Genome-Wide Identification and Expression Analysis of BrGeBP Genes Reveal Their Potential Roles in Cold and Drought Stress Tolerance in Brassica rapa
Abstract
1. Introduction
2. Results
2.1. Identification, Physicochemical Characterization of GeBP Family Genes
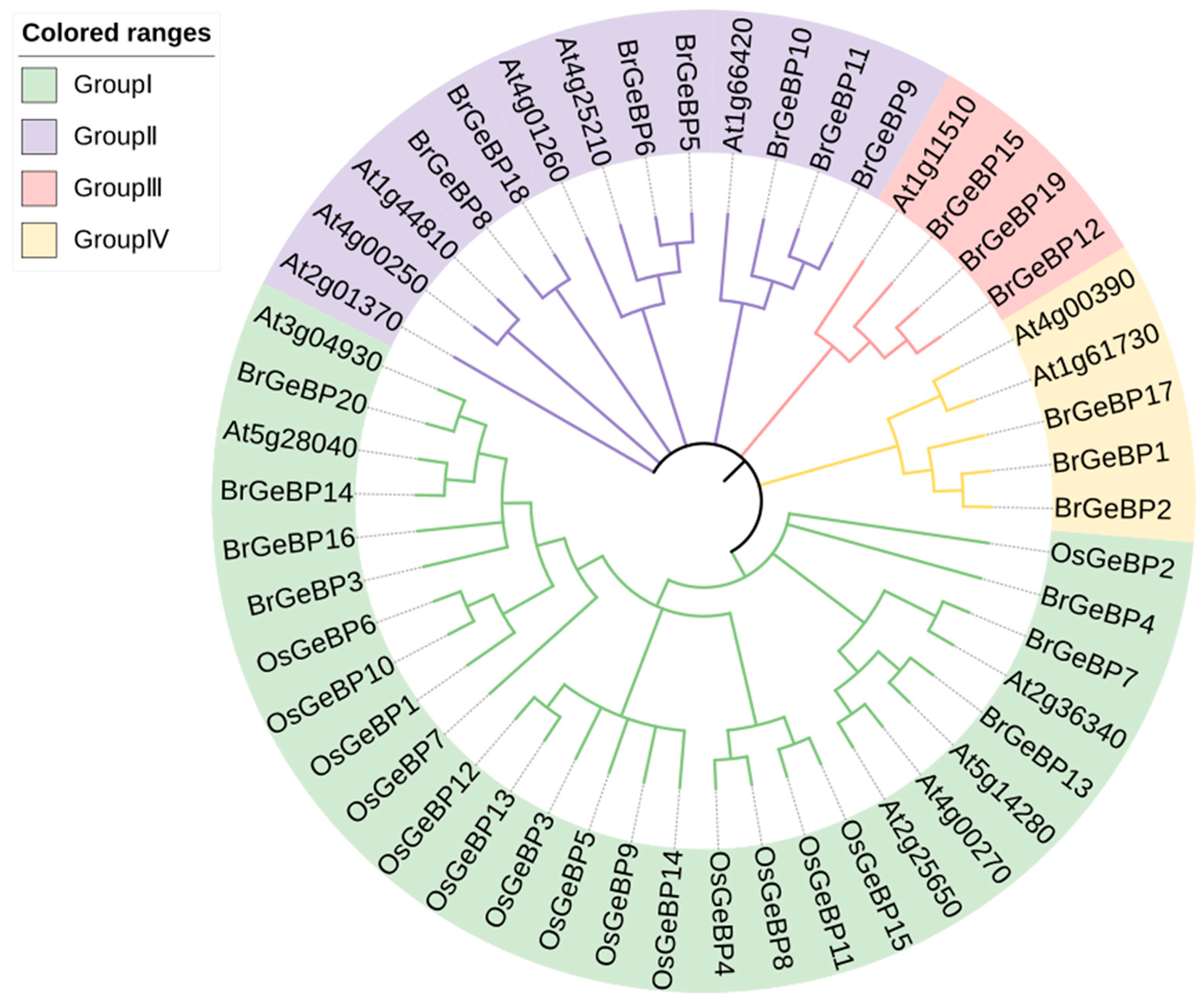
2.2. Phylogenetic Relationships
2.3. Gene Structure and Conserved Motif Analysis
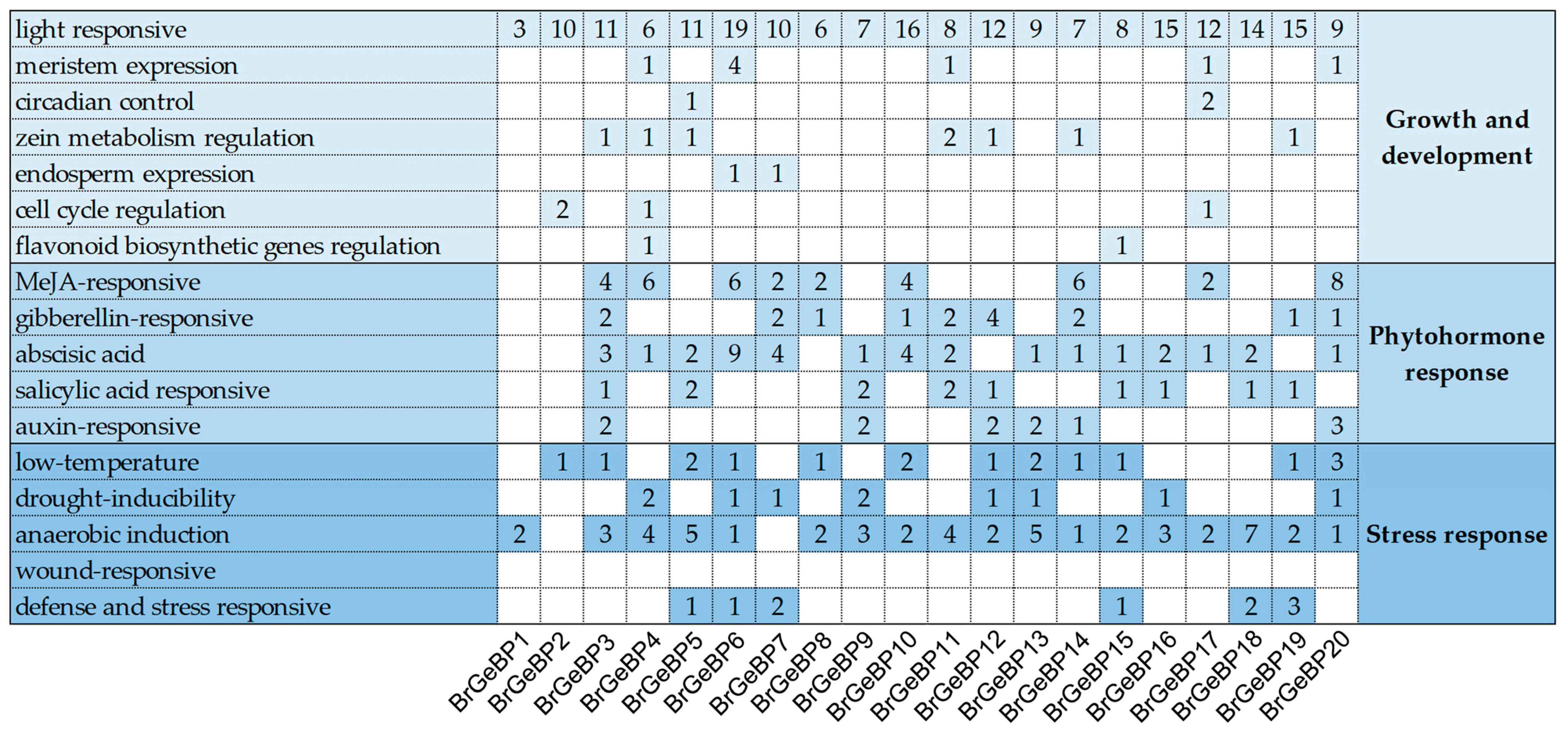
2.4. Analysis of Promoter Cis-Elements of BrGeBPs
2.5. Gene Expression Analysis of the BrGeBPs
2.5.1. Analysis of Tissue-Specific Expression of BrGeBPs
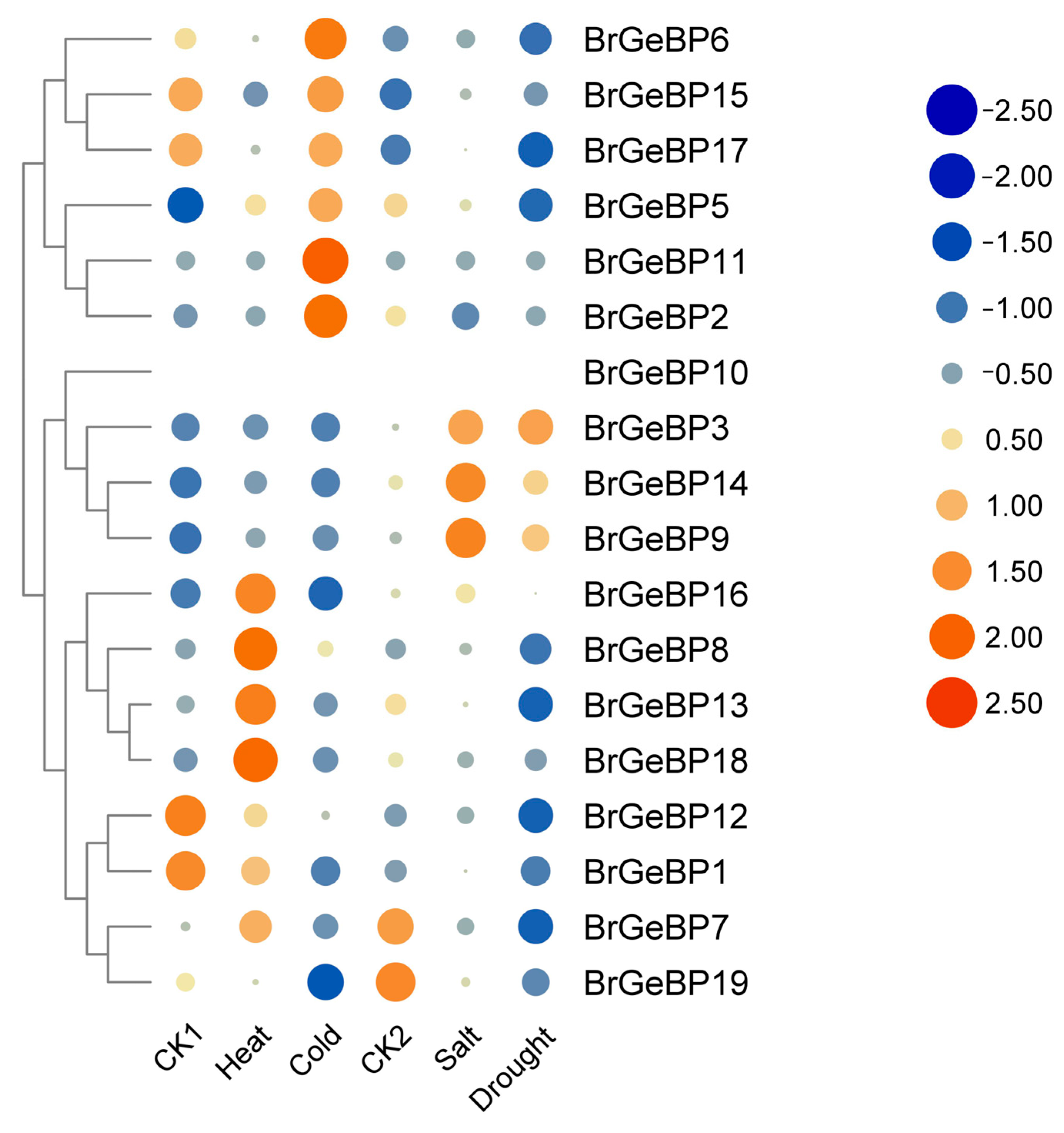
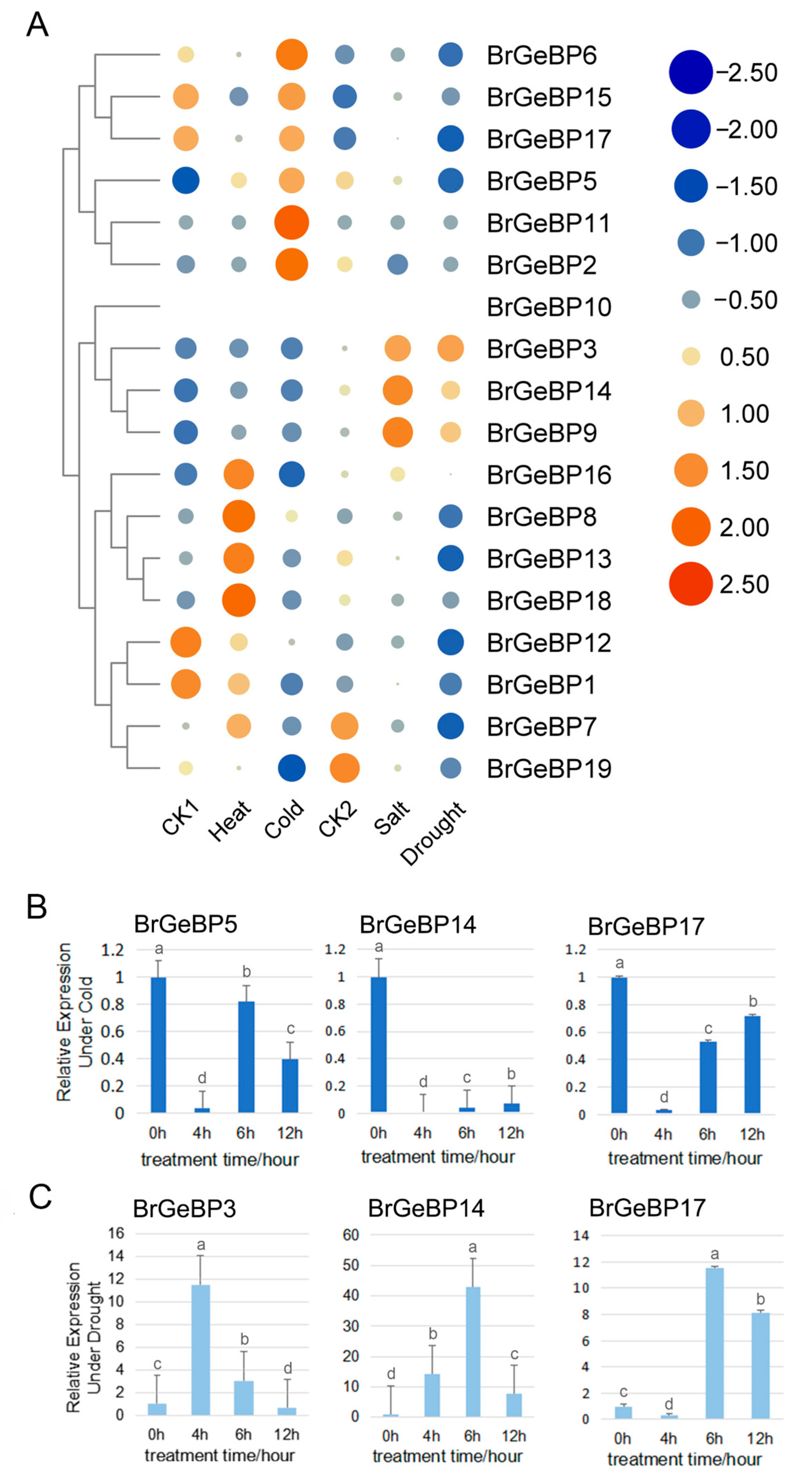
2.5.2. Expression Patterns in Response to Abiotic Stress Analysis
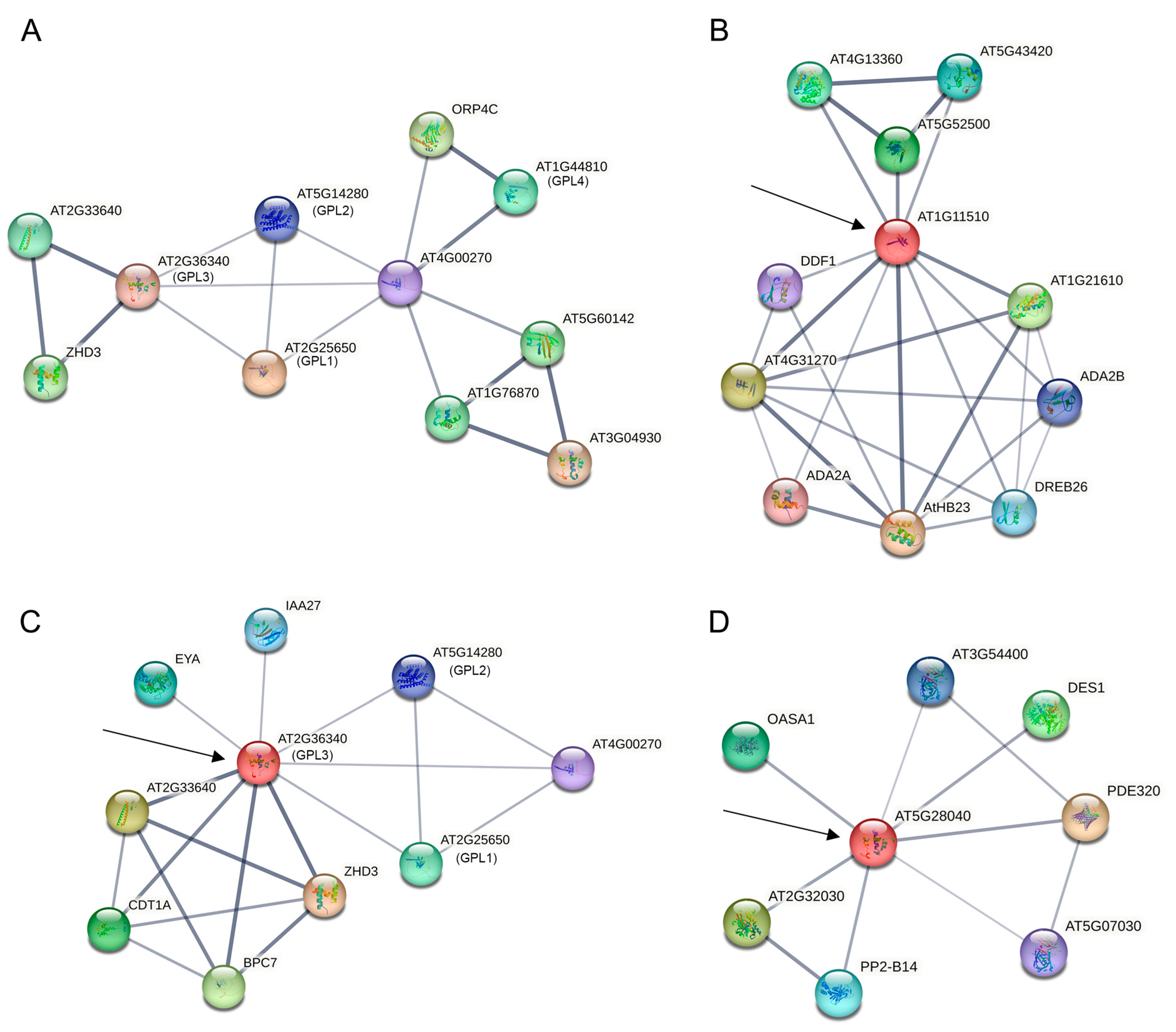
2.6. Prediction of Protein–Protein Interaction
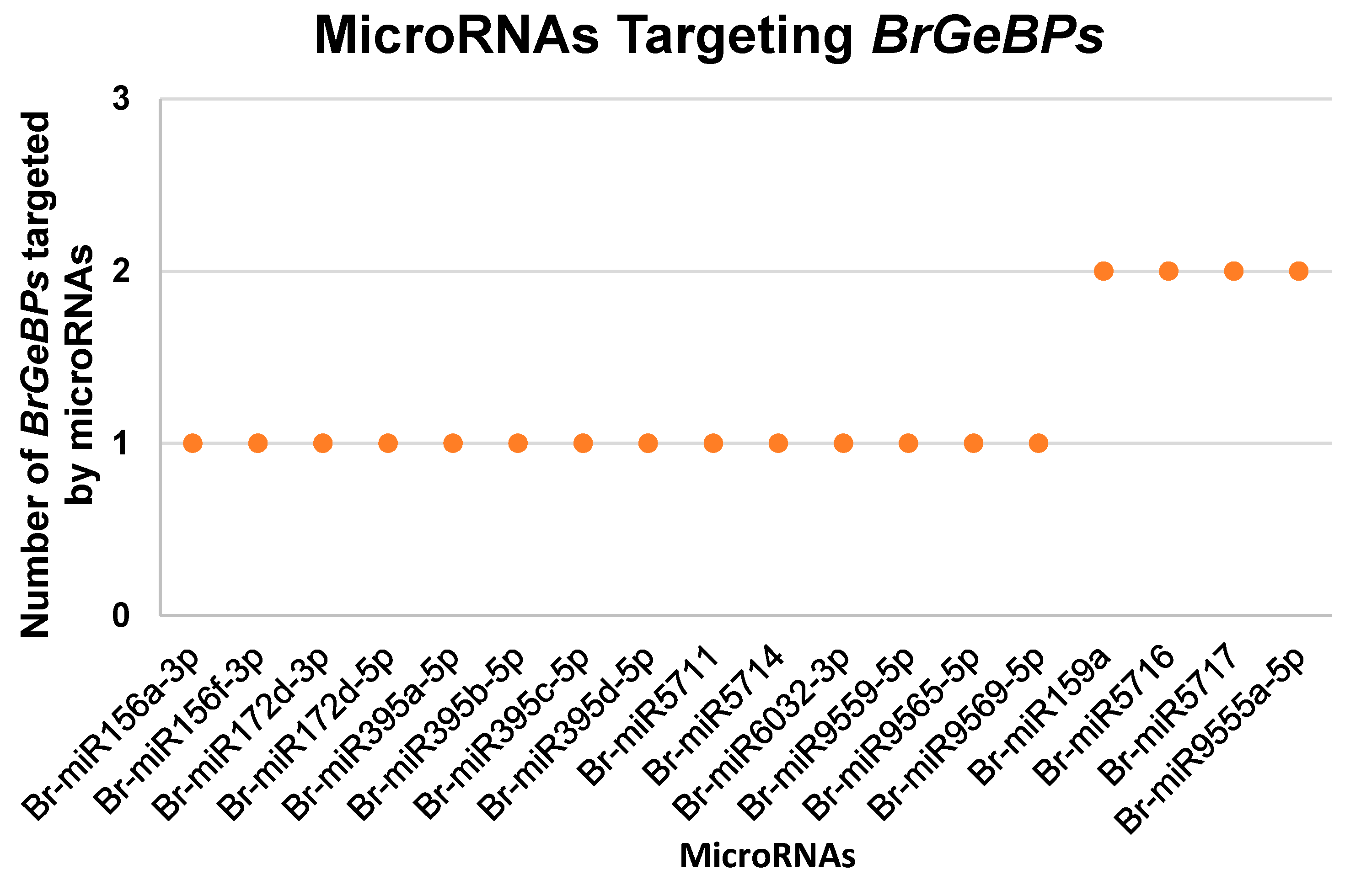
2.7. Prediction of microRNAs Targeting BrGeBPs
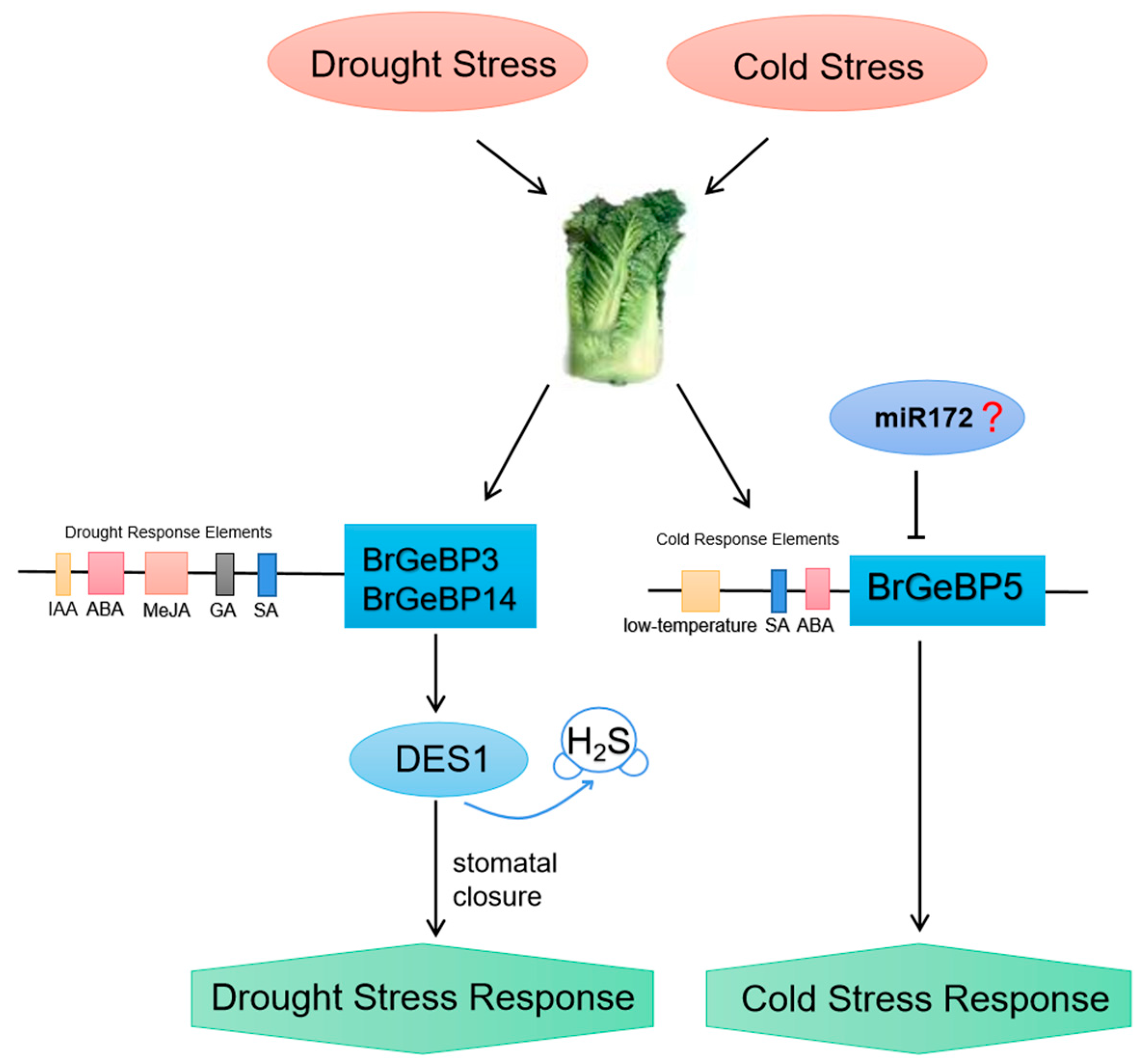
3. Discussion
4. Materials and Methods
4.1. Identification and Physicochemical Characterization of GeBP Family Genes
4.2. Phylogenetic Relationships and Synteny Analysis
4.3. Gene Structure and Analysis of Conserved Motif and Cis-Elements
4.4. Gene Expression Analysis
4.5. Plant Material and Stress Treatments
4.6. Total RNA Extraction and qRT-PCR
4.7. Statistical Analysis
4.8. Prediction of Protein–Protein Interaction
4.9. Prediction of microRNAs Targeting BrGeBPs Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Curaba, J.; Herzog, M.; Vachon, G. GeBP, the first member of a new gene family in Arabidopsis, encodes a nuclear protein with DNA-binding activity and is regulated by KNAT1. Plant J. 2003, 33, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, J.; Song, H.; Zhang, Z.; Chen, X.; Wang, A. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Sun, R.; Yang, N.; Liu, Z.; Xia, Y.; Sun, Y.; Gao, Q.; Pu, J.; Dang, Z.; Zhang, H. Identification and expression analysis of mango GeBP transcription factor family genes. Non-Wood For. Res. 2021, 39, 137–147. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Liu, C.; Zhang, F.; Wei, J.; Li, B. Genome-Wide Characterization and Expression Analysis of GeBP Family Genes in Soybean. Plants 2022, 11, 1848. [Google Scholar] [CrossRef]
- Shan, X.; Yang, K.; Shi, J.; Zhu, C.; Gao, Z. Genome-wide identification and expression analysis of GeBP transcription factorgene family in moso bamboo. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2020, 44, 41–48. [Google Scholar] [CrossRef]
- Chevalier, F.; Perazza, D.; Laporte, F.; Henanff, G.L.; Hornitschek, P.; Bonneville, J.M.; Herzog, M.; Vachon, G. GeBP and GeBP-like proteins are noncanonical leucine-zipper transcription factors that regulate cytokinin response in Arabidopsis. Plant Physiol. 2008, 146, 1142–1154. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef]
- Ray, S.; Dansana, P.K.; Giri, J.; Deveshwar, P.; Arora, R.; Agarwal, P.; Khurana, J.P.; Kapoor, S.; Tyagi, A.K. Modulation of transcription factor and metabolic pathway genes in response to water-deficit stress in rice. Funct. Integr. Genom. 2011, 11, 157–178. [Google Scholar] [CrossRef]
- Liu, R.; Li, H.; Qiao, Z.; Liu, H.; Zhao, L.; Wang, X.; Zhang, Z.; Zhang, S.; Song, L.; You, C. Genome-wide analysis of MdGeBP family and functional identification of MdGeBP3 in Malus domestica. Environ. Exp. Botany 2023, 208, 105262. [Google Scholar] [CrossRef]
