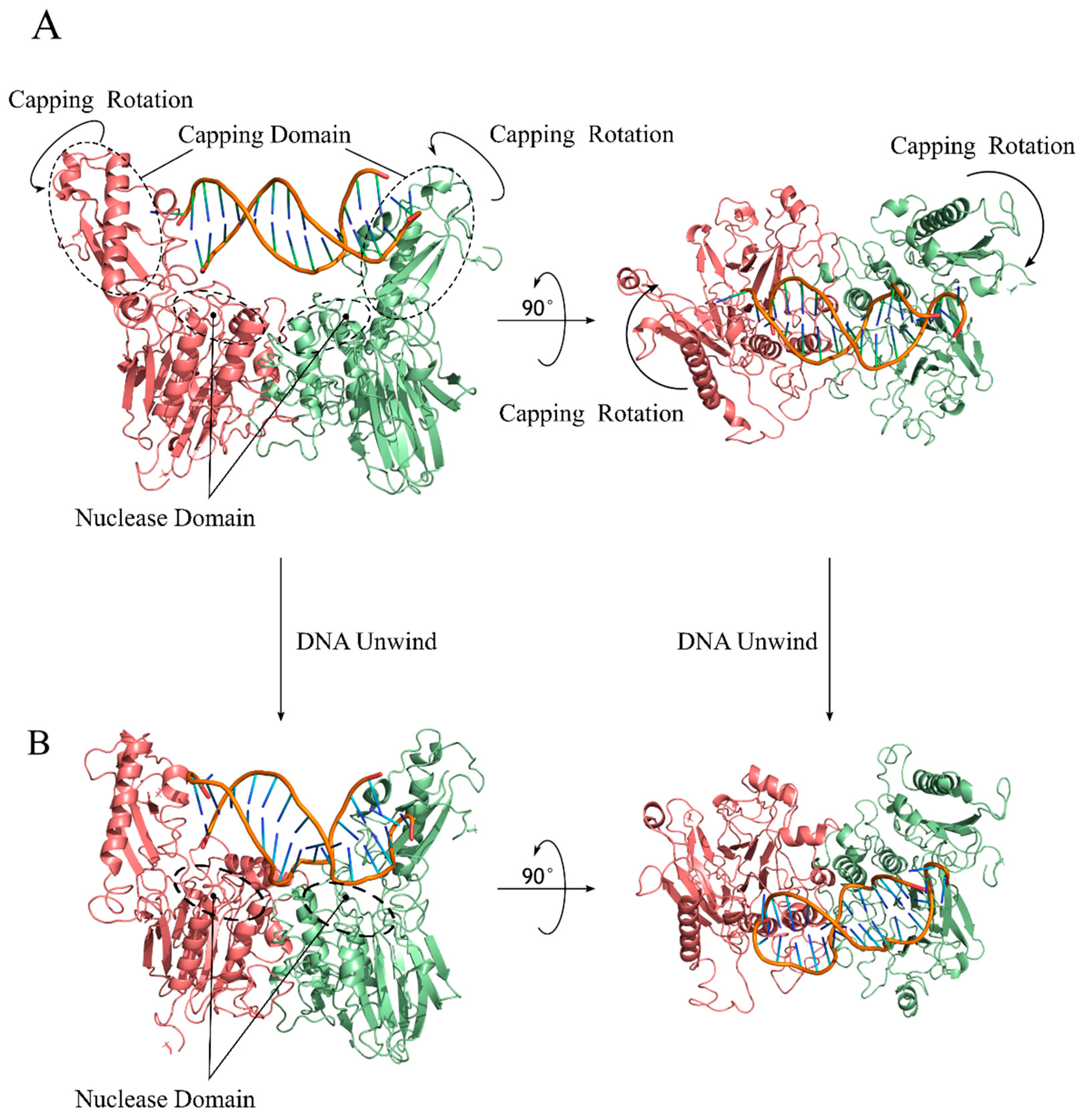
4.2. Cutting State Is Competent for DNA End Recognition and Nuclease Activity
4.3. Regulation of Mre11/Rad50 Complex Functions via Interaction with Different Partners
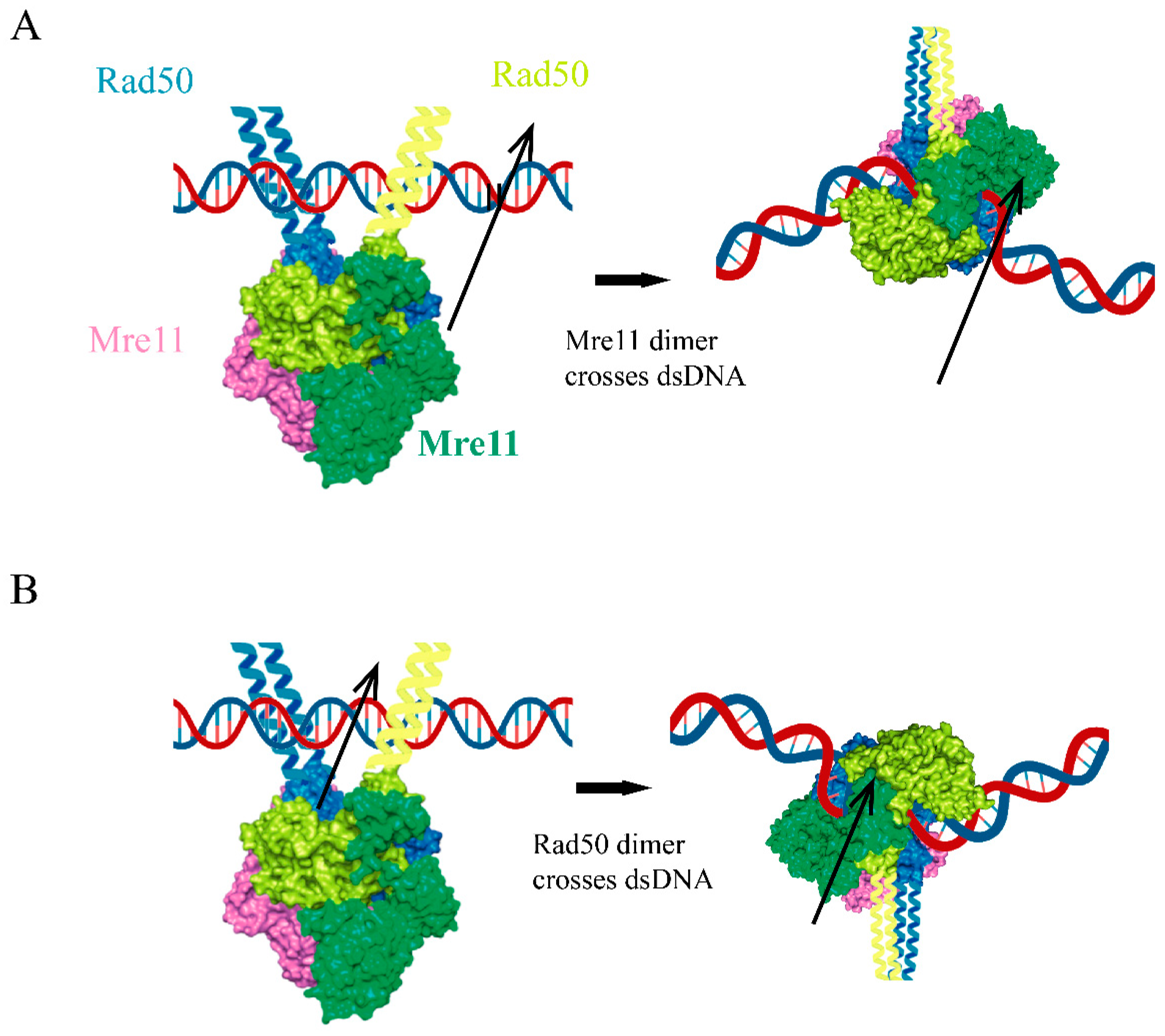
4.4. The Topology of Mre11/Rad50 DNA Binding
4.5. The Dynamic Path from Resting State to Cutting State in DNA-Bound MR Complex Is Still under Debate
4.6. The Mechanism of Tel1/ATM Activation Is Still under Investigation
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Bonetti, D.; Colombo, C.V.; Clerici, M.; Longhese, M.P. Processing of DNA Ends in the Maintenance of Genome Stability. Front. Genet. 2018, 9, 390. [Google Scholar] [PubMed] [Green Version]
- Dupuy, P.; Sauviac, L.; Bruand, C. Stress-Inducible NHEJ in Bacteria: Function in DNA Repair and Acquisition of Heterologous DNA. Nucleic Acids Res. 2019, 47, 1335–1349. [Google Scholar] [CrossRef]
- Kieffer, S.R.; Lowndes, N.F. Immediate-Early, Early, and Late Responses to DNA Double Stranded Breaks. Front. Genet. 2022, 13, 793884. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Jeggo, P.A. Canonical DNA Non-Homologous End-Joining; Capacity versus Fidelity. Br. J. Radiol. 2020, 93, 20190966. [Google Scholar] [CrossRef]
- Nickoloff, J.A.; Sharma, N.; Taylor, L.; Allen, S.J.; Hromas, R. Nucleases and Co-Factors in DNA Replication Stress Responses. DNA 2022, 2, 68–85. [Google Scholar] [CrossRef] [PubMed]
- Tisi, R.; Vertemara, J.; Zampella, G.; Longhese, M.P. Functional and Structural Insights into the MRX/MRN Complex, a Key Player in Recognition and Repair of DNA Double-Strand Breaks. Comput. Struct. Biotechnol. J. 2020, 18, 1137–1152. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.Y.; Toh, M.T.; Wang, X. NBS1 Deficiency Promotes Genome Instability by Affecting DNA Damage Signaling Pathway and Impairing Telomere Integrity. Cell Biochem. Funct. 2012, 30, 233–242. [Google Scholar]
- Zabolotnaya, E.; Mela, I.; Henderson, R.M.; Robinson, N.P. Turning the Mre11/Rad50 DNA Repair Complex on Its Head: Lessons from SMC Protein Hinges, Dynamic Coiled-Coil Movements and DNA Loop-Extrusion? Biochem. Soc. Trans. 2020, 48, 2359–2376. [Google Scholar]
- Reginato, G.; Cejka, P. The MRE11 Complex: A Versatile Toolkit for the Repair of Broken DNA. DNA Repair 2020, 91–92, 102869. [Google Scholar]
- Hailemariam, S.; Kumar, S.; Burgers, P.M. Activation of Tel1ATM Kinase Requires Rad50 ATPase and Long Nucleosome-Free DNA but No DNA Ends. J. Biol. Chem. 2019, 294, 10120–10130. [Google Scholar]
