Transcriptome Analysis Reveals the Effect of PdhR in Plesiomonas shigelloides
Abstract
1. Introduction
2. Results
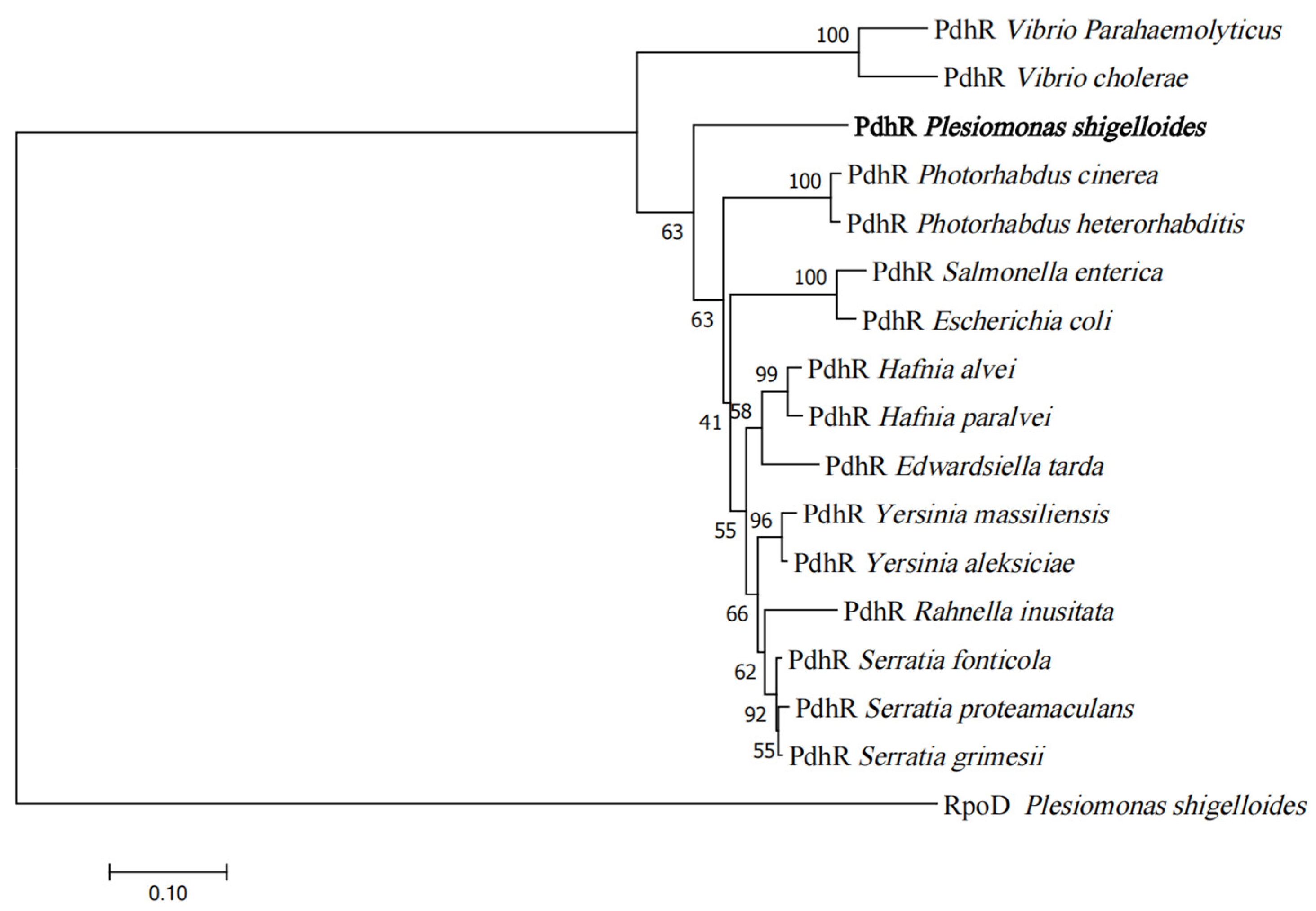
2.1. Phylogenetic Analysis of PdhR
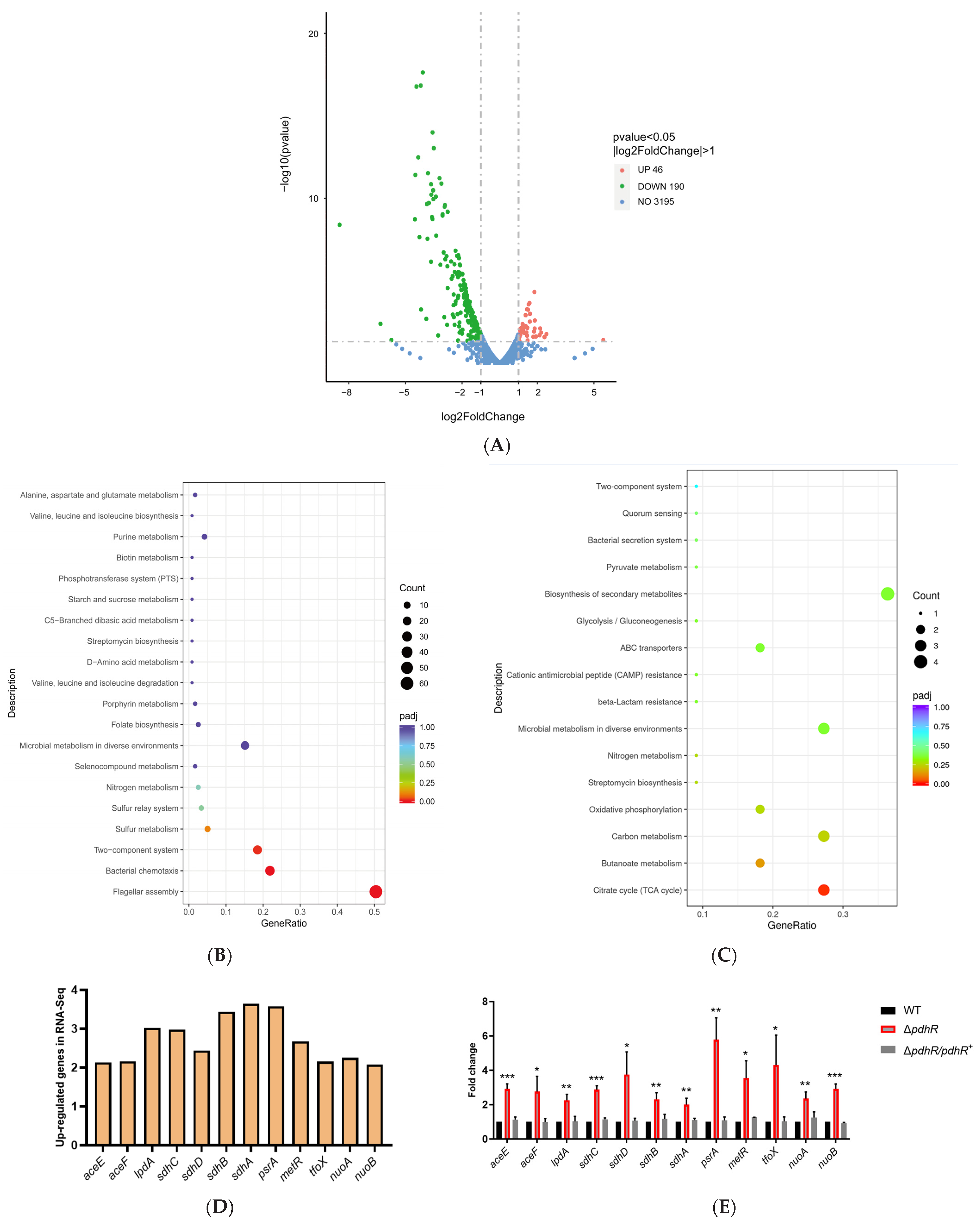
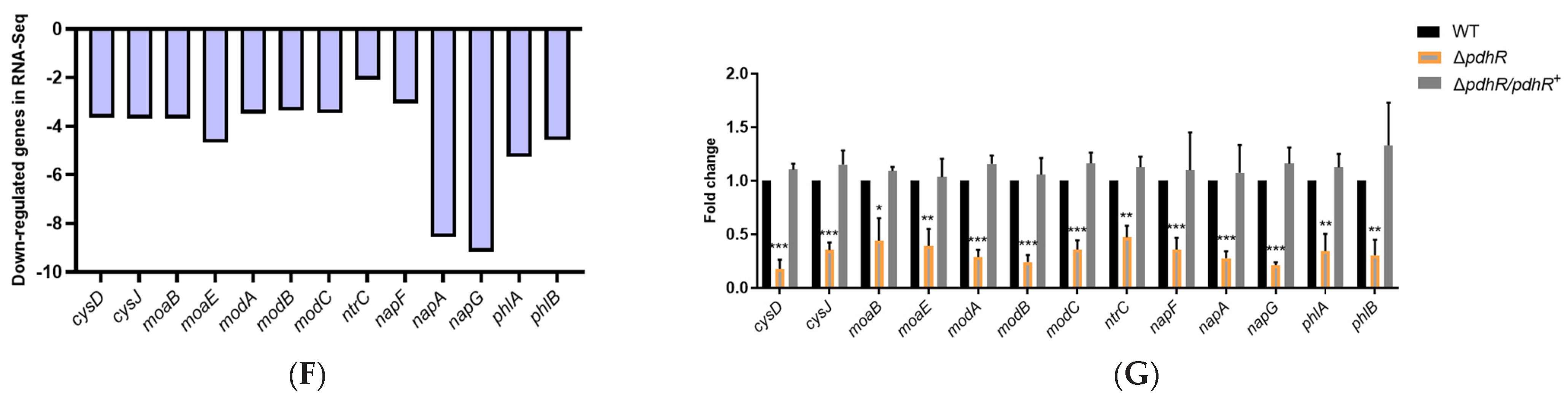
2.2. Transcriptome Sequencing Revealed Gene Expression Related to PdhR of the P. shigelloides
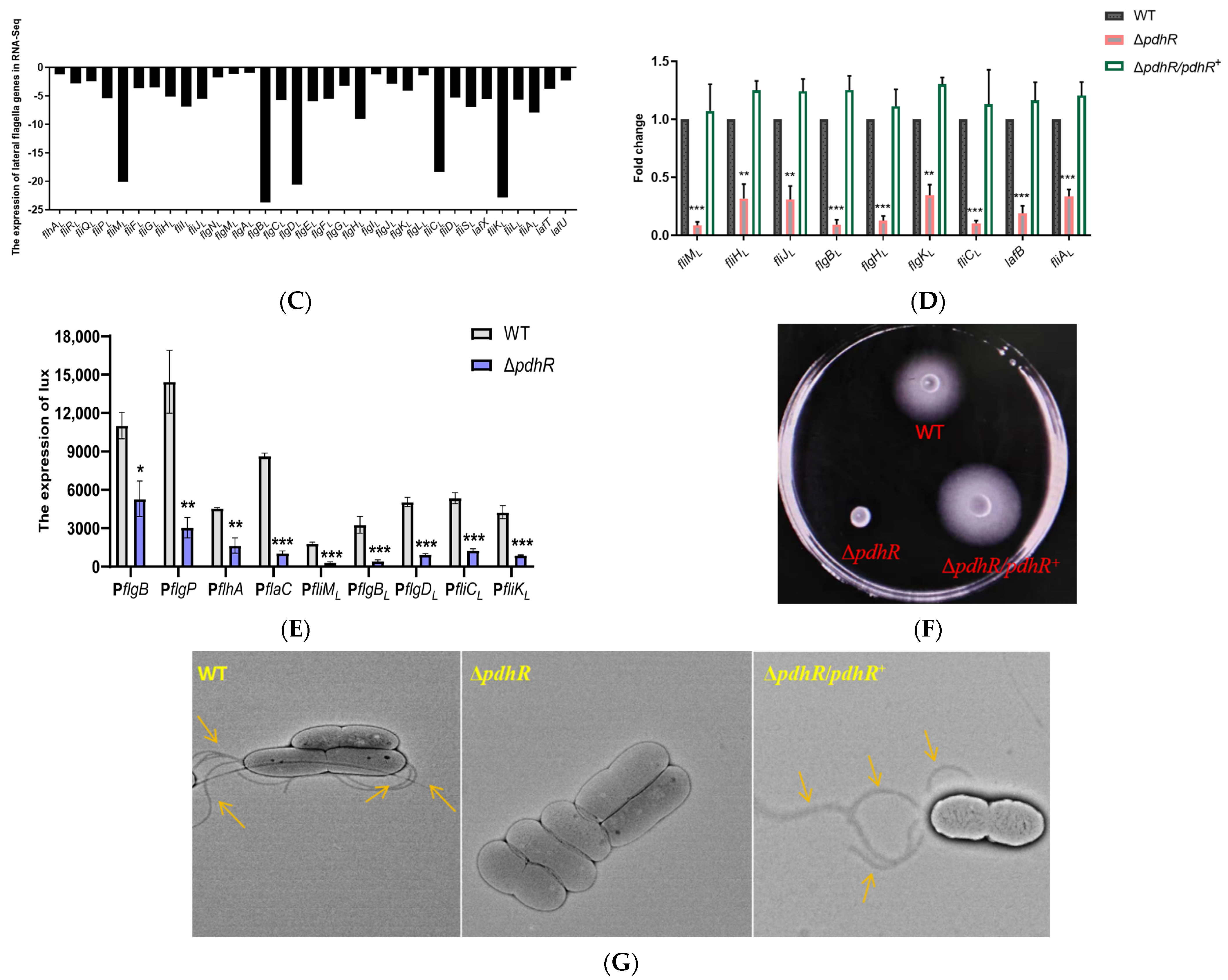
2.3. PdhR Influences Motility and Flagellar Synthesis by Positively Regulating the Expression of Flagellar Genes in P. shigelloides
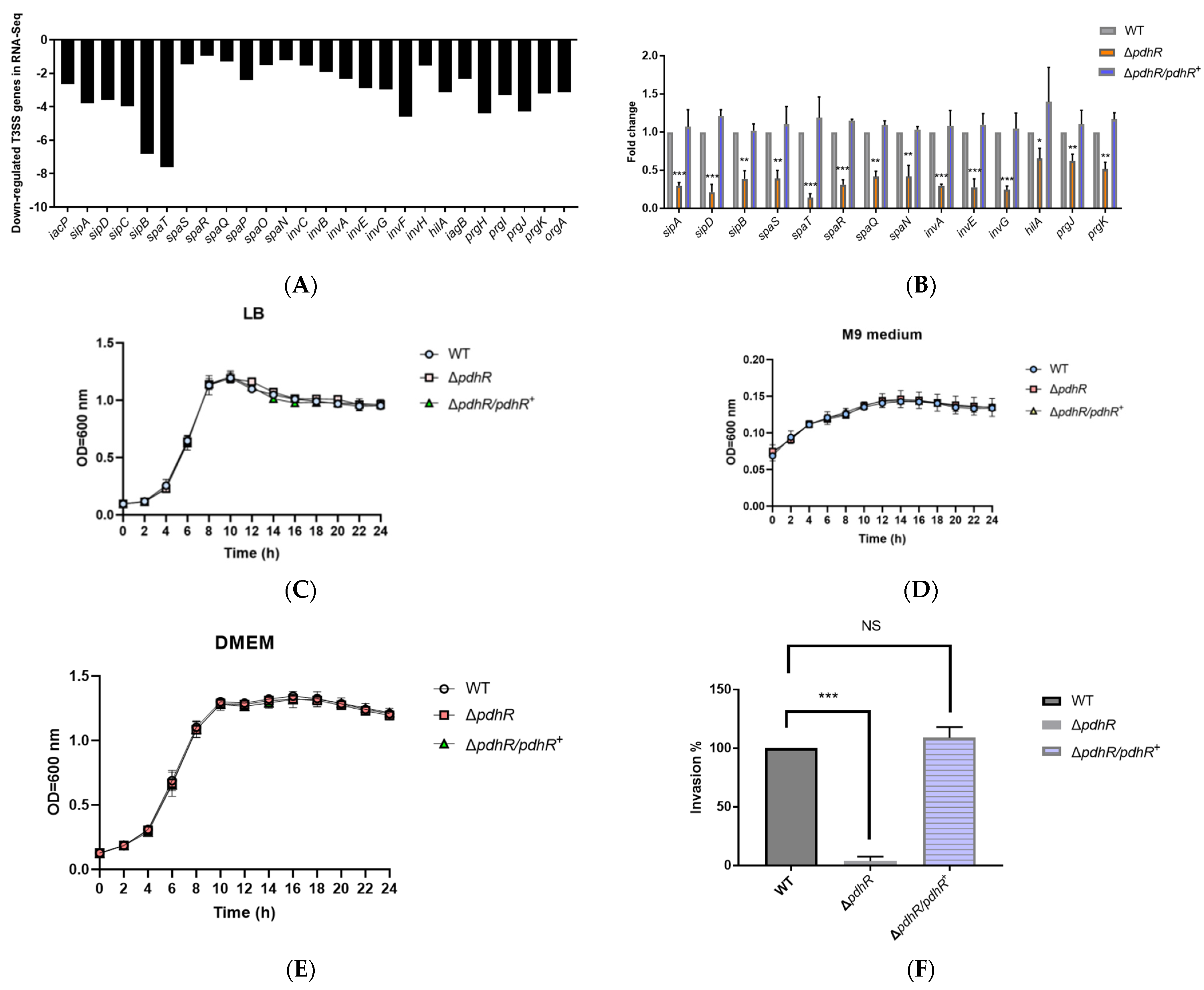
2.4. PdhR Promotes P. shigelloides’ Ability to Infect Caco-2 Cells by Positively Regulating T3SS Expression
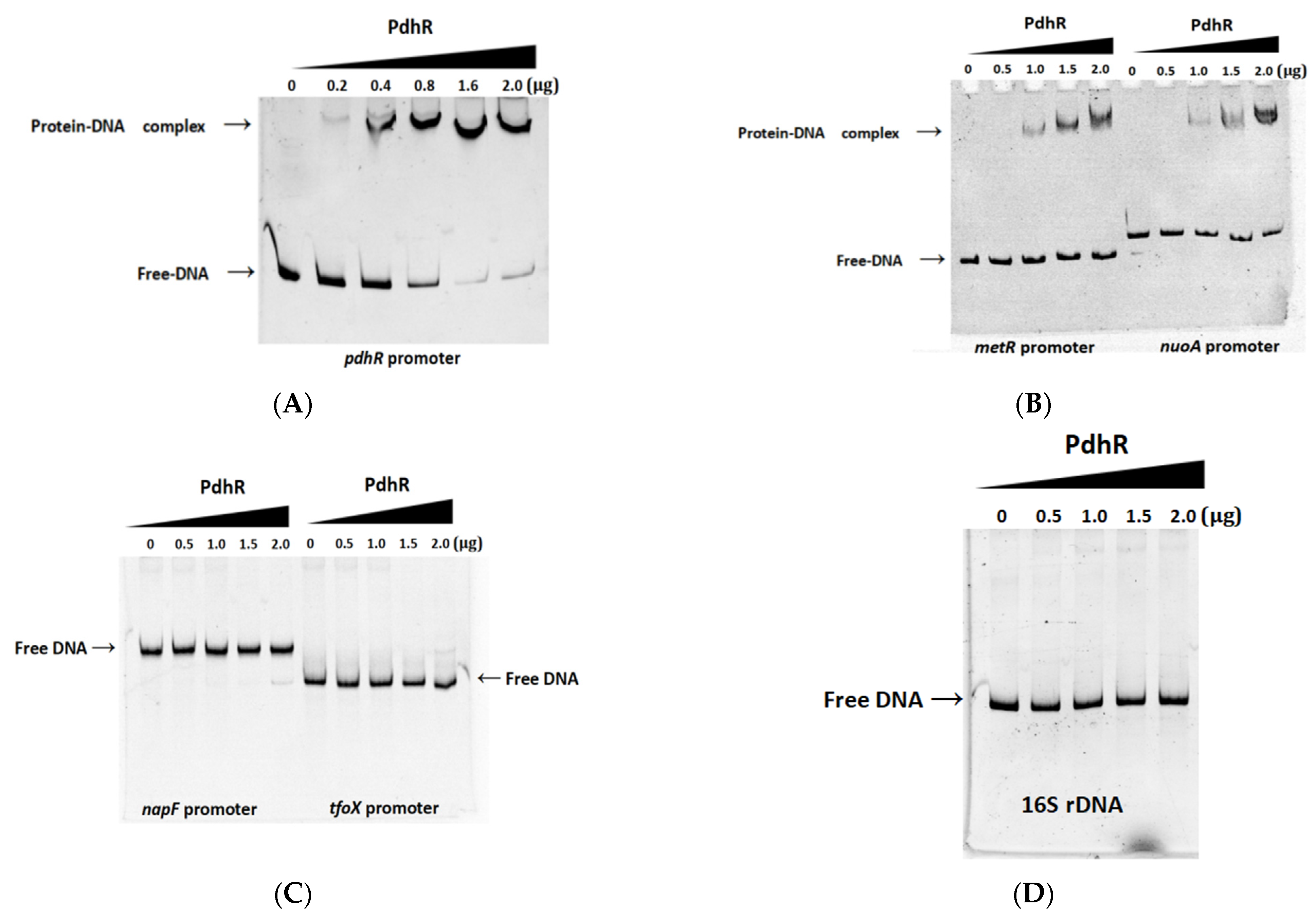
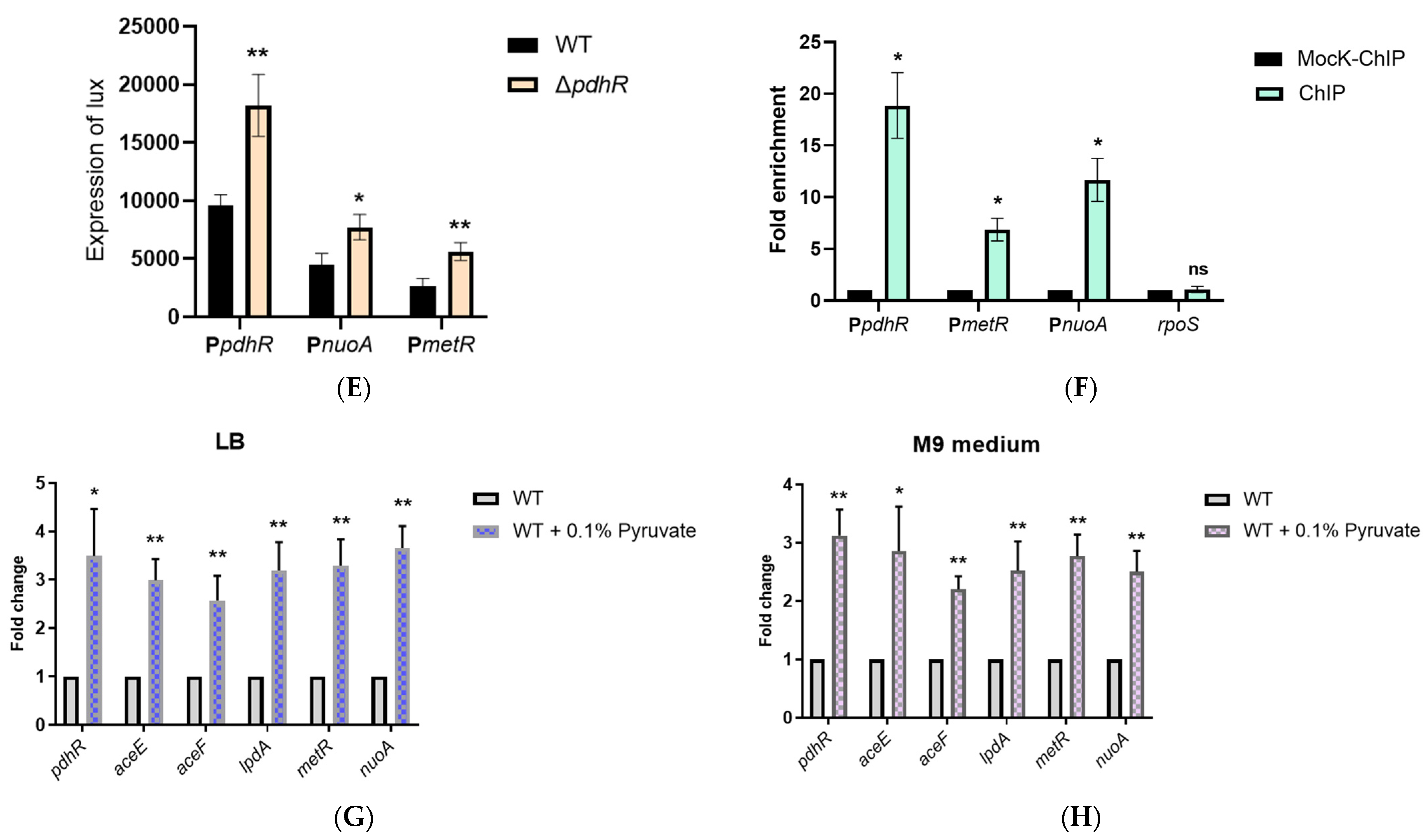
2.5. Pyruvate-Sensing PdhR Directly Negatively Regulates the Expression of pdhR-aceEF, metR, and nuoA
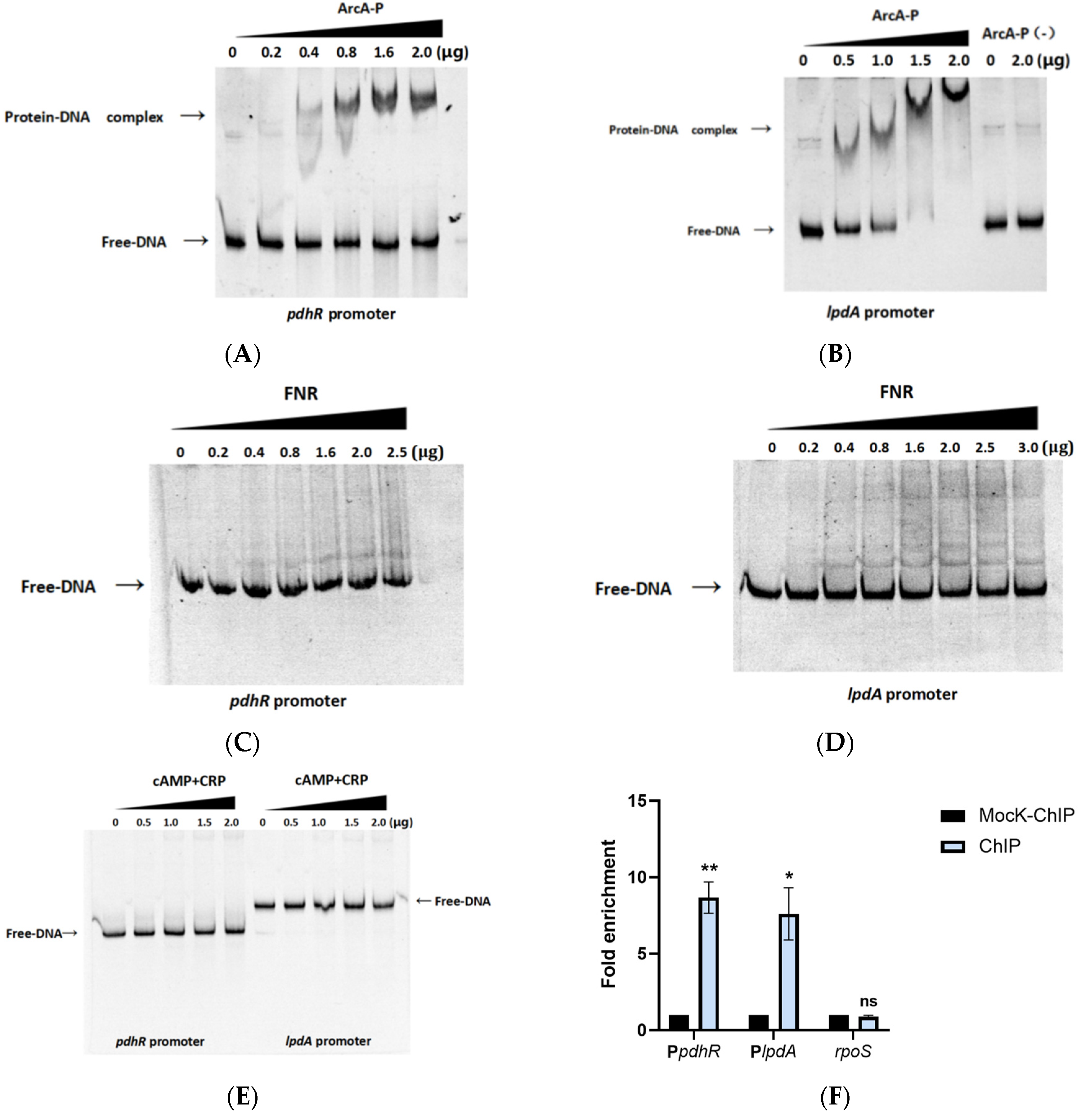
2.6. pdhR Is Directly Negatively Regulated by ArcA and Indirectly Positively Regulated by FNR and CRP
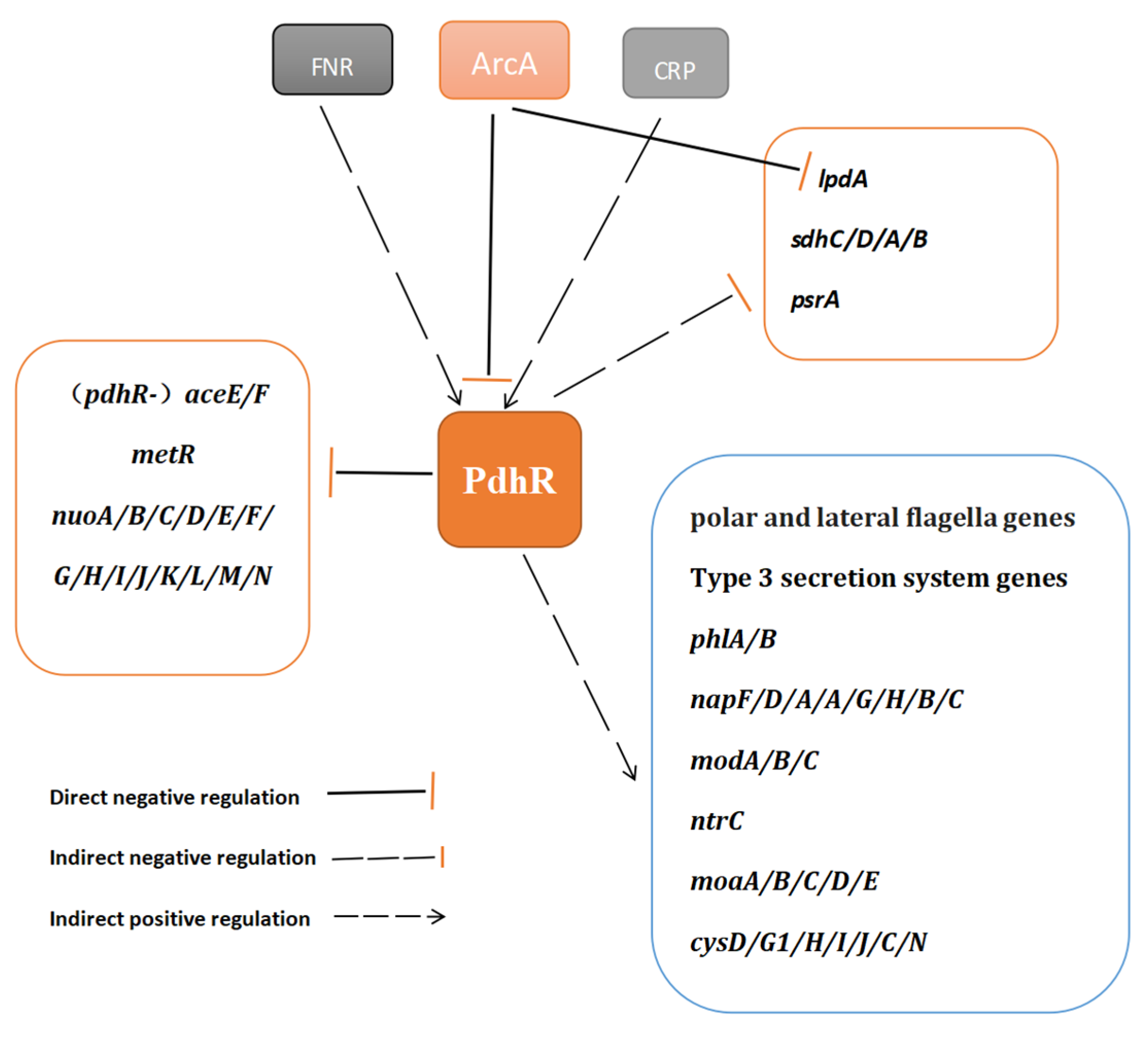
2.7. The Potential Regulatory Pathways of PdhR in P. shigelloides
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Growth Conditions
4.2. Genes Deletion and Complementation
4.3. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.4. RNA Sequencing (RNA-Seq)
4.5. Motility Assay and Transmission Electron Microscopy (TEM) of Flagella
4.6. Expression and Purification of Proteins and Electrophoretic Mobility Shift Assays (EMSAs)
4.7. Chromatin Immunoprecipitation and Quantitative PCR (ChIP-qPCR)
4.8. Luminescence Screening Assay
4.9. Growth Assay
4.10. Invasion Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Puah, S.M.; Puthucheary, S.D.A.; Chua, K.H. Virulence Profiles among Gastrointestinal and Extraintestinal Clinical Isolates of Plesiomonas shigelloides. Jpn. J. Infect. Dis. 2022, 75, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Mandal, B.K.; Whale, K.; Morson, B.C. Acute colitis due to Plesiomonas shigelloides. Br. Med. J. (Clin. Res. Ed.) 1982, 285, 1539–1540. [Google Scholar] [CrossRef] [PubMed]