- Khare, D.; Mitsuda, N.; Lee, S.; Song, W.; Hwang, D.; Ohme-Takagi, M.; Martinoia, E.; Lee, Y.; Hwang, J. Root avoidance of toxic metals requires the GeBP-LIKE 4 transcription factor in Arabidopsis thaliana. New Phytol. 2017, 213, 1257–1273. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, Q.; He, Y.; Liu, W.; Xu, Y.; Liu, K.; Xian, F.; Li, J.; Hu, J. Genome-wide identification, expansion mechanism and expression profiling analysis of GLABROUS1 enhancer-binding protein (GeBP) gene family in Gramineae crops. Int. J. Mol. Sci. 2021, 22, 8758. [Google Scholar] [CrossRef]
- Perazza, D.; Laporte, F.; Balagué, C.; Chevalier, F.; Remo, S.; Bourge, M.; Larkin, J.; Herzog, M.; Vachon, G. GeBP/GPL Transcription Factors Regulate a Subset of CPR5-Dependent Processes. Plant Physiol. 2011, 157, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Kamei, A.; Seki, M.; Umezawa, T.; Ishida, J.; Satou, M.; Akiyama, K.; Zhu, J.; Shinozaki, K. Analysis of gene expression profiles in Arabidopsis salt overly sensitive mutants sos2-1 and sos3-1. Plant Cell Environ. 2005, 28, 1267–1275. [Google Scholar] [CrossRef]
- Bassa, C.; Mila, I.; Bouzayen, M.; Audran-Delalande, C. Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol. 2012, 53, 1583–1595. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, J.; Shen, J.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Fu, L.; Li, Z.; Yang, J.; et al. Hydrogen sulfide-linked persulfidation of ABI4 controls ABA responses through the transactivation of MAPKKK18 in Arabidopsis. Mol. Plant 2021, 14, 921–936. [Google Scholar] [CrossRef] [PubMed]