- Gnügge, R.; Reginato, G.; Cejka, P.; Symington, L.S. Sequence and Chromatin Features Guide DNA Double-Strand Break Resection Initiation. Mol. Cell 2023, 83, 1237–1250.e15. [Google Scholar] [PubMed]
- Lee, J.-H.; Paull, T.T. ATM Activation by DNA Double-Strand Breaks through the Mre11-Rad50-Nbs1 Complex. Science 2005, 308, 551–554. [Google Scholar] [CrossRef]
- Nakada, D.; Matsumoto, K.; Sugimoto, K. ATM-Related Tel1 Associates with Double-Strand Breaks through an Xrs2-Dependent Mechanism. Genes Dev. 2003, 17, 1957–1962. [Google Scholar] [CrossRef] [Green Version]
- You, Z.; Chahwan, C.; Bailis, J.; Hunter, T.; Russell, P. ATM Activation and Its Recruitment to Damaged DNA Require Binding to the C Terminus of Nbs1. Mol. Cell. Biol. 2005, 25, 5363–5379. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, D.; Hayashihara, K.; Shima, H.; Higashide, M.; Terasawa, M.; Gasser, S.M.; Shinohara, M. The MRX Complex Ensures NHEJ Fidelity through Multiple Pathways Including Xrs2-FHA-Dependent Tel1 Activation. PLoS Genet. 2016, 12, e1005942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tannous, E.A.; Burgers, P.M. Novel Insights into the Mechanism of Cell Cycle Kinases Mec1(ATR) and Tel1(ATM). Crit. Rev. Biochem. Mol. Biol. 2021, 56, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Casari, E.; Gnugnoli, M.; Rinaldi, C.; Pizzul, P.; Colombo, C.V.; Bonetti, D.; Longhese, M.P. To Fix or Not to Fix: Maintenance of Chromosome Ends Versus Repair of DNA Double-Strand Breaks. Cells 2022, 11, 3224. [Google Scholar] [CrossRef]
- Herdendorf, T.J.; Nelson, S.W. Catalytic Mechanism of Bacteriophage T4 Rad50 ATP Hydrolysis. Biochemistry 2014, 53, 5647–5660. [Google Scholar]
- Deshpande, R.A.; Williams, G.J.; Limbo, O.; Williams, R.S.; Kuhnlein, J.; Lee, J.-H.; Classen, S.; Guenther, G.; Russell, P.; Tainer, J.A.; et al. ATP-Driven Rad50 Conformations Regulate DNA Tethering, End Resection, and ATM Checkpoint Signaling. EMBO J. 2014, 33, 482–500. [Google Scholar]
- Moncalian, G.; Lengsfeld, B.; Bhaskara, V.; Hopfner, K.-P.; Karcher, A.; Alden, E.; Tainer, J.A.; Paull, T.T. The rad50 Signature Motif: Essential to ATP Binding and Biological Function. J. Mol. Biol. 2004, 335, 937–951. [Google Scholar] [CrossRef]
- Boswell, Z.K.; Rahman, S.; Canny, M.D.; Latham, M.P. A Dynamic Allosteric Pathway Underlies Rad50 ABC ATPase Function in DNA Repair. Sci. Rep. 2018, 8, 1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Käshammer, L.; Saathoff, J.-H.; Lammens, K.; Gut, F.; Bartho, J.; Alt, A.; Kessler, B.; Hopfner, K.-P. Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex. Mol. Cell 2019, 76, 382–394. [Google Scholar]