- McNeeley, D.; Ivy, P.; Craft, J.C.; Cohen, I. Plesiomonas: Biology of the organism and diseases in children. Pediatr. Infect. Dis. 1984, 3, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, T.; Kinoshita, Y.; Shimada, T.; Sakazaki, R. Two epidemics of diarrhoeal disease possibly caused by Plesiomonas shigelloides. J. Hyg. 1978, 80, 275–280. [Google Scholar] [CrossRef]
- Yan, J.; Guo, X.; Li, J.; Li, Y.; Sun, H.; Li, A.; Cao, B. RpoN is required for the motility and contributes to the killing ability of Plesiomonas shigelloides. BMC Microbiol. 2022, 22, 299. [Google Scholar] [CrossRef]
- Göhler, A.-K.; Kökpinar, Ö.; Schmidt-Heck, W.; Geffers, R.; Guthke, R.; Rinas, U.; Schuster, S.; Jahreis, K.; Kaleta, C. More than just a metabolic regulator—Elucidation and validation of new targets of PdhR in Escherichia coli. BMC Syst. Biol. 2011, 5, 197. [Google Scholar] [CrossRef]
- Quail, M.A.; Guest, J.R. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol. Microbiol. 1995, 15, 519–529. [Google Scholar] [CrossRef]
- Stephens, P.E.; Darlison, M.G.; Lewis, H.M.; Guest, J.R. The Pyruvate Dehydrogenase Complex of Escherichia coli K12. Nucleotide Sequence Encoding the Pyruvate Dehydrogenase Component. Eur. J. Biochem. 1983, 133, 155–162. [Google Scholar] [CrossRef]
- Haydon, D.J.; Guest, J.R. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 1991, 63, 291–295. [Google Scholar] [CrossRef]
- Ogasawara, H.; Ishida, Y.; Yamada, K.; Yamamoto, K.; Ishihama, A. PdhR (Pyruvate Dehydrogenase Complex Regulator) Controls the Respiratory Electron Transport System in Escherichia coli. J. Bacteriol. 2007, 189, 5534–5541. [Google Scholar] [CrossRef]
- Sauer, U.; Eikmanns, B.J. The PEP–pyruvate–oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 2005, 29, 765–794. [Google Scholar] [CrossRef]
- Shimada, T.; Nakazawa, K.; Tachikawa, T.; Saito, N.; Niwa, T.; Taguchi, H.; Tanaka, K. Acetate overflow metabolism regulates a major metabolic shift after glucose depletion in Escherichia coli. FEBS Lett. 2021, 595, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, L.; Georgellis, D.; Green, J.; Guest, J.R. Co-regulation of lipoamide dehydrogenase and 2-oxoglutarate dehydrogenase synthesis in Escherichia coli: Characterisation of an ArcA binding site in the lpd promoter. FEMS Microbiol. Lett. 1998, 169, 403–408. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaleta, C.; Göhler, A.; Schuster, S.; Jahreis, K.; Guthke, R.; Nikolajewa, S. Integrative inference of gene-regulatory networks in Escherichia coli using information theoretic concepts and sequence analysis. BMC Syst. Biol. 2010, 4, 116. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Cronan, J.E. PdhR, the pyruvate dehydrogenase repressor, does not regulate lipoic acid synthesis. Res. Microbiol. 2014, 165, 429–438. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wyborn, N.R.; Messenger, S.L.; Henderson, R.A.; Sawers, G.; Roberts, R.E.; Attwood, M.M.; Green, J. Expression of the Escherichia coli yfiD gene responds to intracellular pH and reduces the accumulation of acidic metabolic end products. Microbiology 2002, 148, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Blankenhorn, D.; Phillips, J.; Slonczewski, J.L. Acid- and Base-Induced Proteins during Aerobic and Anaerobic Growth of Escherichia coli Revealed by Two-Dimensional Gel Electrophoresis. J. Bacteriol. 1999, 181, 2209–2216. [Google Scholar] [CrossRef]
- Calhoun, M.W.; Gennis, R.B. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J. Bacteriol. 1993, 175, 3013–3019. [Google Scholar] [CrossRef]
- Calhoun, M.W.; Oden, K.L.; Gennis, R.B.; de Mattos, M.J.; Neijssel, O.M. Energetic efficiency of Escherichia coli: Effects of mutations in components of the aerobic respiratory chain. J. Bacteriol. 1993, 175, 3020–3025. [Google Scholar] [CrossRef][Green Version]
- Ingledew, W.J.; Poole, R.K. The respiratory chains of Escherichia coli. Microbiol. Rev. 1984, 48, 222–271. [Google Scholar] [CrossRef]
- Aguilera, L.; Campos, E.; Giménez, R.; Badía, J.; Aguilar, J.; Baldoma, L. Dual role of LldR in regulation of the lldPRD operon, involved in L-lactate metabolism in Escherichia coli. J. Bacteriol. 2008, 190, 2997–3005. [Google Scholar] [CrossRef] [PubMed]