- García-Cano, E.; Hak, H.; Magori, S.; Lazarowitz, S.G.; Citovsky, V. The Agrobacterium F-Box Protein Effector VirF Destabilizes the Arabidopsis GLABROUS1 Enhancer/Binding Protein-Like Transcription Factor VFP4, a Transcriptional Activator of Defense Response Genes. Mol. Plant Microbe Interact. 2018, 31, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Chen, Q.; Wang, S.; Lers, A. Downregulation of GeBP-like α factor by MiR827 suggests their involvement in senescence and phosphate homeostasis. BMC Biol. 2021, 19, 90. [Google Scholar] [CrossRef]
- Cui, C.; Wang, J.J.; Zhao, J.H.; Fang, Y.Y.; He, X.F.; Guo, H.S.; Duan, C.G. A Brassica miRNA Regulates Plant Growth and Immunity through Distinct Modes of Action. Mol. Plant 2020, 13, 231–245. [Google Scholar] [CrossRef]
- Wang, W.; Liu, D.; Zhang, X.; Chen, D.; Cheng, Y.; Shen, F. Plant MicroRNAs in Cross-Kingdom Regulation of Gene Expression. Int. J. Mol. Sci. 2018, 19, 2007. [Google Scholar] [CrossRef]
- Liang, G.; Yang, F.; Yu, D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010, 62, 1046–1057. [Google Scholar] [CrossRef]
- Keshabarz, H.; Sanavy, S.A.M.; Moghadan, R.S.G. Impact of Foliar Application with Salicylic Acid on Biochemical Characters of Canola Plants under Cold Stress Condition. Not. Sci. Biologicae. 2016, 8, 98–105. [Google Scholar] [CrossRef]
- Cheng, F.; Lu, J.; Gao, M.; Shi, K.; Kong, Q.; Huang, Y.; Bie, Z. Redox Signaling and CBF-Responsive Pathway Are Involved in Salicylic Acid-Improved Photosynthesis and Growth under Chilling Stress in Watermelon. Front. Plant Sci. 2016, 7, 1519. [Google Scholar] [CrossRef]
- Ahmad, F.; Singh, A.; Kamal, A. Salicylic acid–mediated defense mechanisms to abiotic stress tolerance. In Plant Signaling Molecules; Woodhead Publishing: Sawston, UK, 2019; pp. 355–369. [Google Scholar] [CrossRef]