- Gut, F.; Käshammer, L.; Lammens, K.; Bartho, J.D.; Boggusch, A.-M.; van de Logt, E.; Kessler, B.; Hopfner, K.-P. Structural Mechanism of Endonucleolytic Processing of Blocked DNA Ends and Hairpins by Mre11-Rad50. Mol. Cell 2022, 82, 3513–3522.e6. [Google Scholar] [PubMed]
- Seifert, F.U.; Lammens, K.; Stoehr, G.; Kessler, B.; Hopfner, K.-P. Structural Mechanism of ATP-Dependent DNA Binding and DNA End Bridging by Eukaryotic Rad50. EMBO J. 2016, 35, 759–772. [Google Scholar] [PubMed] [Green Version]
- Hopfner, K.P.; Karcher, A.; Shin, D.S.; Craig, L.; Arthur, L.M.; Carney, J.P.; Tainer, J.A. Structural Biology of Rad50 ATPase: ATP-Driven Conformational Control in DNA Double-Strand Break Repair and the ABC-ATPase Superfamily. Cell 2000, 101, 789–800. [Google Scholar]
- Williams, G.J.; Williams, R.S.; Williams, J.S.; Moncalian, G.; Arvai, A.S.; Limbo, O.; Guenther, G.; SilDas, S.; Hammel, M.; Russell, P.; et al. ABC ATPase Signature Helices in Rad50 Link Nucleotide State to Mre11 Interface for DNA Repair. Nat. Struct. Mol. Biol. 2011, 18, 423–431. [Google Scholar]
- Boswell, Z.K.; Canny, M.D.; Buschmann, T.A.; Sang, J.; Latham, M.P. Adjacent Mutations in the Archaeal Rad50 ABC ATPase D-Loop Disrupt Allosteric Regulation of ATP Hydrolysis through Different Mechanisms. Nucleic Acids Res. 2020, 48, 2457–2472. [Google Scholar] [CrossRef] [Green Version]
- Cassani, C.; Vertemara, J.; Bassani, M.; Marsella, A.; Tisi, R.; Zampella, G.; Longhese, M.P. The ATP-Bound Conformation of the Mre11-Rad50 Complex Is Essential for Tel1/ATM Activation. Nucleic Acids Res. 2019, 47, 3550–3567. [Google Scholar]
- Park, Y.B.; Hohl, M.; Padjasek, M.; Jeong, E.; Jin, K.S.; Krężel, A.; Petrini, J.H.J.; Cho, Y. Eukaryotic Rad50 Functions as a Rod-Shaped Dimer. Nat. Struct. Mol. Biol. 2017, 24, 248–257. [Google Scholar]
- Hohl, M.; Kochańczyk, T.; Tous, C.; Aguilera, A.; Krężel, A.; Petrini, J.H.J. Interdependence of the Rad50 Hook and Globular Domain Functions. Mol. Cell 2015, 57, 479–491. [Google Scholar]