- Faith, J.J.; Hayete, B.; Thaden, J.T.; Mogno, I.; Wierzbowski, J.; Cottarel, G.; Kasif, S.; Collins, J.J.; Gardner, T.S. Large-Scale Mapping and Validation of Escherichia coli Transcriptional Regulation from a Compendium of Expression Profiles. PLoS Biol. 2007, 5, e8. [Google Scholar] [CrossRef] [PubMed]
- Anzai, T.; Imamura, S.; Ishihama, A.; Shimada, T. Expanded roles of pyruvate-sensing PdhR in transcription regulation of the Escherichia coli K-12 genome: Fatty acid catabolism and cell motility. Microb. Genom. 2020, 6, e000442. [Google Scholar] [CrossRef]
- Green, J.; Paget, M.S. Bacterial redox sensors. Nat. Rev. Microbiol. 2004, 2, 954–966. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Weber, K.D.; Qiu, Y.; Kiley, P.J.; Blattner, F.R. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 2005, 187, 1135–1160. [Google Scholar] [CrossRef] [PubMed]
- Marshall, F.A.; Messenger, S.L.; Wyborn, N.R.; Guest, J.R.; Wing, H.; Busby, S.J.; Green, J. A novel promoter architecture for microaerobic activation by the anaerobic transcription factor FNR. Mol. Microbiol. 2001, 39, 747–753. [Google Scholar] [CrossRef]
- Salmon, K.; Hung, S.P.; Mekjian, K.; Baldi, P.; Hatfield, G.W.; Gunsalus, R.P. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 2003, 278, 29837–29855. [Google Scholar] [CrossRef]
- Trotter, E.W.; Rolfe, M.D.; Hounslow, A.M.; Craven, C.J.; Williamson, M.P.; Sanguinetti, G.; Poole, R.K.; Green, J. Reprogramming of Escherichia coli K-12 Metabolism during the Initial Phase of Transition from an Anaerobic to a Micro-Aerobic Environment. PLoS ONE 2011, 6, e25501. [Google Scholar] [CrossRef]
- Iuchi, S.; Lin, E.C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 1988, 85, 1888–1892. [Google Scholar] [CrossRef]
- Quail, M.A.; Haydon, D.J.; Guest, J.R. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol. Microbiol. 1994, 12, 95–104. [Google Scholar] [CrossRef]
- Gutierrez-Ríos, R.M.; Freyre-Gonzalez, J.A.; Resendis, O.; Collado-Vides, J.; Saier, M.; Gosset, G. Identification of regulatory network topological units coordinating the genome-wide transcriptional response to glucose in Escherichia coli. BMC Microbiol. 2007, 7, 53. [Google Scholar] [CrossRef]