- Tottempudi, K.P. Mechanisms of chilling-induced oxidative stress injury and tolerance in developing maize seedlings: Changes in antioxidant system, oxidation of proteins and lipids, and protease activities. Plant J. 1996, 10, 1017–1026. [Google Scholar] [CrossRef]
- Guo, W.; Chen, R.; Gong, Z.; Yin, Y.; Ahmed, S.; He, Y. Exogenous abscisic acid increases antioxidant enzymes and related gene expression in pepper (Capsicum annuum) leaves subjected to chilling stress. Genet. Mol. Res. 2012, 11, 4063–4080. [Google Scholar] [CrossRef] [PubMed]
- Ó’Maoiléidigh, D.S.; van Driel, A.D.; Singh, A.; Sang, Q.; Le Bec, N.; Vincent, C.; Vayssieres, A.; Branchat, M.R.; Severing, E.; Gallegos, R.M.; et al. Systematic analyses of the MIR172 family members of Arabidopsis define their distinct roles in regulation of APETALA2 during floral transition. PLoS Biol. 2021, 19, e3001043. [Google Scholar] [CrossRef] [PubMed]
- Shohat, H.; Cheriker, H.; Kilambi, H.V.; Illouz Eliaz, N.; Blum, S.; Amsellem, Z.; Tarkowská, D.; Aharoni, A.; Eshed, Y.; Weiss, D. Inhibition of gibberellin accumulation by water deficiency promotes fast and long-term ‘drought avoidance’ responses in tomato. New Phytol. 2021, 232, 1985–1998. [Google Scholar] [CrossRef]
- Yoshida, T.; Christmann, A.; Yamaguchi-Shinozaki, K.; Grill, E.; Fernie, A.R. Revisiting the Basal Role of ABA—Roles Outside of Stress. Trends Plant Sci. 2019, 24, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Hassan, M.; Li, Z.; Peng, Y. Indole-3-acetic acid improves drought tolerance of white clover via activating auxin, abscisic acid and jasmonic acid related genes and inhibiting senescence genes. BMC Plant Biol. 2020, 20, 150. [Google Scholar] [CrossRef]