- Hohl, M.; Kwon, Y.; Galván, S.M.; Xue, X.; Tous, C.; Aguilera, A.; Sung, P.; Petrini, J.H.J. The Rad50 Coiled-Coil Domain Is Indispensable for Mre11 Complex Functions. Nat. Struct. Mol. Biol. 2011, 18, 1124–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatebe, H.; Lim, C.T.; Konno, H.; Shiozaki, K.; Shinohara, A.; Uchihashi, T.; Furukohri, A. Rad50 Zinc Hook Functions as a Constitutive Dimerization Module Interchangeable with SMC Hinge. Nat. Commun. 2020, 11, 370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Herrero, F.; de Jager, M.; Dekker, N.H.; Kanaar, R.; Wyman, C.; Dekker, C. Mesoscale Conformational Changes in the DNA-Repair Complex Rad50/Mre11/Nbs1 upon Binding DNA. Nature 2005, 437, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Rotheneder, M.; Stakyte, K.; van de Logt, E.; Bartho, J.D.; Lammens, K.; Fan, Y.; Alt, A.; Kessler, B.; Jung, C.; Roos, W.P.; et al. Cryo-EM Structure of the Mre11-Rad50-Nbs1 Complex Reveals the Molecular Mechanism of Scaffolding Functions. Mol. Cell 2022, 83, 167–185.e9. [Google Scholar] [CrossRef]
- Tran, J.B.; Padjasek, M.; Krężel, A. Relations between Structure and Zn(II) Binding Affinity Shed Light on the Mechanisms of Rad50 Hook Domain Functioning and Its Phosphorylation. Int. J. Mol. Sci. 2022, 23, 11140. [Google Scholar] [CrossRef]
- van Noort, J.; van Der Heijden, T.; de Jager, M.; Wyman, C.; Kanaar, R.; Dekker, C. The Coiled-Coil of the Human Rad50 DNA Repair Protein Contains Specific Segments of Increased Flexibility. Proc. Natl. Acad. Sci. USA 2003, 100, 7581–7586. [Google Scholar] [CrossRef]
- Zabolotnaya, E.; Mela, I.; Williamson, M.J.; Bray, S.M.; Yau, S.K.; Papatziamou, D.; Edwardson, J.M.; Robinson, N.P.; Henderson, R.M. Modes of Action of the Archaeal Mre11/Rad50 DNA-Repair Complex Revealed by Fast-Scan Atomic Force Microscopy. Proc. Natl. Acad. Sci. USA 2020, 117, 14936–14947. [Google Scholar] [CrossRef]
- Furuse, M.; Nagase, Y.; Tsubouchi, H.; Murakami-Murofushi, K.; Shibata, T.; Ohta, K. Distinct Roles of Two Separable in Vitro Activities of Yeast Mre11 in Mitotic and Meiotic Recombination. EMBO J. 1998, 17, 6412–6425. [Google Scholar] [CrossRef] [Green Version]
- Shibata, A.; Moiani, D.; Arvai, A.S.; Perry, J.; Harding, S.M.; Genois, M.-M.; Maity, R.; van Rossum-Fikkert, S.; Kertokalio, A.; Romoli, F.; et al. DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities. Mol. Cell 2014, 53, 7–18. [Google Scholar] [CrossRef] [Green Version]
- Gobbini, E.; Cassani, C.; Vertemara, J.; Wang, W.; Mambretti, F.; Casari, E.; Sung, P.; Tisi, R.; Zampella, G.; Longhese, M.P. The MRX Complex Regulates Exo1 Resection Activity by Altering DNA End Structure. EMBO J. 2018, 37, e98588. [Google Scholar] [CrossRef]