- Roy, S.; Vivoli Vega, M.; Ames, J.R.; Britten, N.; Kent, A.; Evans, K.; Isupov, M.N.; Harmer, N.J.; GoVV Consortium. The ROK kinase N-acetylglucosamine kinase uses a sequential random enzyme mechanism with successive conformational changes upon each substrate binding. J. Biol. Chem. 2023, 299, 103033. [Google Scholar] [CrossRef]
- Wang, R.; Qian, J.; Ji, D.; Liu, X.; Dong, R. Transcriptome Analysis Reveals Effect of Dietary Probiotics on Immune Response Mechanism in Southern Catfish (Silurus meridionalis) in Response to Plesiomonas shigelloides. Animals 2023, 13, 449. [Google Scholar] [CrossRef]
- Maciejewska, A.; Bednarczyk, B.; Lugowski, C.; Lukasiewicz, J. Structural Studies of the Lipopolysaccharide Isolated from Plesiomonas shigelloides O22:H3 (CNCTC 90/89). Int. J. Mol. Sci. 2020, 21, 6788. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; An, C.; Bao, Y.; Zhao, X.; Jernigan, R.L.; Lithio, A.; Nettleton, D.; Li, L.; Wurtele, E.S.; Nolan, L.K.; et al. ArcA Controls Metabolism, Chemotaxis, and Motility Contributing to the Pathogenicity of Avian Pathogenic Escherichia coli. Infect. Immun. 2015, 83, 3545–3554. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.R.; Fink, R.C.; Vazquez-Torres, A.; Porwollik, S.; Jones-Carson, J.; McClelland, M.; Hassan, H.M. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol. 2011, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, J.; Guo, X.; Wang, X.; Liu, F.; Cao, B. The global regulators ArcA and CytR collaboratively modulate Vibrio cholerae motility. BMC Microbiol. 2022, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, D.; Guo, T.; Ran, T.; Wang, W.; Xu, D. Differential roles for ArcA and ArcB homologues in swarming motility in Serratia marcescens FS14. Antonie Van Leeuwenhoek 2017, 111, 609–617. [Google Scholar] [CrossRef]
- Rahmatelahi, H.; El-Matbouli, M.; Menanteau-Ledouble, S. Delivering the pain: An overview of the type III secretion system with special consideration for aquatic pathogens. Vet. Res. 2021, 52, 146. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.; Li, L.; Ma, H.; Riely, B.K.; Liu, B.; Li, H. The critical role of MetR/MetB/MetC/MetX in cysteine and methionine metabolism, fungal development and virulence of Alternaria alternata. Appl. Environ. Microbiol. 2021, 87, e01911-20. [Google Scholar] [CrossRef]
