Knocking Out OsRLK7-1 Impairs Rice Growth and Development but Enhances Its Resistance to Planthoppers
Abstract
:1. Introduction
2. Results
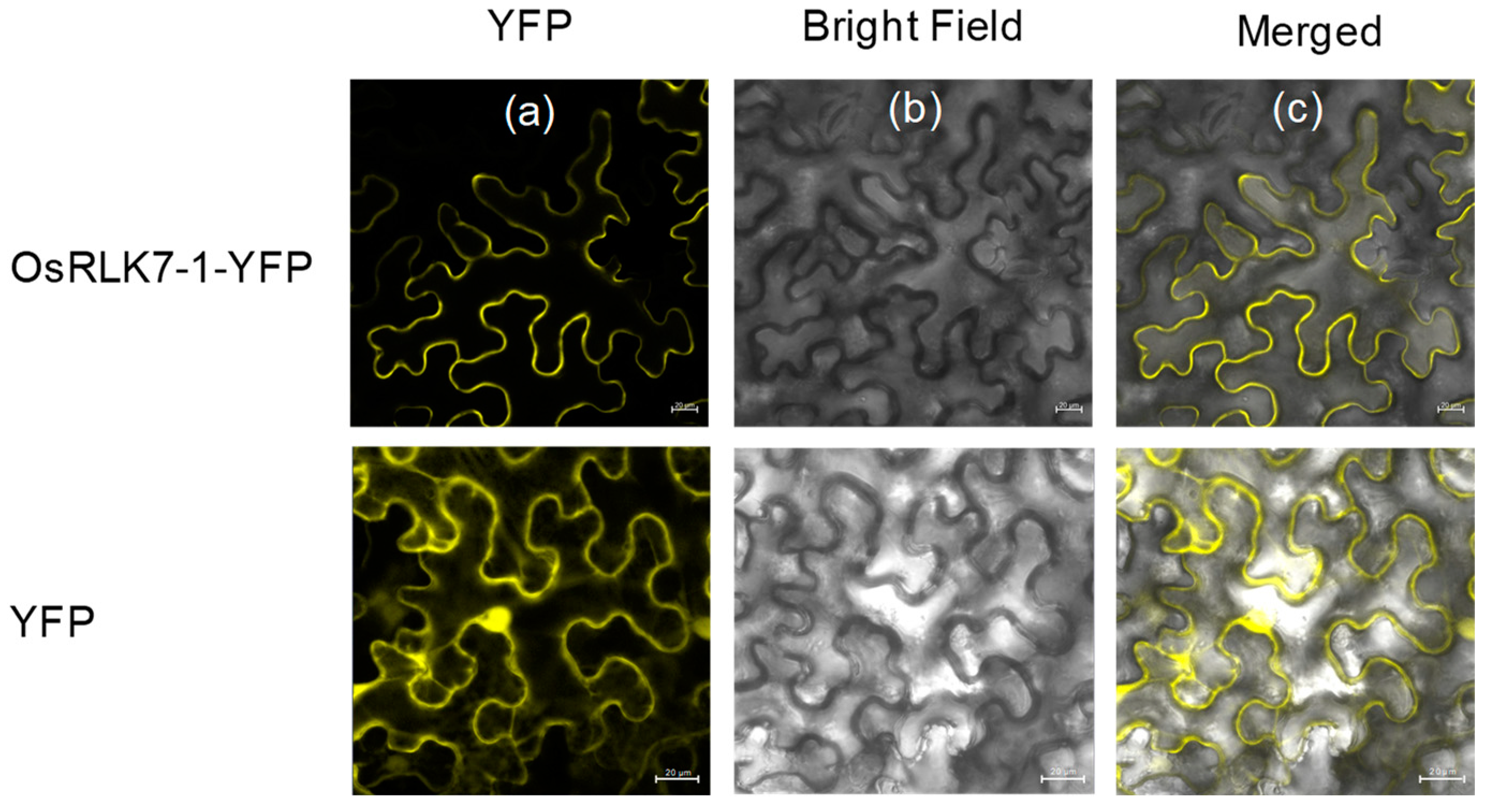
2.1. The Isolation and Characterization of OsRLK7-1