- Rahman, S.; Beikzadeh, M.; Canny, M.D.; Kaur, N.; Latham, M.P. Mutation of Conserved Mre11 Residues Alter Protein Dynamics to Separate Nuclease Functions. J. Mol. Biol. 2020, 432, 3289–3308. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Beikzadeh, M.; Latham, M.P. Biochemical and Structural Characterization of Analogs of MRE11 Breast Cancer-Associated Mutant F237C. Sci. Rep. 2021, 11, 7089. [Google Scholar] [CrossRef] [PubMed]
- Cannon, B.; Kuhnlein, J.; Yang, S.-H.; Cheng, A.; Schindler, D.; Stark, J.M.; Russell, R.; Paull, T.T. Visualization of Local DNA Unwinding by Mre11/Rad50/Nbs1 Using Single-Molecule FRET. Proc. Natl. Acad. Sci. USA 2013, 110, 18868–18873. [Google Scholar] [CrossRef]
- Lim, H.S.; Kim, J.S.; Park, Y.B.; Gwon, G.H.; Cho, Y. Crystal Structure of the Mre11-Rad50-ATPγS Complex: Understanding the Interplay between Mre11 and Rad50. Genes Dev. 2011, 25, 1091–1104. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Sung, S.; Kim, Y.; Li, F.; Gwon, G.; Jo, A.; Kim, A.-K.; Kim, T.; Song, O.-K.; Lee, S.E.; et al. ATP-Dependent DNA Binding, Unwinding, and Resection by the Mre11/Rad50 Complex. EMBO J. 2016, 35, 743–758. [Google Scholar] [CrossRef] [Green Version]
- Krogh, B.O.; Llorente, B.; Lam, A.; Symington, L.S. Mutations in Mre11 Phosphoesterase Motif I That Impair Saccharomyces Cerevisiae Mre11-Rad50-Xrs2 Complex Stability in Addition to Nuclease Activity. Genetics 2005, 171, 1561–1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canny, M.D.; Latham, M.P. LRET-Derived HADDOCK Structural Models Describe the Conformational Heterogeneity Required for DNA Cleavage by the Mre11-Rad50 DNA Damage Repair Complex. eLife 2022, 11, e69579. [Google Scholar] [CrossRef]
- Oh, J.; Symington, L.S. Role of the Mre11 Complex in Preserving Genome Integrity. Genes 2018, 9, 589. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.S.; Dodson, G.E.; Limbo, O.; Yamada, Y.; Williams, J.S.; Guenther, G.; Classen, S.; Glover, J.N.M.; Iwasaki, H.; Russell, P.; et al. Nbs1 Flexibly Tethers Ctp1 and Mre11-Rad50 to Coordinate DNA Double-Strand Break Processing and Repair. Cell 2009, 139, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.; Al-Zain, A.; Cannavo, E.; Cejka, P.; Symington, L.S. Xrs2 Dependent and Independent Functions of the Mre11-Rad50 Complex. Mol. Cell 2016, 64, 405–415. [Google Scholar] [CrossRef] [Green Version]