- Chen, S.; Jia, H.; Wang, X.; Shi, C.; Wang, X.; Ma, P.; Wang, J.; Ren, M.; Li, J. Hydrogen Sulfide Positively Regulates Abscisic Acid Signaling through Persulfidation of SnRK2.6 in Guard Cells. Mol. Plant 2020, 13, 732–744. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Liu, J.; Liu, T.; Xue, S. Hydrogen sulfide (H2S) signaling in plant development and stress responses. aBIOTECH 2021, 2, 32–63. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Appel, R.D.; Bairoch, A.; Hochstrasser, D.F. A new generation of information retrieval tools for biologists: The example of the ExPASy WWW server. Trends Biochem. Sci. 1994, 19, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomic Data. In Plant Genomics Databases: Methods and Protocols; Humana Press: New York, NY, USA, 2017; Volume 1533, pp. 1–31. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]

| Gene ID | Gene Name | Chromosome (Chr) | Start | End | pI | Molecular Weight (Average) | Protein Length (aa) | Subcellular Location | A. thaliana Homologous GeBP Genes |
|---|---|---|---|---|---|---|---|---|---|
| Bra031402 | BrGeBP1 | A01 | 17755753 | 17756751 | 5.09 | 36,940.88 | 332 | nucl: 13 | At4g00390 |
| Bra021317 | BrGeBP2 | A01 | 22344927 | 22345853 | 8.89 | 34,587.78 | 308 | nucl: 14 | At4g00390 |
| Bra040116 | BrGeBP3 | A01 | 28356166 | 28357348 | 5.17 | 40,831.34 | 371 | nucl: 8, cyto: 3, vacu: 2 | At5g28040 |
| Bra013842 | BrGeBP4 | A01 | 8032900 | 8036434 | 8.95 | 69,620.11 | 609 | chlo: 9, nucl: 2, extr: 2 | At5g25140 |
| Bra013873 | BrGeBP5 | A01 | 8175027 | 8176103 | 7.53 | 39,311.39 | 358 | nucl: 11, chlo: 1, cyto: 1 | At4g25210 |
| Bra019183 | BrGeBP6 | A03 | 25822483 | 25823583 | 5.72 | 40,527.59 | 366 | nucl: 12, cyto: 1 | At4g25210 |