- Gai, Y.; Liu, B.; Ma, H.; Li, L.; Chen, X.; Moenga, S.; Riely, B.; Fayyaz, A.; Wang, M.; Li, H. The methionine biosynthesis regulator AaMetR contributes to oxidative stress tolerance and virulence in Alternaria alternata. Microbiol. Res. 2019, 219, 94–109. [Google Scholar] [CrossRef] [PubMed]
- de Melo, A.T.; Martho, K.F.; Roberto, T.N.; Nishiduka, E.S.; Machado, J.; Brustolini, O.J.B.; Tashima, A.K.; Vasconcelos, A.T.; Vallim, M.A.; Pascon, R.C. The regulation of the sulfur amino acid biosynthetic pathway in Cryptococcus neoformans: The relationship of Cys3, Calcineurin, and Gpp2 phosphatases. Sci. Rep. 2019, 9, 11923. [Google Scholar] [CrossRef] [PubMed]
- Weissbach, H.; Brot, N. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 1991, 5, 1593–1597. [Google Scholar] [CrossRef]
- Urbanowski, M.L.; Stauffer, G.V. Role of homocysteine in metR-mediated activation of the metE and metH genes in Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1989, 171, 3277–3281. [Google Scholar] [CrossRef]
- Lorenz, E.; Stauffer, G.V. RNA polymerase, PurR and MetR interactions at the glyA promoter of Escherichia coli. Microbiology 1996, 142 Pt 7, 1819–1824. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, X.-Y.; Redfield, B.; Maxon, M.; Weissbach, H.; Brot, N. The effect of homocysteine on metR regulation of metE, metR and metH expression in vitro. Biochem. Biophys. Res. Commun. 1989, 163, 79–83. [Google Scholar] [CrossRef]
- Pan, X.; Sun, C.; Tang, M.; You, J.; Osire, T.; Zhao, Y.; Xu, M.; Zhang, X.; Shao, M.; Yang, S.; et al. LysR-Type Transcriptional Regulator MetR Controls Prodigiosin Production, Methionine Biosynthesis, Cell Motility, H2O2 Tolerance, Heat Tolerance, and Exopolysaccharide Synthesis in Serratia marcescens. Appl. Environ. Microbiol. 2020, 86, e02241-19. [Google Scholar] [CrossRef]
