Towards Anticancer and Antibacterial Agents: Design and Synthesis of 1,2,3-Triazol-quinobenzothiazine Derivatives
Abstract
:1. Introduction
2. Results and Discussion
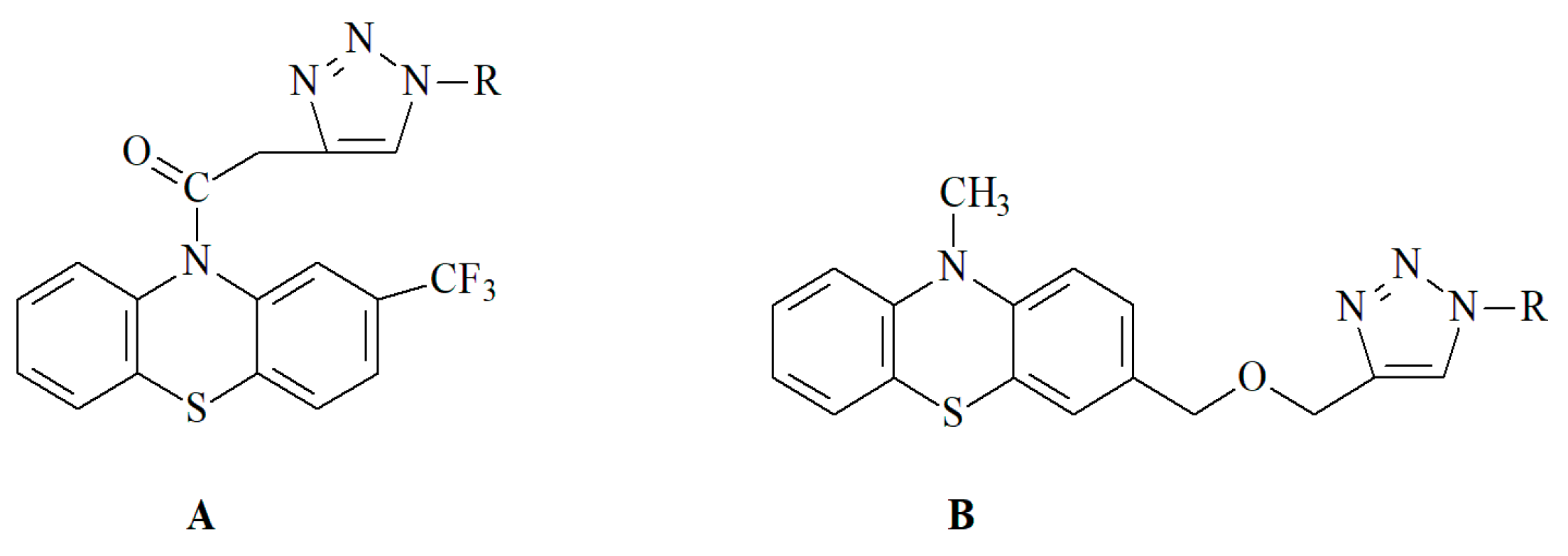
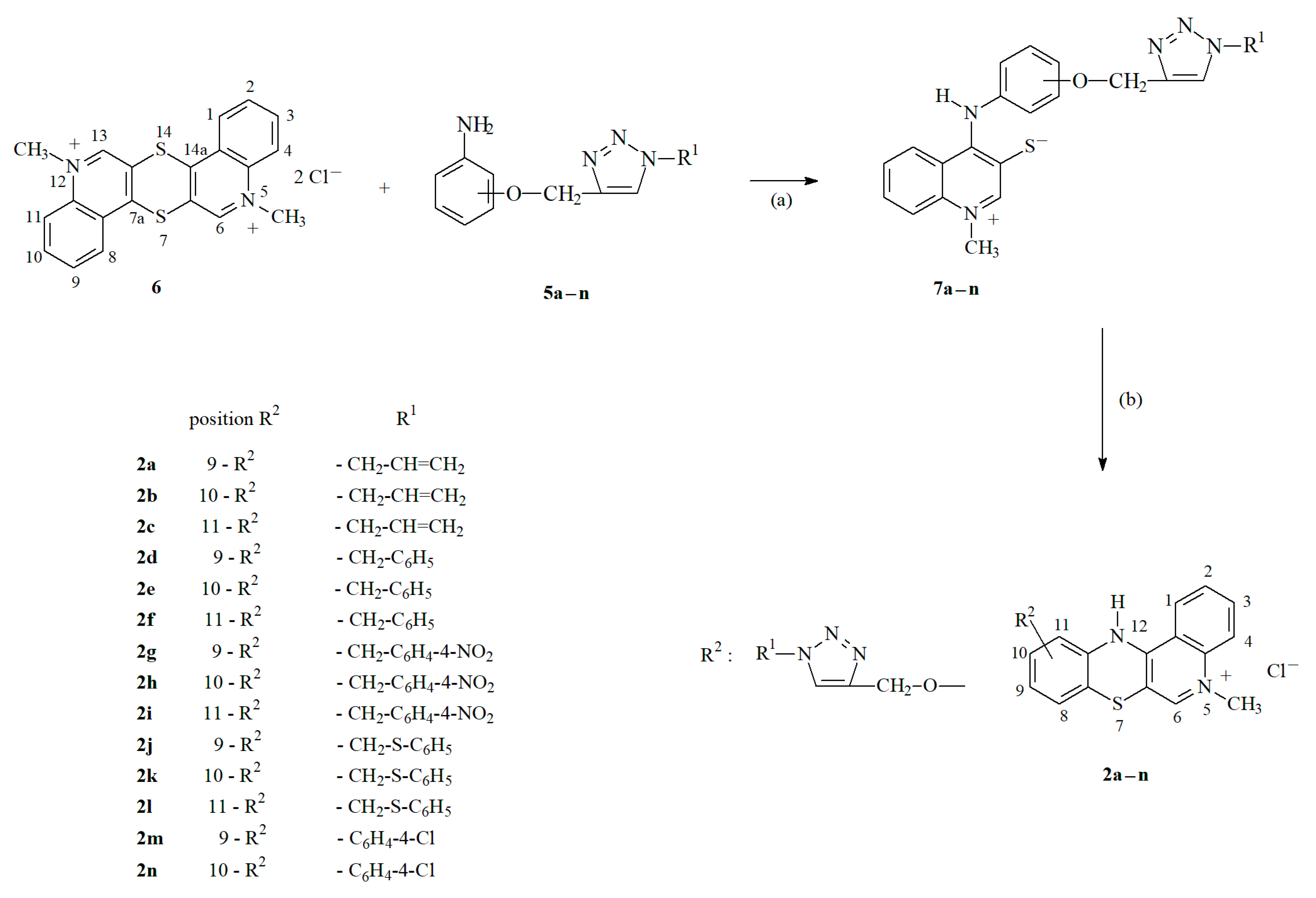
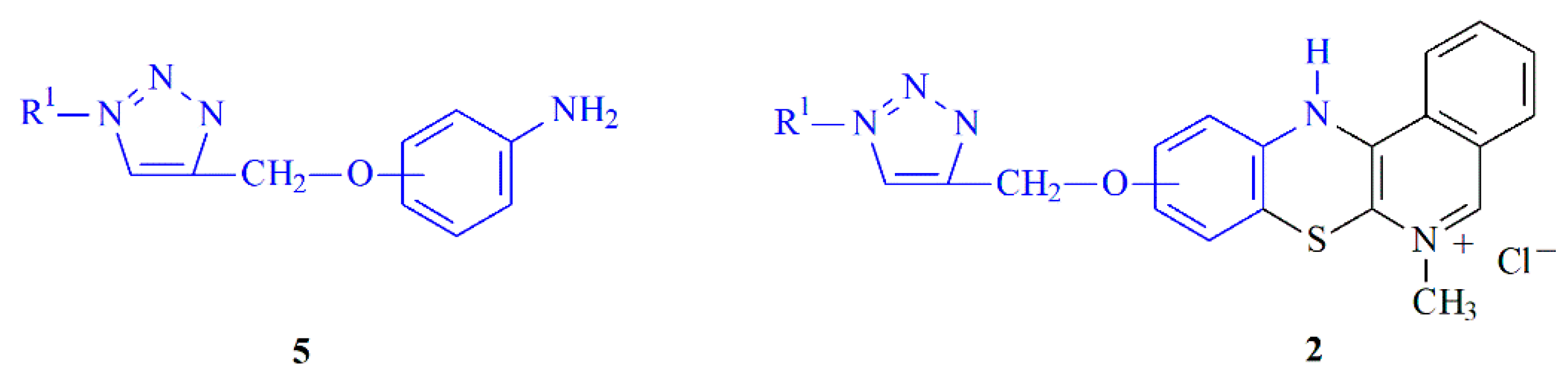
2.1. Chemistry—Design and Synthesis
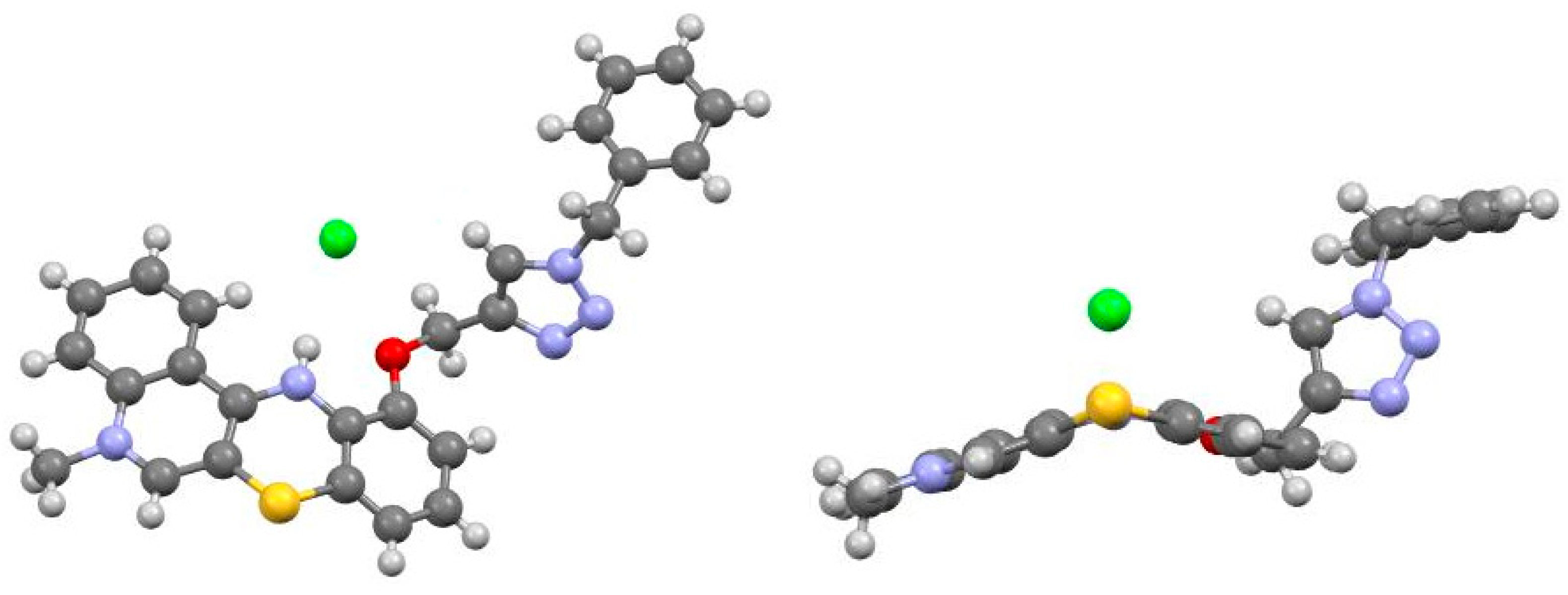
2.2. X-ray Structural Analysis
2.3. In Vitro Cytotoxic Activity
2.4. In Vitro Antimicrobial Activity
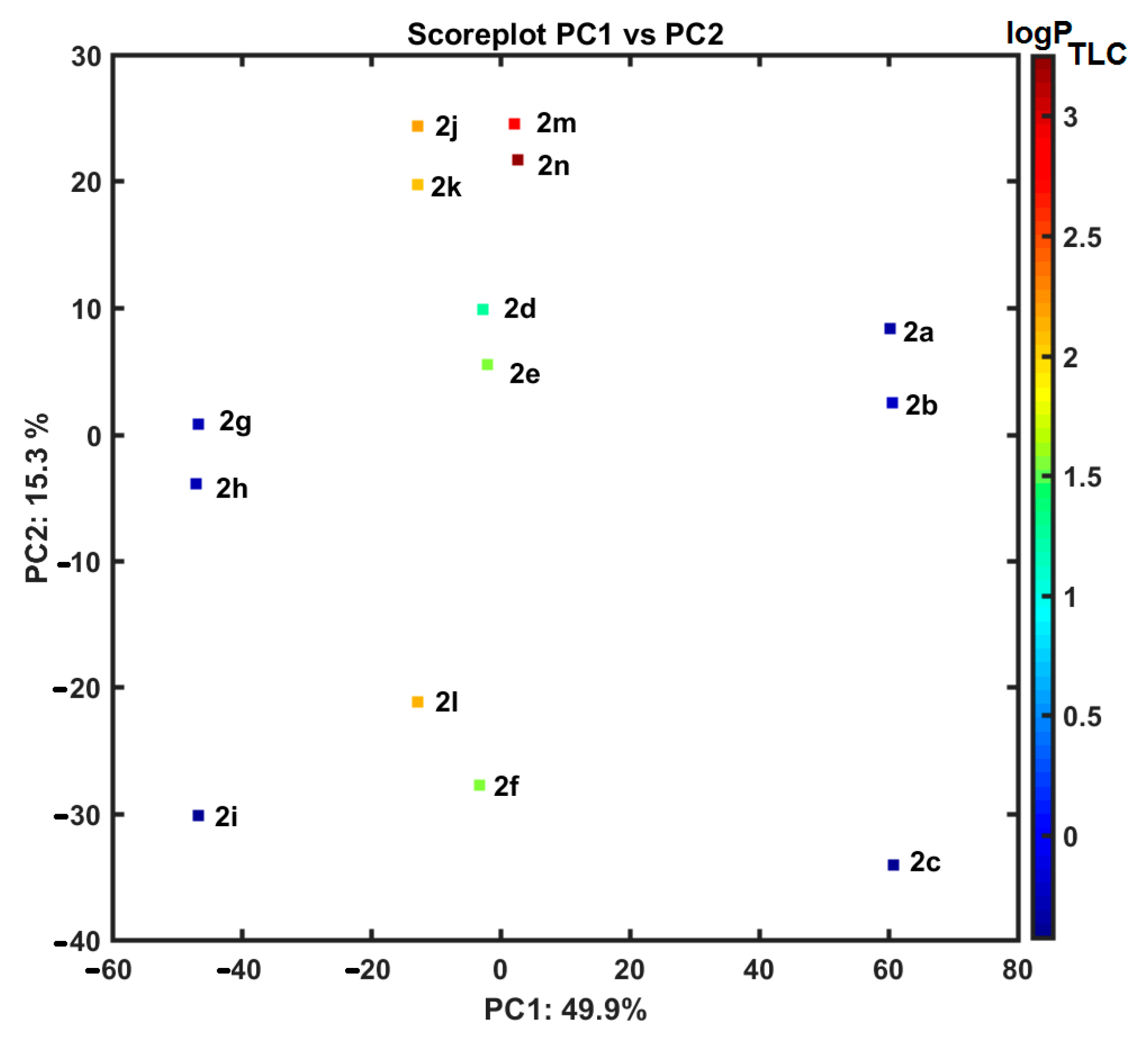
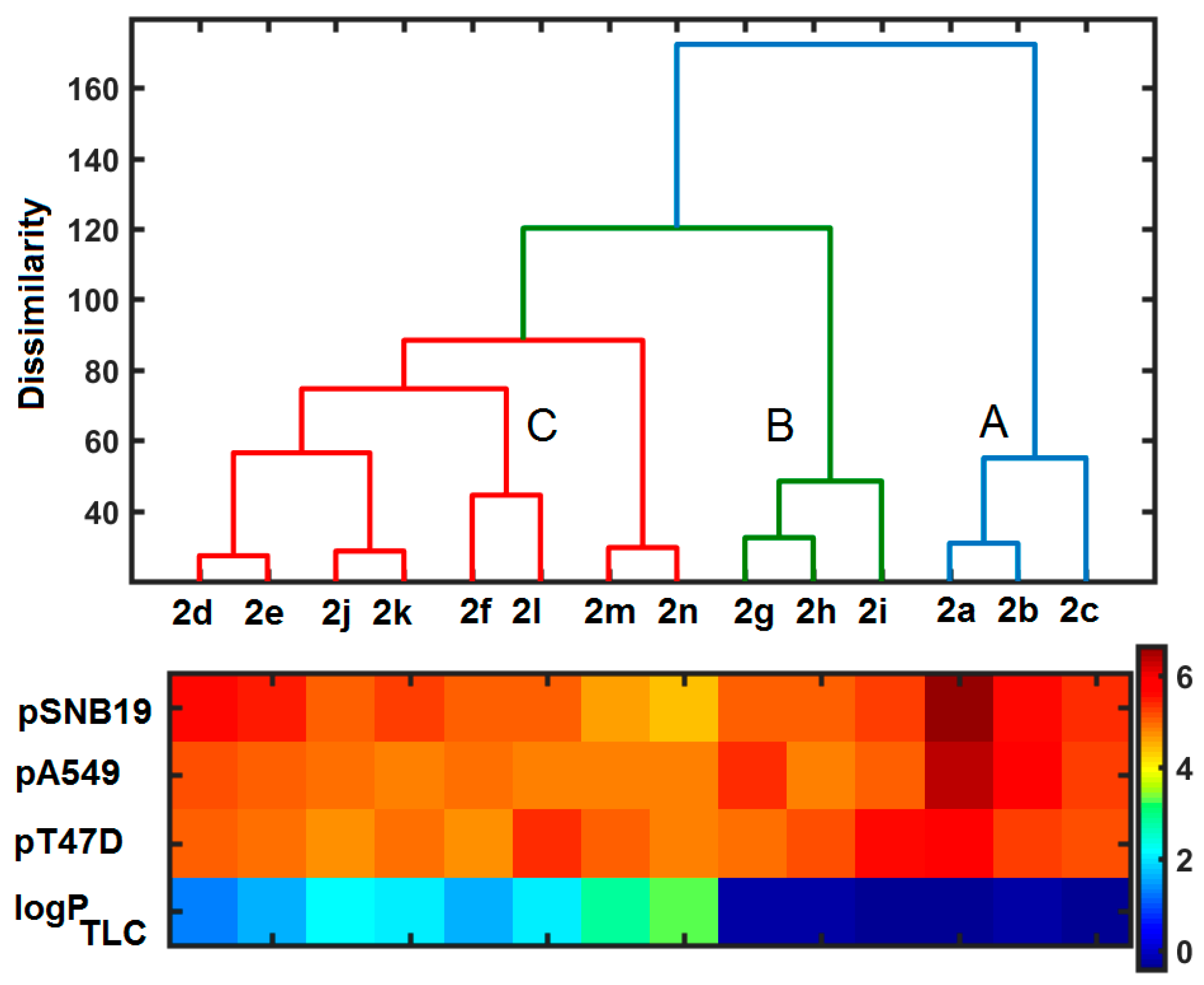
2.5. Similarity-Related Property Assessment
2.6. ClogP Estimation versus Experimental Lipophilicity
3. Materials and Methods
3.1. Chemistry
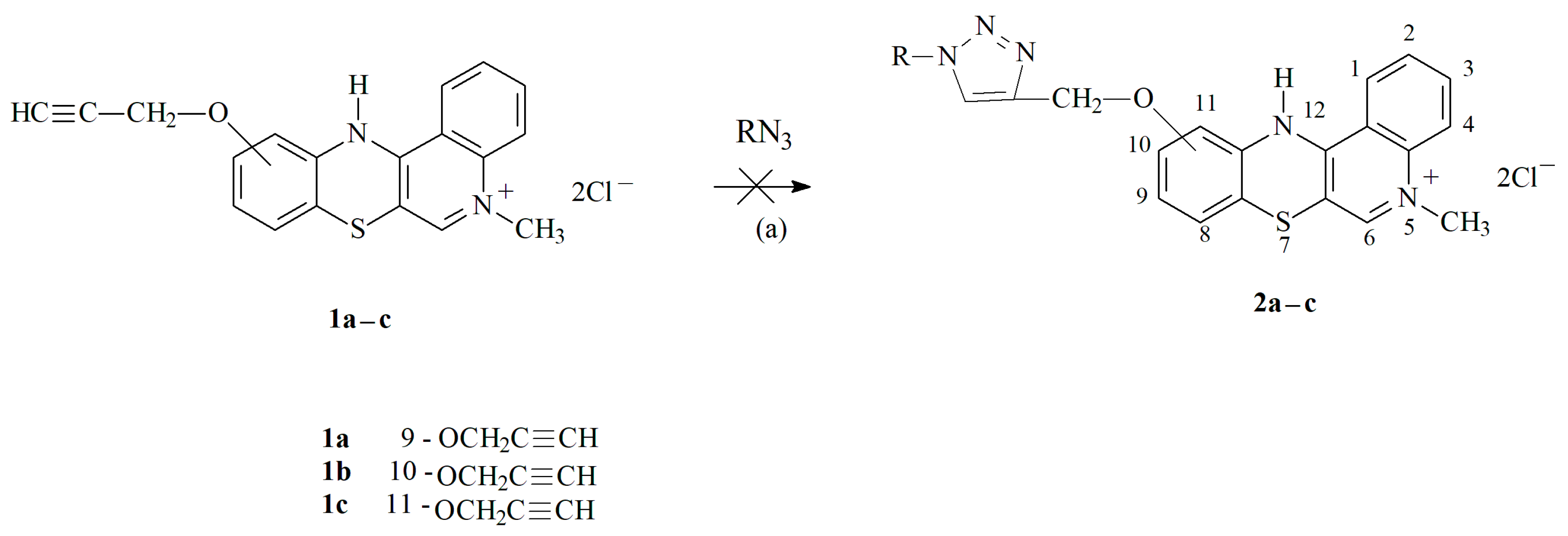
3.1.1. Synthesis of Propargiloxy-5-Methylo-12H-quino[3,4-b][1,4]benzothiazine Derivatives 2a–c
3.1.2. Synthesis of Aniline Derivatives of 1,2,3-triazole 5a–n
Procedure A: Preparation of Derivatives 5a–c
Procedure B: Preparation of Derivatives 5d–l
Procedure C: Preparation of Derivatives 5m–n
3.1.3. Synthesis of 1,2,3-Triazole Derivatives of 5-Methyl-12H-quino[3,4-b][1,4]benzothiazinium Chlorides 2a–n
Procedure A
Procedure B
3.2. X-ray Structural Analysis
3.3. Biological Evaluation
3.3.1. Cell Culture
3.3.2. Proliferation Assay
3.3.3. In Vitro Antibacterial Evaluation
3.3.4. In Vitro Antimycobacterial Evaluation
3.3.5. Determination of Minimum Bactericidal Concentrations
3.3.6. MTT Assay
3.3.7. TLC-Based Lipophilicity Determination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Posso, M.C.; Domingues, F.C.; Ferreira, S.; Silvestre, S. Development of phenothiazine hybrids with potential medicinal interest: A review. Molecules 2022, 27, 276. [Google Scholar] [CrossRef]
- Varga, B.; Csonka, Á.; Csonka, A.; Molnár, J.; Amaral, L.; Spengler, G. Possible biological and clinical applications of Ppenothiazines. Anticancer Res. 2017, 37, 5983–5993. [Google Scholar] [PubMed]