- Cannavo, E.; Reginato, G.; Cejka, P. Stepwise 5′ DNA End-Specific Resection of DNA Breaks by the Mre11-Rad50-Xrs2 and Sae2 Nuclease Ensemble. Proc. Natl. Acad. Sci. USA 2019, 116, 5505–5513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zdravković, A.; Daley, J.M.; Dutta, A.; Niwa, T.; Murayama, Y.; Kanamaru, S.; Ito, K.; Maki, T.; Argunhan, B.; Takahashi, M.; et al. A Conserved Ctp1/CtIP C-Terminal Peptide Stimulates Mre11 Endonuclease Activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2016287118. [Google Scholar] [CrossRef] [PubMed]
- Öz, R.; Howard, S.M.; Sharma, R.; Törnkvist, H.; Ceppi, I.; Kk, S.; Kristiansson, E.; Cejka, P.; Westerlund, F. Phosphorylated CtIP Bridges DNA to Promote Annealing of Broken Ends. Proc. Natl. Acad. Sci. USA 2020, 117, 21403–21412. [Google Scholar] [PubMed]
- Ferrari, M.; Dibitetto, D.; De Gregorio, G.; Eapen, V.V.; Rawal, C.C.; Lazzaro, F.; Tsabar, M.; Marini, F.; Haber, J.E.; Pellicioli, A. Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break. PLoS Genet. 2015, 11, e1004928. [Google Scholar]
- Mojumdar, A.; Adam, N.; Cobb, J.A. Nej1 Interacts with Sae2 at DNA Double-Stranded Breaks to Inhibit DNA Resection. J. Biol. Chem. 2022, 298, 101937. [Google Scholar]
- Clerici, M.; Mantiero, D.; Lucchini, G.; Longhese, M.P. The Saccharomyces Cerevisiae Sae2 Protein Negatively Regulates DNA Damage Checkpoint Signalling. EMBO Rep. 2006, 7, 212–218. [Google Scholar]
- Alani, E.; Padmore, R.; Kleckner, N. Analysis of Wild-Type and rad50 Mutants of Yeast Suggests an Intimate Relationship between Meiotic Chromosome Synapsis and Recombination. Cell 1990, 61, 419–436. [Google Scholar] [CrossRef]
- Zhang, L.-L.; Wu, Z.; Zhou, J.-Q. Tel1 and Rif2 Oppositely Regulate Telomere Protection at Uncapped Telomeres in Saccharomyces Cerevisiae. J. Genet. Genom. 2018, 45, 467–476. [Google Scholar]
- Hailemariam, S.; De Bona, P.; Galletto, R.; Hohl, M.; Petrini, J.H.; Burgers, P.M. The Telomere-Binding Protein Rif2 and ATP-Bound Rad50 Have Opposing Roles in the Activation of Yeast Tel1 Kinase. J. Biol. Chem. 2019, 294, 18846–18852. [Google Scholar] [CrossRef]
- Cassani, C.; Gobbini, E.; Wang, W.; Niu, H.; Clerici, M.; Sung, P.; Longhese, M.P. Tel1 and Rif2 Regulate MRX Functions in End-Tethering and Repair of DNA Double-Strand Breaks. PLoS Biol. 2016, 14, e1002387. [Google Scholar] [CrossRef] [Green Version]
- Marsella, A.; Gobbini, E.; Cassani, C.; Tisi, R.; Cannavo, E.; Reginato, G.; Cejka, P.; Longhese, M.P. Sae2 and Rif2 Regulate MRX Endonuclease Activity at DNA Double-Strand Breaks in Opposite Manners. Cell Rep. 2021, 34, 108906. [Google Scholar] [CrossRef] [PubMed]