- Castro-Guerrero, N.; Sinha, P.K.; Torres-Bacete, J.; Matsuno-Yagi, A.; Yagi, T. Pivotal roles of three conserved carboxyl residues of the NuoC (30k) segment in the structural integrity of proton-translocating NADH-quinone oxidoreductase from Escherichia coli. Biochemistry 2010, 49, 10072–10080. [Google Scholar] [CrossRef]
- Lynch, A.S.; Lin, E.C. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: Characterization of DNA binding at target promoters. J. Bacteriol. 1996, 178, 6238–6249. [Google Scholar] [CrossRef]
- Gunsalus, R.; Park, S.-J. Aerobic-anaerobic gene regulation in Escherichia coli: Control by the ArcAB and Fnr regulons. Res. Microbiol. 1994, 145, 437–450. [Google Scholar] [CrossRef]
- Nyström, T.; Larsson, C.; Gustafsson, L. Bacterial defense against aging: Role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J. 1996, 15, 3219–3228. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Feng, L.; Yang, B.; Zhang, W.; Wang, P.; Jiang, X.; Wang, L. Signal transduction pathway mediated by the novel regulator LoiA for low oxygen tension induced Salmonella Typhimurium invasion. PLoS Pathog. 2017, 13, e1006429. [Google Scholar] [CrossRef] [PubMed]
- Xi, D.; Jing, F.; Liu, Q.; Cao, B. Plesiomonas shigelloides sipD mutant, generated by an efficient gene transfer system, is less invasive. J. Microbiol. Methods 2019, 159, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Wilhelms, M.; Fulton, K.M.; Twine, S.M.; Tomás, J.M.; Merino, S. Differential Glycosylation of Polar and Lateral Flagellins in Aeromonas hydrophila AH-3. J. Biol. Chem. 2012, 287, 27851–27862. [Google Scholar] [CrossRef]
- Jiang, F.; Huang, X.; Barbieri, N.L.; Logue, C.M.; Nolan, L.K.; Li, G. Citrate utilization under anaerobic environment in Escherichia coli is under direct control of Fnr and indirect control of ArcA and Fnr via CitA-CitB system. Environ. Microbiol. 2021, 23, 1496–1509. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, W.; Hou, X.; Ma, S.; Wang, X.; Yan, X.; Yang, B.; Huang, D.; Liu, B.; Feng, L. Nitric oxide is a host cue for Salmonella Typhimurium systemic infection in mice. Commun. Biol. 2023, 6, 501. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Zhang, H.Z.; Liang, W.L.; Zhang, L.J.; Zhu, J.; Kan, B. Plasticity of regulation of mannitol phosphotransferase system operon by CRP-cAMP complex in Vibrio cholerae. Biomed. Environ. Sci. 2013, 26, 831–840. [Google Scholar]