- Mitchell, S.C. Phenothiazine: The parent molecule. Curr. Drug Targ. 2006, 7, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Baldessarini, R.J.; Tarazi, F.I. Pharmacotherapy of psychosis and mania. In Goodman and Gilman’s the Pharmacological Basis of Therapeutics, 11th ed.; Brunton, L.L., Ed.; McGraw-Hill Medical Publishing Division: New York, NY, USA, 2006; pp. 461–500. [Google Scholar]
- Mosnaim, A.D.; Ranade, V.V.; Wolf, M.E.; Puente, J.; Valenzuela, M.A. Phenothiazine molecule provides the basic chemical structure for various classes of pharmacotherapeutic agents. Am. J. Therapeut. 2006, 13, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Zieba, A.; Czuba, Z.; Król, W. In vitro antimicrobial activity of novel azaphenothiazine derivatives. Acta Pol. Pharm. Drug Res. 2012, 69, 1149–1152. [Google Scholar]
- Kisiel-Nawrot, E.; Latocha, M.; Bak, A.; Kozik, V.; Jampilek, J.; Zieba, A. Anticancer efficacy of antibacterial quinobenzothiazines. Appl. Sci. 2023, 13, 2886. [Google Scholar] [CrossRef]
- Zięba, A.; Maślankiewicz, A.; Suwińska, K. 1-Alkyl-4-(arylamino)quinolinium-3-thiolates and 7-alkyl-12H-quino[3,4-b]-1,4-benzothiazinium salts. Eur. J. Org. Chem. 2000, 16, 2947–2953. [Google Scholar] [CrossRef]
- Empel, A.; Bak, A.; Kozik, V.; Latocha, M.; Cizek, A.; Jampilek, J.; Suwinska, K.; Sochanik, A.; Zieba, A. Towards property profiling: Synthesis and SAR probing of new tetracyclic diazaphenothiazine analogues. Int. J. Mol. Sci. 2021, 22, 12826. [Google Scholar] [CrossRef]
- Hein, J.E.; Fokin, V.V. Copper-catalyzed azide–alkyne cycloaddition (CuAAC) and beyond: New reactivity of copper(I) acetylides. Chem. Soc. Rev. 2010, 39, 1302–1315. [Google Scholar] [CrossRef]
- Khar, R.; Sharma, P.C.; Yar, M.S. Pharmacological significance of triazole scaffold. J. Enzyme Inhib. Med. Chem. 2011, 26, 1–21. [Google Scholar] [CrossRef]
- Thirumurugan, P.; Matosiuk, D.; Jóźwiak, K. Click chemistry for drug development and diverse chemical-biology application. Chem. Rev. 2013, 113, 4905–49792. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Ahmad, S.; Alam, M.S. Bioactive triazoles: A potential review. J. Chem. Pharm. Res. 2012, 4, 5157–5164. [Google Scholar]