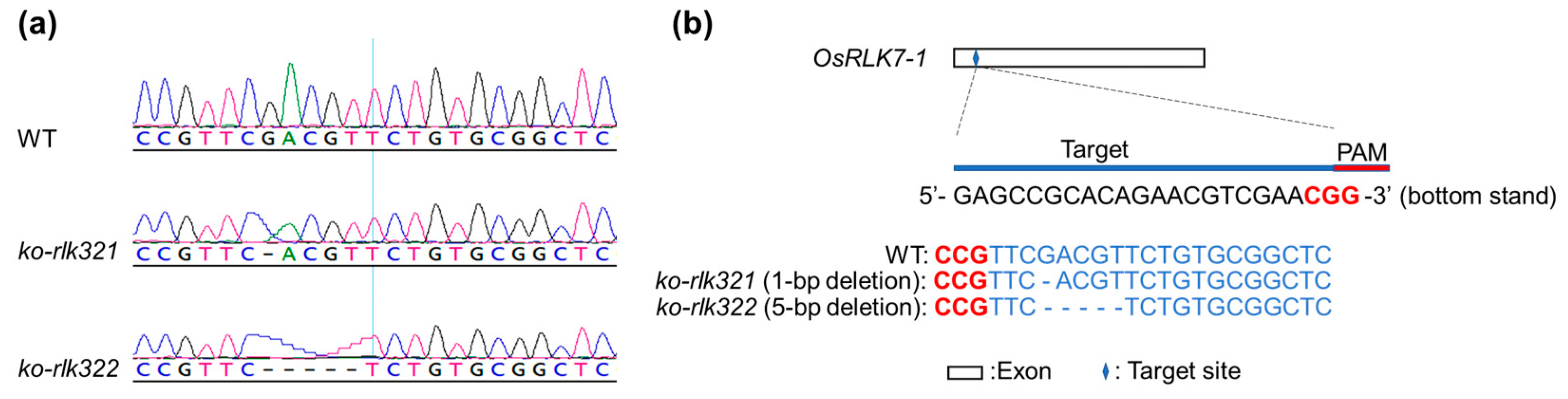
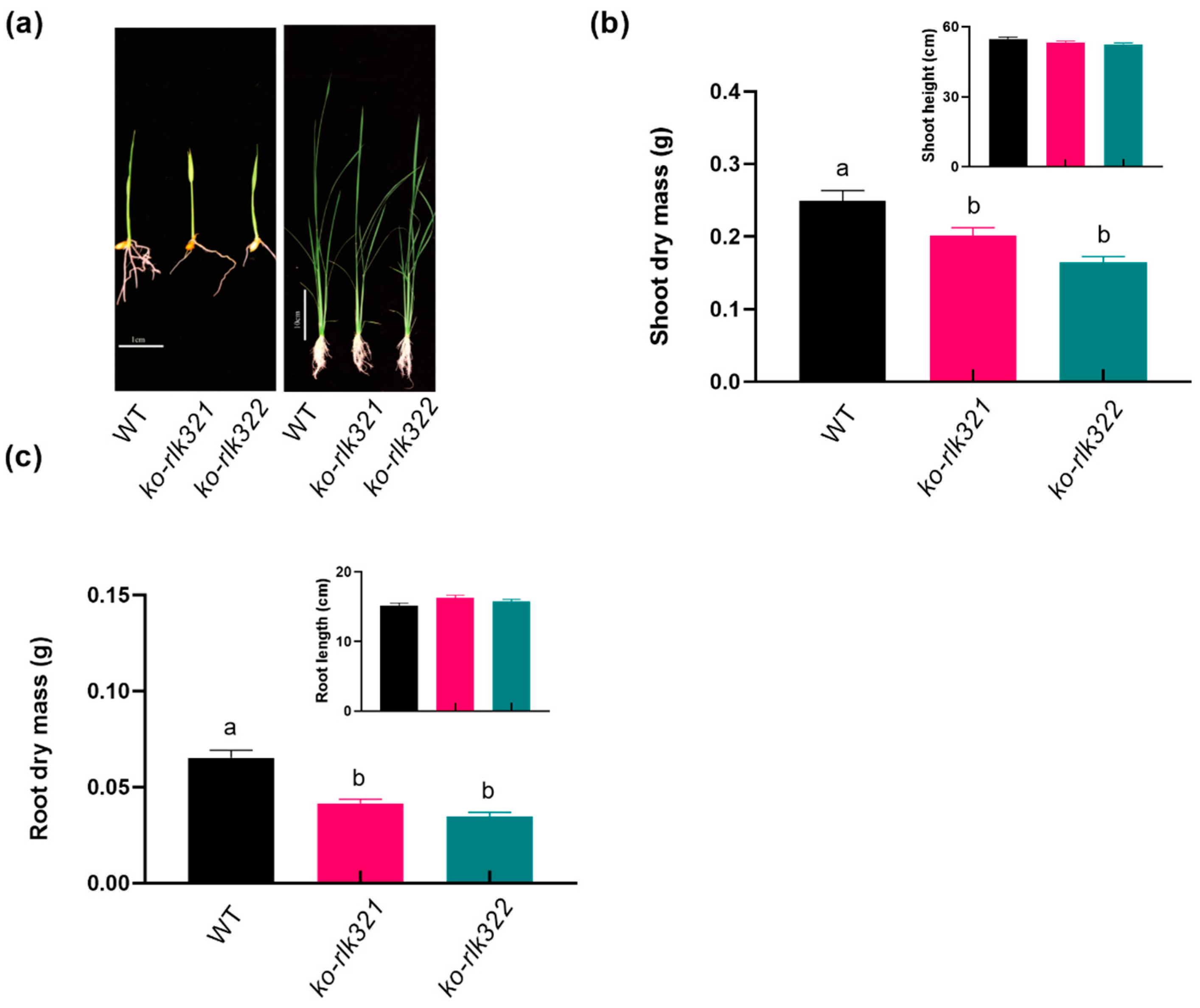
2.2. Knockout of OsRLK7-1 in Rice