| Bra017244 | BrGeBP7 | A04 | 15774327 | 15776242 | 4.56 | 43,160.65 | 377 | nucl: 10, cyto: 3 | At2g36340 |
| Bra032717 | BrGeBP8 | A04 | 5236288 | 5237286 | 5.31 | 36,619.28 | 332 | nucl: 13 | At1g44810 |
| Bra028105 | BrGeBP9 | A04 | 5768566 | 5771723 | 8.98 | 72,816.9 | 640 | nucl: 13 | At1g61730 |
| Bra028106 | BrGeBP10 | A04 | 5772763 | 5773737 | 6.17 | 33,634.76 | 299 | nucl: 14 | At1g11510 |
| Bra028108 | BrGeBP11 | A04 | 5776855 | 5777754 | 5.91 | 33,917.13 | 299 | nucl: 14 | At1g61730 |
| Bra025445 | BrGeBP12 | A04 | 8693888 | 8694811 | 5.61 | 34,648.02 | 307 | nucl: 13 | At1g11510 |
| Bra039500 | BrGeBP13 | A05 | 9536224 | 9537362 | 5.71 | 34,161.25 | 300 | nucl: 10, cyto: 2, plas: 2 | At5g14280 |
| Bra009986 | BrGeBP14 | A06 | 18469313 | 18470557 | 4.87 | 44,964.03 | 414 | nucl: 12, cyto: 1 | At5g28040 |
| Bra019847 | BrGeBP15 | A06 | 4019905 | 4020975 | 7.66 | 39,168.47 | 356 | nucl: 14 | At1g11510 |
| Bra036116 | BrGeBP16 | A09 | 2641324 | 2642491 | 4.69 | 38,400.59 | 358 | nucl: 12, cysk: 1 | At5g28040 |
| Bra027078 | BrGeBP17 | A09 | 8253097 | 8254239 | 5.15 | 42,108.78 | 380 | nucl: 13 | At1g61730 |
| Bra034488 | BrGeBP18 | Scaffold000096 | 356960 | 357928 | 5.35 | 35,634.26 | 322 | nucl: 13 | At1g61730 |
| Bra038583 | BrGeBP19 | Scaffold000149 | 250721 | 251980 | 5.08 | 46,353.94 | 419 | nucl: 10, cyto: 4 | At1g61730 |
| Bra040416 | BrGeBP20 | Scaffold000203 | 97200 | 98477 | 4.84 | 46,795.99 | 425 | nucl: 13 | At3g04930 |
| MicroRNAs | MicroRNA Targeting BrGeBPs | |
|---|---|---|
| Br-miR156a-3p | BrGeBP9 | |
| Br-miR156f-3p | BrGeBP9 | |
| Br-miR172d-3p | BrGeBP15 | |
| Br-miR172d-5p | BrGeBP5 | |
| Br-miR395a-5p | BrGeBP4 | |
| Br-miR395b-5p | BrGeBP4 | |
| Br-miR395c-5p | BrGeBP4 | |