- Lau, Y.H.; Rutledge, P.J.; Watkinson, M.; Todd, M.H. Chemical sensors that incorporate click-derived triazoles. Chem. Soc. Rev. 2011, 40, 2848–2866. [Google Scholar] [CrossRef] [PubMed]
- El-Sagheer, A.H.; Brown, T. Click nucleic acid ligation: Applications in biology and nanotechnology. Acc. Chem. Res. 2012, 45, 1258–1267. [Google Scholar] [CrossRef] [PubMed]
- Sangshetti, J.N.; Shinde, D.B. Synthesis of some novel 3-(1-(1-substitutedpiperidin-4-yl)-1H-1,2,3-triazol-4-yl)-5-substituted phenyl-1,2,4-oxadiazoles as antifungal agents. Eur. J. Med. Chem. 2011, 46, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Sindhu, J.; Khurana, J.M.; Sharma, C.; Aneja, K.R. Syntheses, biological evaluation and photophysical studies of novel 1,2,3-triazole linked azo dyes. RSC Adv. 2014, 4, 5915. [Google Scholar] [CrossRef]
- Chandrika, P.M.; Yakaiah, T.; Gayatri, G.; Kumar, K.P.; Narsaiah, B.; Murthy, U.S.N.; Rao, A.R.R. Click chemistry: Studies on the synthesis of novel fluorous tagged triazol-4-yl substituted quinazoline derivatives and their biological evaluation–Theoretical and experimental validation. Eur. J. Med. Chem. 2010, 45, 78–84. [Google Scholar] [CrossRef]
- Buckle, D.R.; Rockell, C.J.; Smith, H.; Spicer, B.A. Studies on 1,2,3-triazoles. Synthesis and antiallergic properties of 9-oxo-1H,9H-benzothiopyrano[2,3-d]-1,2,3-triazoles and their S-oxides. J. Med. Chem 1984, 27, 223–227. [Google Scholar] [CrossRef]
- Montagu, A.; Roy, V.; Balzarini, J.; Snoeck, R.; Andrei, G.; Agrofoglio, L.A. Synthesis of new C5-(1-substituted-1,2,3-triazol-4 or 5-yl)-20-deoxyuridines and their antiviral evaluation. Eur. J. Med. Chem. 2011, 46, 778–786. [Google Scholar] [CrossRef]