- Roisné-Hamelin, F.; Pobiega, S.; Jézéquel, K.; Miron, S.; Dépagne, J.; Veaute, X.; Busso, D.; Du, M.-H.L.; Callebaut, I.; Charbonnier, J.-B.; et al. Mechanism of MRX Inhibition by Rif2 at Telomeres. Nat. Commun. 2021, 12, 2763. [Google Scholar] [CrossRef] [PubMed]
- Khayat, F.; Cannavo, E.; Alshmery, M.; Foster, W.R.; Chahwan, C.; Maddalena, M.; Smith, C.; Oliver, A.W.; Watson, A.T.; Carr, A.M.; et al. Inhibition of MRN Activity by a Telomere Protein Motif. Nat. Commun. 2021, 12, 3856. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, D.; Rinaldi, C.; Vertemara, J.; Notaro, M.; Pizzul, P.; Tisi, R.; Zampella, G.; Longhese, M.P. DNA Binding Modes Influence Rap1 Activity in the Regulation of Telomere Length and MRX Functions at DNA Ends. Nucleic Acids Res. 2020, 48, 2424–2441. [Google Scholar] [CrossRef]
- Whelan, D.R.; Rothenberg, E. Super-Resolution Mapping of Cellular Double-Strand Break Resection Complexes during Homologous Recombination. Proc. Natl. Acad. Sci. USA 2021, 118, 2021963118. [Google Scholar] [CrossRef] [PubMed]
- Lisby, M.; Barlow, J.H.; Burgess, R.C.; Rothstein, R. Choreography of the DNA Damage Response: Spatiotemporal Relationships among Checkpoint and Repair Proteins. Cell 2004, 118, 699–713. [Google Scholar]
- Wang, Y.-L.; Zhao, W.-W.; Bai, S.-M.; Feng, L.-L.; Bie, S.-Y.; Gong, L.; Wang, F.; Wei, M.-B.; Feng, W.-X.; Pang, X.-L.; et al. MRNIP Condensates Promote DNA Double-Strand Break Sensing and End Resection. Nat. Commun. 2022, 13, 2638. [Google Scholar] [CrossRef]
- Hoencamp, C.; Rowland, B.D. Genome Control by SMC Complexes. Nat. Rev. Mol. Cell Biol. 2023; in press. [Google Scholar] [CrossRef]
- Myler, L.R.; Gallardo, I.F.; Soniat, M.M.; Deshpande, R.A.; Gonzalez, X.B.; Kim, Y.; Paull, T.T.; Finkelstein, I.J. Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol. Cell 2017, 67, 891–898.e4. [Google Scholar] [CrossRef] [Green Version]
- Kissling, V.M.; Reginato, G.; Bianco, E.; Kasaciunaite, K.; Tilma, J.; Cereghetti, G.; Schindler, N.; Lee, S.S.; Guérois, R.; Luke, B.; et al. Mre11-Rad50 Oligomerization Promotes DNA Double-Strand Break Repair. Nat. Commun. 2022, 13, 2374. [Google Scholar] [CrossRef]
- Haering, C.H.; Löwe, J.; Hochwagen, A.; Nasmyth, K. Molecular Architecture of SMC Proteins and the Yeast Cohesin Complex. Mol. Cell 2002, 9, 773–788. [Google Scholar] [CrossRef]