| Br-miR395d-5p | BrGeBP4 | |
| Br-miR5711 | BrGeBP19 | |
| Br-miR5714 | BrGeBP5 | |
| Br-miR6032-3p | BrGeBP19 | |
| Br-miR9559-5p | BrGeBP20 | |
| Br-miR9565-5p | BrGeBP20 | |
| Br-miR9569-5p | BrGeBP20 | |
| Br-miR159a | BrGeBP8 | BrGeBP18 |
| Br-miR5716 | BrGeBP9 | BrGeBP10 |
| Br-miR5717 | BrGeBP4 | BrGeBP13 |
| Br-miR9555a-5p | BrGeBP13 | BrGeBP7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Wu, X.; Wang, Z.; Zhang, X.; Chen, L.; Duan, Q.; Huang, J. Genome-Wide Identification and Expression Analysis of BrGeBP Genes Reveal Their Potential Roles in Cold and Drought Stress Tolerance in Brassica rapa. Int. J. Mol. Sci. 2023, 24, 13597. https://doi.org/10.3390/ijms241713597
Wang R, Wu X, Wang Z, Zhang X, Chen L, Duan Q, Huang J. Genome-Wide Identification and Expression Analysis of BrGeBP Genes Reveal Their Potential Roles in Cold and Drought Stress Tolerance in Brassica rapa. International Journal of Molecular Sciences. 2023; 24(17):13597. https://doi.org/10.3390/ijms241713597
Chicago/Turabian StyleWang, Ruolan, Xiaoyu Wu, Ziwen Wang, Xiaoyu Zhang, Luhan Chen, Qiaohong Duan, and Jiabao Huang. 2023. "Genome-Wide Identification and Expression Analysis of BrGeBP Genes Reveal Their Potential Roles in Cold and Drought Stress Tolerance in Brassica rapa" International Journal of Molecular Sciences 24, no. 17: 13597. https://doi.org/10.3390/ijms241713597
APA StyleWang, R., Wu, X., Wang, Z., Zhang, X., Chen, L., Duan, Q., & Huang, J. (2023). Genome-Wide Identification and Expression Analysis of BrGeBP Genes Reveal Their Potential Roles in Cold and Drought Stress Tolerance in Brassica rapa. International Journal of Molecular Sciences, 24(17), 13597. https://doi.org/10.3390/ijms241713597