- Cheng, H.; Wan, J.; Lin, M.I.; Liu, Y.; Lu, X.; Liu, J.; Xu, Y.; Chen, J.; Tu, Z.; Cheng, Y.S.E.; et al. Design, synthesis, and in vitro biological evaluation of 1H-1,2,3-triazole-4-carboxamide derivatives as new anti-influenza agents targeting virus nucleoprotein. J. Med. Chem. 2012, 55, 2144–2153. [Google Scholar] [CrossRef] [PubMed]
- Boechat, N.; Ferreira, V.F.; Ferreira, S.B.; Ferreira, M.L.G.; Silva, F.C.; Bastos, M.M.; Costa, M.S.; Lourenco, M.C.S.; Pinto, A.C.; Krettli, A.U.; et al. Novel 1,2,3-triazole derivatives for use against Mycobacterium tuberculosis H37Rv (ATCC 27294) Strain. J. Med. Chem. 2011, 54, 5988–5999. [Google Scholar] [CrossRef]
- Pulipati, L.; Yogeeswari, P.; Sriram, D.; Kantevari, S. Click-based synthesis and antitubercular evaluation of novel dibenzo[b,d]thiophene-1,2,3-triazoles with piperidine, piperazine, morpholine and thiomorpholine appendages. Bioorg. Med. Chem. Lett. 2016, 26, 2649–2654. [Google Scholar] [CrossRef] [PubMed]
- Angajala, K.K.; Vianala, S.; Macha, R.; Raghavender, M.; Thupuran, M.K.; Pathi, P.J. Synthesis, anti-inflammatory, bactericidal activities and docking studies of novel 1,2,3-triazoles derived from ibuprofen using click chemistry. Springer Plus. 2016, 5, 423. [Google Scholar] [CrossRef]
- Mutschler, E.; Geisslinger, G.; Kroemer, H.K.; Ruth, P.; Schafer-Korting, M. Farmakologia i Toksykologia, 3rd ed.; MedPharma Polska: Wrocław, Poland, 2013; pp. 143–385. [Google Scholar]
- Addla, D.; Jallapally, A.; Divya Gurram, D.; Yogeeswari, P.; Sriram, D.; Kantevari, S. Rational design, synthesis and antitubercular evaluation of novel 2-(trifluoromethyl)phenothiazine-[1,2,3]triazole hybrids. Bioorg. Med. Chem. Lett. 2014, 24, 233–236. [Google Scholar] [CrossRef]
- Reddyrajula, R.; Dalimba, U.; Kumar, S.M. Molecular hybridization approach for phenothiazine incorporated 1,2,3-triazole hybrids as promising antimicrobial agents: Design, synthesis, molecular docking and in silico ADME studies. Eur. J. Med. Chem. 2019, 168, 263–282. [Google Scholar] [CrossRef] [PubMed]