- Wu, P.; Wang, Q.; Yang, Q.; Feng, X.; Liu, X.; Sun, H.; Yan, J.; Kang, C.; Liu, B.; Liu, Y.; et al. A Novel Role of the Two-Component System Response Regulator UvrY in Enterohemorrhagic Escherichia coli O157:H7 Pathogenicity Regulation. Int. J. Mol. Sci. 2023, 24, 2297. [Google Scholar] [CrossRef]
- Yan, J.; Li, Y.; Guo, X.; Wang, X.; Liu, F.; Li, A.; Cao, B. The effect of ArcA on the growth, motility, biofilm formation, and virulence of Plesiomonas shigelloides. BMC Microbiol. 2021, 21, 266. [Google Scholar] [CrossRef]
- Schubert, R.H.; Holz-Bremer, A. Cell Adhesion of Plesiomonas shigelloides. Zentralbl. Hyg. Umweltmed. 1999, 202, 383–388. [Google Scholar] [CrossRef]
- Yang, S.; Xi, D.; Wang, X.; Li, Y.; Li, Y.; Yan, J.; Cao, B. Vibrio cholerae VC1741 (PsrA) enhances the colonization of the pathogen in infant mice intestines in the presence of the long-chain fatty acid, oleic acid. Microb. Pathog. 2020, 147, 104443. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, J.; Yang, B.; Xue, X.; Li, J.; Li, Y.; Li, A.; Ding, P.; Cao, B. Transcriptome Analysis Reveals the Effect of PdhR in Plesiomonas shigelloides. Int. J. Mol. Sci. 2023, 24, 14473. https://doi.org/10.3390/ijms241914473
Yan J, Yang B, Xue X, Li J, Li Y, Li A, Ding P, Cao B. Transcriptome Analysis Reveals the Effect of PdhR in Plesiomonas shigelloides. International Journal of Molecular Sciences. 2023; 24(19):14473. https://doi.org/10.3390/ijms241914473
Chicago/Turabian StyleYan, Junxiang, Bin Yang, Xinke Xue, Jinghao Li, Yuehua Li, Ang Li, Peng Ding, and Boyang Cao. 2023. "Transcriptome Analysis Reveals the Effect of PdhR in Plesiomonas shigelloides" International Journal of Molecular Sciences 24, no. 19: 14473. https://doi.org/10.3390/ijms241914473
APA StyleYan, J., Yang, B., Xue, X., Li, J., Li, Y., Li, A., Ding, P., & Cao, B. (2023). Transcriptome Analysis Reveals the Effect of PdhR in Plesiomonas shigelloides. International Journal of Molecular Sciences, 24(19), 14473. https://doi.org/10.3390/ijms241914473