- Williams, R.M.; Yates, L.A.; Zhang, X. Structures and Regulations of ATM and ATR, Master Kinases in Genome Integrity. Curr. Opin. Struct. Biol. 2020, 61, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Lammens, K.; Bemeleit, D.J.; Möckel, C.; Clausing, E.; Schele, A.; Hartung, S.; Schiller, C.B.; Lucas, M.; Angermüller, C.; Söding, J.; et al. The Mre11:Rad50 Structure Shows an ATP-Dependent Molecular Clamp in DNA Double-Strand Break Repair. Cell 2011, 145, 54–66. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.; Lee, S.J.; Rothstein, R.; Symington, L.S. Xrs2 and Tel1 Independently Contribute to MR-Mediated DNA Tethering and Replisome Stability. Cell Rep. 2018, 25, 1681–1692.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupré, A.; Boyer-Chatenet, L.; Gautier, J. Two-Step Activation of ATM by DNA and the Mre11-Rad50-Nbs1 Complex. Nat. Struct. Mol. Biol. 2006, 13, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, K.; Kwon, Y.; Sung, P.; Sugimoto, K. Correction for Fukunaga et al. “Activation of Protein Kinase Tel1 through Recognition of Protein-Bound DNA Ends”. Mol. Cell. Biol. 2017, 37, e00263-17. [Google Scholar] [CrossRef] [Green Version]
- Frigerio, C.; Di Nisio, E.; Galli, M.; Colombo, C.V.; Negri, R.; Clerici, M. The Chromatin Landscape around DNA Double-Strand Breaks in Yeast and Its Influence on DNA Repair Pathway Choice. Int. J. Mol. Sci. 2023, 24, 3248. [Google Scholar] [CrossRef]
- Forey, R.; Barthe, A.; Tittel-Elmer, M.; Wery, M.; Barrault, M.-B.; Ducrot, C.; Seeber, A.; Krietenstein, N.; Szachnowski, U.; Skrzypczak, M.; et al. A Role for the Mre11-Rad50-Xrs2 Complex in Gene Expression and Chromosome Organization. Mol. Cell 2021, 81, 183–197.e6. [Google Scholar] [CrossRef]
- Priya, B.; Ravi, S.; Kirubakaran, S. Targeting ATM and ATR for Cancer Therapeutics: Inhibitors in Clinic. Drug Discov. Today 2023, 28, 103662. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vertemara, J.; Tisi, R. Dynamic Properties of the DNA Damage Response Mre11/Rad50 Complex. Int. J. Mol. Sci. 2023, 24, 12377. https://doi.org/10.3390/ijms241512377
Vertemara J, Tisi R. Dynamic Properties of the DNA Damage Response Mre11/Rad50 Complex. International Journal of Molecular Sciences. 2023; 24(15):12377. https://doi.org/10.3390/ijms241512377
Chicago/Turabian StyleVertemara, Jacopo, and Renata Tisi. 2023. "Dynamic Properties of the DNA Damage Response Mre11/Rad50 Complex" International Journal of Molecular Sciences 24, no. 15: 12377. https://doi.org/10.3390/ijms241512377
APA StyleVertemara, J., & Tisi, R. (2023). Dynamic Properties of the DNA Damage Response Mre11/Rad50 Complex. International Journal of Molecular Sciences, 24(15), 12377. https://doi.org/10.3390/ijms241512377