- Nycz-Emper, A.; Bober, K.; Wyszomirski, M.; Kisiel, E.; Zięba, A. The application of CA and PCA to the evaluation of lipophilicity and physicochemical properties of tetracyclic diazaphenothiazine derivatives. J. Anal. Methods Chem. 2019, 20, 8131235. [Google Scholar]
- Zieba, A.; Wojtyczka, R.D.; Idzik, D.; Kepa, M. Synthesis and in vitro antimicrobial activity of 1-methyl-3-sulfonylthio-4-aminoquinolinium chlorides. Acta Pol. Pharm. 2013, 70, 163–166. [Google Scholar] [PubMed]
- Zieba, A.; Sochanik, A.; Szurko, A.; Rams, M.; Mrozek, A.; Cmoch, P. Synthesis and in vitro antiproliferative activity of 5-alkyl-12(H)-quino[3,4-b][1,4]benzothiazinium salts. Eur. J. Med. Chem. 2010, 45, 4733–4739. [Google Scholar] [CrossRef]
- Pivetta, T.P.; Vieira, T.; Silva, J.C.; Ribeiro, P.A.; Raposo, M. Phototoxic potential of different DNA intercalators for skin cancer therapy: In vitro screening. Int. J. Mol. Sci. 2023, 24, 5602. [Google Scholar] [CrossRef]
- Zięba, A.; Latocha, M.; Sochanik, A.; Nycz, A.; Kuśmierz, D. Synthesis and in vitro antiproliferative activity of novel phenyl ring-substituted5-alkyl-12(H)-quino[3,4-b][1,4]benzothiazine derivatives. Molecules 2016, 21, 1455. [Google Scholar] [CrossRef]
- Kisiel-Nawrot, E.; Pindjakova, D.; Latocha, M.; Bąk, A.; Kozik, V.; Suwińska, K.; Sochanik, A.; Cizek, A.; Jampilek, J.; Zięba, A. Design, synthesis and antimicrobial properties of new tetracyclic quinobenzothiazine derivatives. Int. J. Mol. Sci. 2022, 23, 15078. [Google Scholar] [CrossRef]
- Siles, R.; Kawasaki, Y.; Ross, P.; Freire, E. Synthesis and biochemical evaluation of triazole/tetrazole-containing sulfonamides against thrombin and related serine proteases. Bioorg. Med. Chem. Lett. 2011, 21, 5305–5309. [Google Scholar] [CrossRef] [PubMed]
- Klaveren, S.; Dernovsek, J.; Jakopin, Z.; Anderluh, M.; Leffler, H.; Nilsson, U.J.; Tomasic, T. Design and synthesis of novel 3-triazolyl-1-thiogalactosides as galectin-1,-3 and -8 inhibitors. RSC Adv. 2022, 12, 18973. [Google Scholar] [CrossRef] [PubMed]
- Dmitrenko, O.; Chaplin, A.; Balbutskaya, A.; Pkhakadze, T.; Alkhovsky, S. In silico genome-scale analysis of molecular mechanisms contributing to the development of a persistent infection with methicillin-resistant Staphylococcus aureus (MRSA) ST239. Int. J. Mol. Sci. 2022, 23, 16086. [Google Scholar] [CrossRef]
- Russo, A.; Picciarella, A.; Russo, R.; d’Ettorre, G.; Ceccarelli, G. Time to effective therapy is an important determinant of survival in bloodstream infections caused by Vancomycin-resistant Enterococcus spp. Int. J. Mol. Sci. 2022, 23, 11925. [Google Scholar] [CrossRef] [PubMed]
- Luukinen, H.; Hammaren, M.M.; Vanha-Aho, L.M.; Parikka, M. Modeling tuberculosis in Mycobacterium marinum infected adult Zebrafish. J. Vis. Exp. 2018, 140, 58299. [Google Scholar]
- Pankey, G.A.; Sabath, L.D. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin. Infect. Dis. 2004, 38, 864–870. [Google Scholar] [CrossRef]
- Portela, C.A.; Smart, K.F.; Tumanov, S.; Cook, G.M.; Villas-Boas, S.G. Global metabolic response of Enterococcus faecalis to oxygen. J. Bacteriol. 2014, 196, 2012–2022. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.d.L.E.; Igrejas, G.; Poeta, P. Enterococci, from harmless bacteria to a pathogen. Microorganisms 2020, 8, 1118. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Salamzade, R.; Selleck, E.; Bryan, N.; Mello, S.S.; Manson, A.L.; Earl, A.M. Genes contributing to the unique biology and intrinsic antibiotic resistance of Enterococcus faecalis. mBio 2020, 11, e02962-20. [Google Scholar] [CrossRef]
- Sundarsingh, J.A.T.; Ranjitha, J.; Rajan, A.; Shankar, V. Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. J. Inf. Public. Health 2020, 13, 1255–1264. [Google Scholar]
- Bedaquilin. DrugBank. Available online: https://go.drugbank.com/drugs/DB08903 (accessed on 6 July 2023).
- Measuring Cell Viability/Cytotoxicity. Dojindo EU GmbH, Munich, Germany. Available online: https://www.dojindo.eu.com/Protocol/Dojindo-Cell-Proliferation-Protocol.pdf (accessed on 6 July 2023).
- Bueno, J. Antitubercular in vitro drug discovery: Tools for begin the search. In Understanding Tuberculosis—New Approaches to Fighting Against Drug Resistance; IntechOpen: Rijeka, Croatia, 2012; pp. 147–168. [Google Scholar]
- Chavan, S.; Nicholls, I.A.; Karlsson, B.C.G.; Rosengren, A.M.; Ballabio, D.; Consonni, V.; Todeschini, R. Towards global QSAR model building for acute toxicity: Munro database case study. Int. J. Mol. Sci. 2014, 15, 18162–18174. [Google Scholar] [CrossRef]
- Silva, L.B.; Ferreira, E.F.B.; Maryam; Espejo-Román, J.M.; Costa, G.V.; Cruz, J.V.; Kimani, N.M.; Costa, J.S.; Bittencourt, J.A.H.M.; Cruz, J.N.; et al. Galantamine based novel acetylcholinesterase enzyme inhibitors: A molecular modeling design approach. Molecules 2023, 28, 1035. [Google Scholar] [CrossRef]
- Andersen, C.M.; Bro, R. Variable selection in regression—A tutorial. J. Chemom. 2010, 24, 728–737. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Cryst. 2015, C71, 3–8. [Google Scholar]
- Nubel, U.; Dordel, J.; Kurt, K.; Strommenger, B.; Westh, H.; Shukla, S.K.; Zemlickova, H.; Leblois, R.; Wirth, T.; Jombart, T.; et al. A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog. 2010, 6, e1000855. [Google Scholar] [CrossRef]
- Gonec, T.; Zadrazilova, I.; Nevin, E.; Kauerova, T.; Pesko, M.; Kos, J.; Oravec, M.; Kollar, P.; Coffey, A.; O’Mahony, J.; et al. Synthesis and Biological Evaluation of N-Alkoxyphenyl-3-hydroxynaphthalene-2-carboxanilides. Molecules 2015, 20, 9767–9787. [Google Scholar] [CrossRef] [PubMed]
- National Committee for Clinical Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; M07; NCCLS: Wayne, PA, USA, 2018. [Google Scholar]
- Schwalbe, R.; Steele-Moore, L.; Goodwin, A.C. Antimicrobial Susceptibility Testing Protocols; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Scandorieiro, S.; de Camargo, L.C.; Lancheros, C.A.; Yamada-Ogatta, S.F.; Nakamura, C.V.; de Oliveira, A.G.; Andrade, C.G.; Duran, N.; Nakazato, G.; Kobayashi, R.K. Synergistic and additive effect of oregano essential oil and biological silver nanoparticles against multidrug-resistant bacterial strains. Front. Microbiol. 2016, 7, 760. [Google Scholar] [CrossRef]
- Guimaraes, A.C.; Meireles, L.M.; Lemos, M.F.; Guimaraes, M.C.C.; Endringer, D.C.; Fronza, M.; Scherer, R. Antibacterial activity of terpenes and terpenoids present in essential oils. Molecules 2019, 24, 2471. [Google Scholar] [CrossRef] [PubMed]
- Abate, G.; Mshana, R.N.; Miorner, H. Evaluation of a colorimetric assay based on 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) for rapid detection of rifampicin resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 1998, 2, 1011–1016. [Google Scholar]
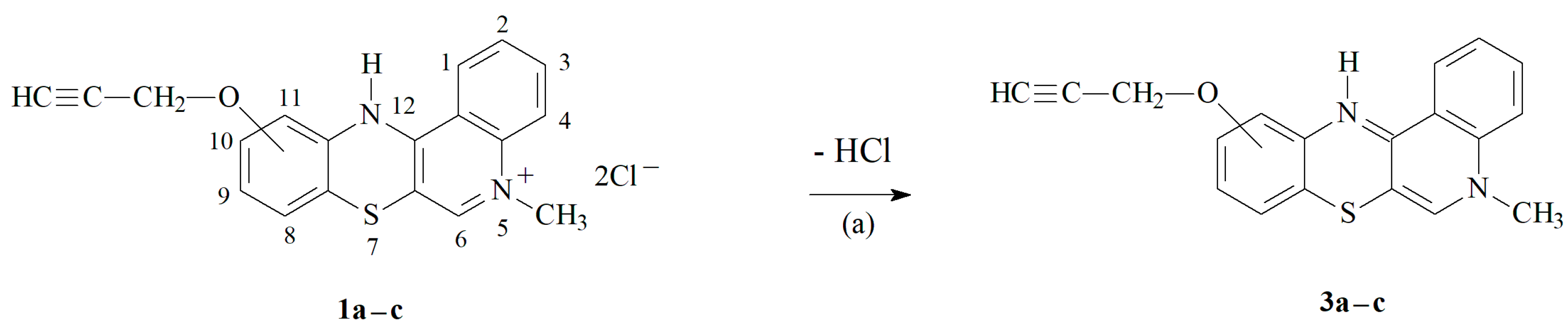



| IC50 [µM] | ||||
|---|---|---|---|---|
| No. | SNB-19 | A549 | T47D | NHDF |
| 5a | >100 | >100 | >100 | >100 |
| 5f | >100 | >100 | >100 | >100 |
| 5h | >100 | >100 | >100 | >100 |
| 5i | >100 | >100 | >100 | >100 |
| CIS | 16.7 ± 1.3 | 3.0 ± 0.2 | 9.0 ± 0.7 | 29.9 ± 3.3 |
| DOX | 1.7 ± 0.2 | 1.3 ± 0.1 | 0.8 ± 0.1 | 5.5 ± 0.5 |
| IC50 [µM] | ||||
|---|---|---|---|---|
| No. | SNB-19 | A549 | T47D | NHDF |
| 2a | 0.23 ± 0.06 | 0.45 ± 0.08 | 2.1 ± 0.5 | 47.8 ± 3.5 |
| 2b | 2.7 ± 0.4 | 1.8 ± 0.2 | 5.3 ± 0.6 | >100 |
| 2c | 4.1 ± 0.5 | 5.7 ± 0.9 | 7.6 ± 0.4 | >100 |
| 2d | 2.5 ± 0.1 | 6.3 ± 0.4 | 8.6 ± 0.7 | >100 |
| 2e | 3.4 ± 0.3 | 8.4 ± 0.6 | 12.5 ± 0.8 | 56.6 ± 4.2 |
| 2f | 8.5 ± 0.5 | 11.5 ± 0.6 | 20.5 ± 1.4 | 30.5 ± 2.1 |
| 2g | 8.4 ± 0.7 | 4.5 ± 0.3 | 11.8 ± 0.5 | 27.6 ± 0.6 |
| 2h | 9.3 ± 0.5 | 14.6 ± 0.6 | 6.3 ± 0.3 | >100 |
| 2i | 4.9 ± 0.3 | 9.0 ± 0.4 | 2.8 ± 0.2 | >100 |
| 2j | 8.8 ± 0.4 | 10.5 ± 0.7 | 19.3 ± 1.2 | 28.7 ± 1.2 |
| 2k | 6.1 ± 0.3 | 16.4 ± 0.9 | 10.6 ± 0.9 | >100 |
| 2l | 9.6 ± 0.8 | 14.1 ± 0.6 | 4.3 ± 0.2 | >100 |
| 2m | 23.4 ± 1.5 | 16.8 ± 1.2 | 8.9 ± 0.9 | >100 |
| 2n | 42.9 ± 1.8 | 14.3 ± 0.8 | 15.4 ± 1.1 | >100 |
| CIS | 16.7 ± 1.3 | 3.0 ± 0.2 | 9.0 ± 0.7 | 29.9 ± 3.3 |
| DOX | 1.7 ± 0.2 | 1.3 ± 0.1 | 0.8 ± 0.1 | 5.5 ± 0.5 |
| No. | MIC [μM]MBC [μM] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SA | MRSA1 | MRSA2 | MRSA3 | EF | VRE1 | VRE2 | VRE3 | MS | MM | |
| 2a | 131 | 262 | 262 | 131 | 262 | 524 | 262 | 262 | 32.7 | 65.5 |
| 131 | NT | 262 | 131 | NT | NT | NT | NT | 32.7 | 65.5 | |
| 2b | 524 | >524 | >524 | 262 | >524 | >524 | >524 | 524 | 65.5 | 65.5 |
| NT | NT | NT | 262 | NT | NT | NT | NT | 65.5 | 65.5 | |
| 2c | 524 | 524 | 524 | 524 | >524 | >524 | >524 | 524 | 65.5 | 131 |
| NT | NT | NT | NT | NT | NT | NT | NT | 65.5 | 131 | |
| 2d | 131 | 131 | 131 | 65.5 | >524 | >524 | >524 | >524 | 32.7 | 32.7 |
| 65.5 | 131 | 131 | 65.5 | NT | NT | NT | NT | 32.7 | 32.7 | |
| 2e | 32.8 | 65.5 | 65.5 | 16.3 | 524 | 524 | 524 | 524 | 32.7 | 32.7 |
| 65.5 | 65.5 | 65.5 | 16.3 | NT | NT | NT | NT | 32.7 | 32.7 | |
| 2f | >524 | >524 | >524 | 524 | >524 | >524 | >524 | >524 | 65.5 | 65.5 |
| NT | NT | NT | NT | NT | NT | NT | NT | 65.5 | 65.5 | |
| 2g | 262 | 262 | 262 | 65.5 | 262 | 524 | 262 | 131 | 32.7 | 65.5 |
| 262 | 262 | NT | 65.5 | NT | NT | NT | NT | 65.5 | 131 | |
| 2h | 131 | 262 | 262 | 131 | >524 | >524 | >524 | >524 | 16.3 | 32.7 |
| 131 | 262 | 262 | 131 | NT | NT | NT | NT | 16.3 | 32.7 | |
| 2i | 524 | 524 | 524 | 524 | >524 | >524 | >524 | 524 | 65.5 | 131 |
| NT | NT | NT | NT | NT | NT | NT | NT | 65.5 | 131 | |
| 2j | 65.5 | 65.5 | 65.5 | 32.7 | 524 | 524 | 524 | 524 | 16.3 | 16.3 |
| 65.5 | 65.5 | 65.5 | 32.7 | NT | NT | NT | NT | 16.3 | 16.3 | |
| 2k | 65.5 | 65.5 | 65.5 | 16.3 | 524 | >524 | 524 | 524 | 32.7 | 32.7 |
| 65.5 | 65.5 | 65.5 | 16.3 | NT | NT | NT | NT | 32.7 | 32.7 | |
| 2l | 65.5 | 65.5 | 65.5 | 32.7 | 524 | 524 | 524 | 524 | 16.3 | 16.3 |
| 65.5 | 131 | 65.5 | 32.7 | NT | NT | NT | NT | 16.3 | 16.3 | |
| 2m | 65.5 | 131 | 131 | 32.7 | 65.5 | >524 | 524 | 524 | 8.19 | 16.3 |
| 65.5 | 131 | 131 | 32.7 | NT | NT | 524 | NT | 8.19 | 16.3 | |
| 2n | >524 | >524 | >524 | 262 | >524 | >524 | >524 | >524 | 16.3 | 32.7 |
| NT | NT | NT | 262 | NT | NT | NT | NT | 16.3 | 32.7 | |
| AMP | 5.72 | >45.8 | >45.8 | 45.8 | 2.81 | 11.5 | 11.5 | 11.5 | – | – |
| 5.72 | >45.8 | >45.8 | 45.8 | 2.81 | 11.5 | 11.5 | 11.5 | |||
| CPX | 3.02 | 24.1 | 192 | 24.1 | 1.51 | 1.51 | 3.02 | 193 | 0.38 | 0.38 |
| 3.02 | 24.1 | 386 | 24.1 | 3.02 | 3.02 | 3.02 | 386 | 0.38 | 0.38 | |
| RIF | – | – | – | – | – | – | – | – | 19.4 | 2.43 |
| – | – | |||||||||
| No. | Conc. | M. smegmatis Respiration Inhibition [%] |
|---|---|---|
| 2h | 1× MIC (1× MBC) | 81.4 |
| 2j | 2× MIC (2× MBC) | 87.6 |
| 2l | 2× MIC (2× MBC) | 87.3 |
| 2m | 1× MIC (1× MBC) | 87.1 |
| 2n | 1× MIC (1× MBC) | 85.9 |
| CPX | 32× MIC (32× MBC) | 85.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kisiel-Nawrot, E.; Pindjakova, D.; Latocha, M.; Bak, A.; Kozik, V.; Suwinska, K.; Cizek, A.; Jampilek, J.; Zięba, A. Towards Anticancer and Antibacterial Agents: Design and Synthesis of 1,2,3-Triazol-quinobenzothiazine Derivatives. Int. J. Mol. Sci. 2023, 24, 13250. https://doi.org/10.3390/ijms241713250
Kisiel-Nawrot E, Pindjakova D, Latocha M, Bak A, Kozik V, Suwinska K, Cizek A, Jampilek J, Zięba A. Towards Anticancer and Antibacterial Agents: Design and Synthesis of 1,2,3-Triazol-quinobenzothiazine Derivatives. International Journal of Molecular Sciences. 2023; 24(17):13250. https://doi.org/10.3390/ijms241713250
Chicago/Turabian StyleKisiel-Nawrot, Ewa, Dominika Pindjakova, Malgorzata Latocha, Andrzej Bak, Violetta Kozik, Kinga Suwinska, Alois Cizek, Josef Jampilek, and Andrzej Zięba. 2023. "Towards Anticancer and Antibacterial Agents: Design and Synthesis of 1,2,3-Triazol-quinobenzothiazine Derivatives" International Journal of Molecular Sciences 24, no. 17: 13250. https://doi.org/10.3390/ijms241713250
APA StyleKisiel-Nawrot, E., Pindjakova, D., Latocha, M., Bak, A., Kozik, V., Suwinska, K., Cizek, A., Jampilek, J., & Zięba, A. (2023). Towards Anticancer and Antibacterial Agents: Design and Synthesis of 1,2,3-Triazol-quinobenzothiazine Derivatives. International Journal of Molecular Sciences, 24(17), 13250. https://doi.org/10.3390/ijms241713250


_Putnam.png)