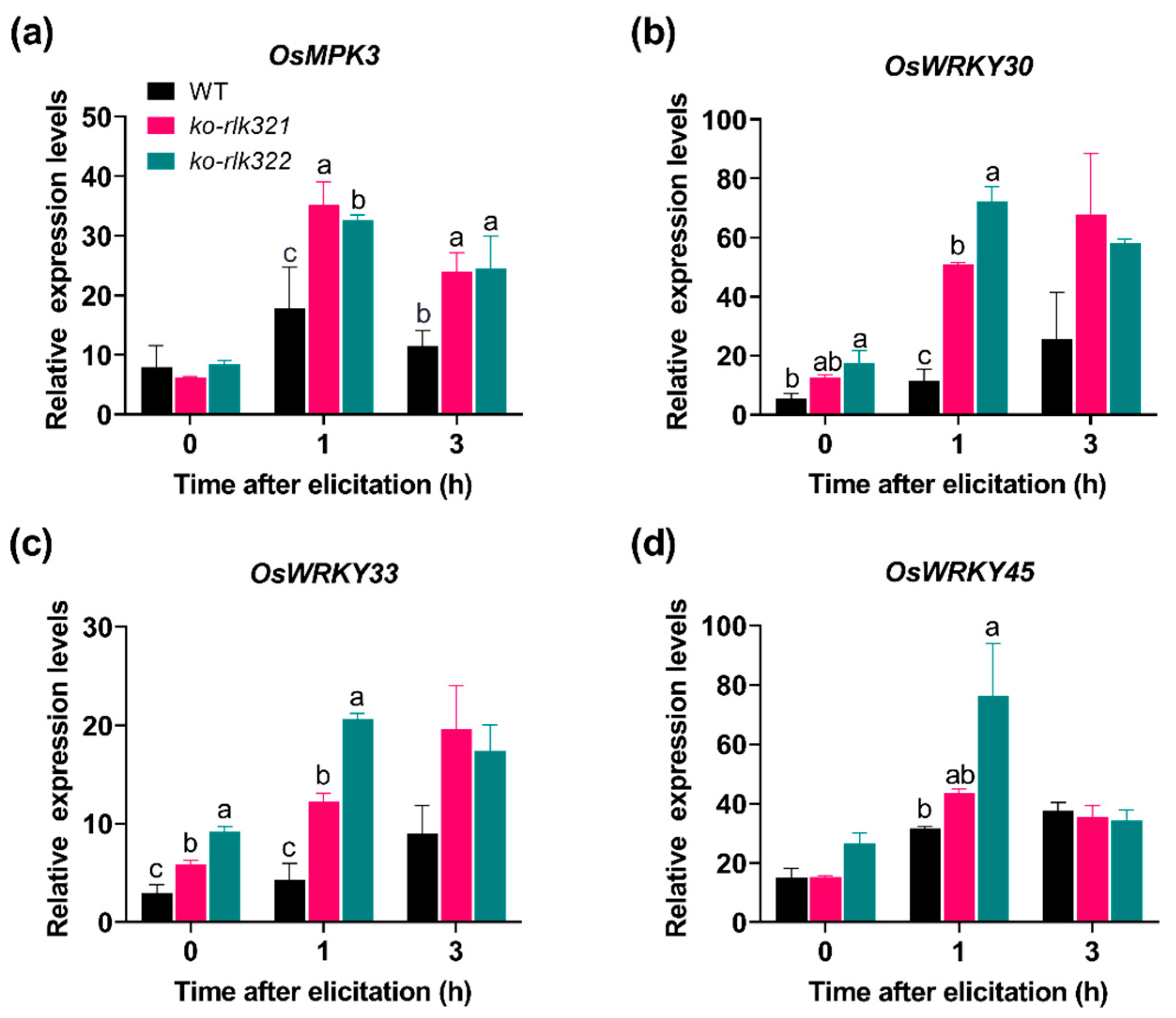
2.3. OsRLK7-1 Negatively Regulates MPKs and WRKYs
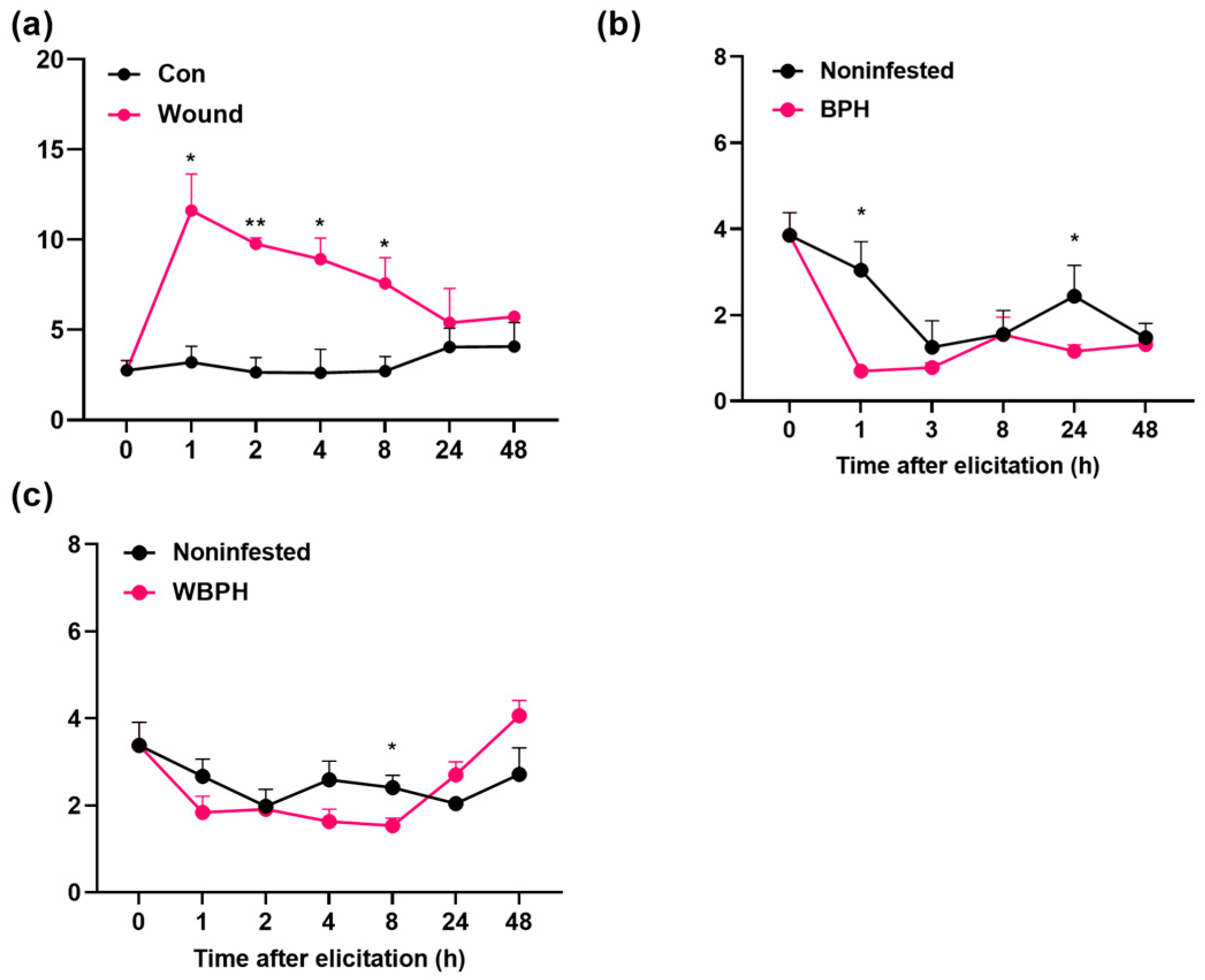
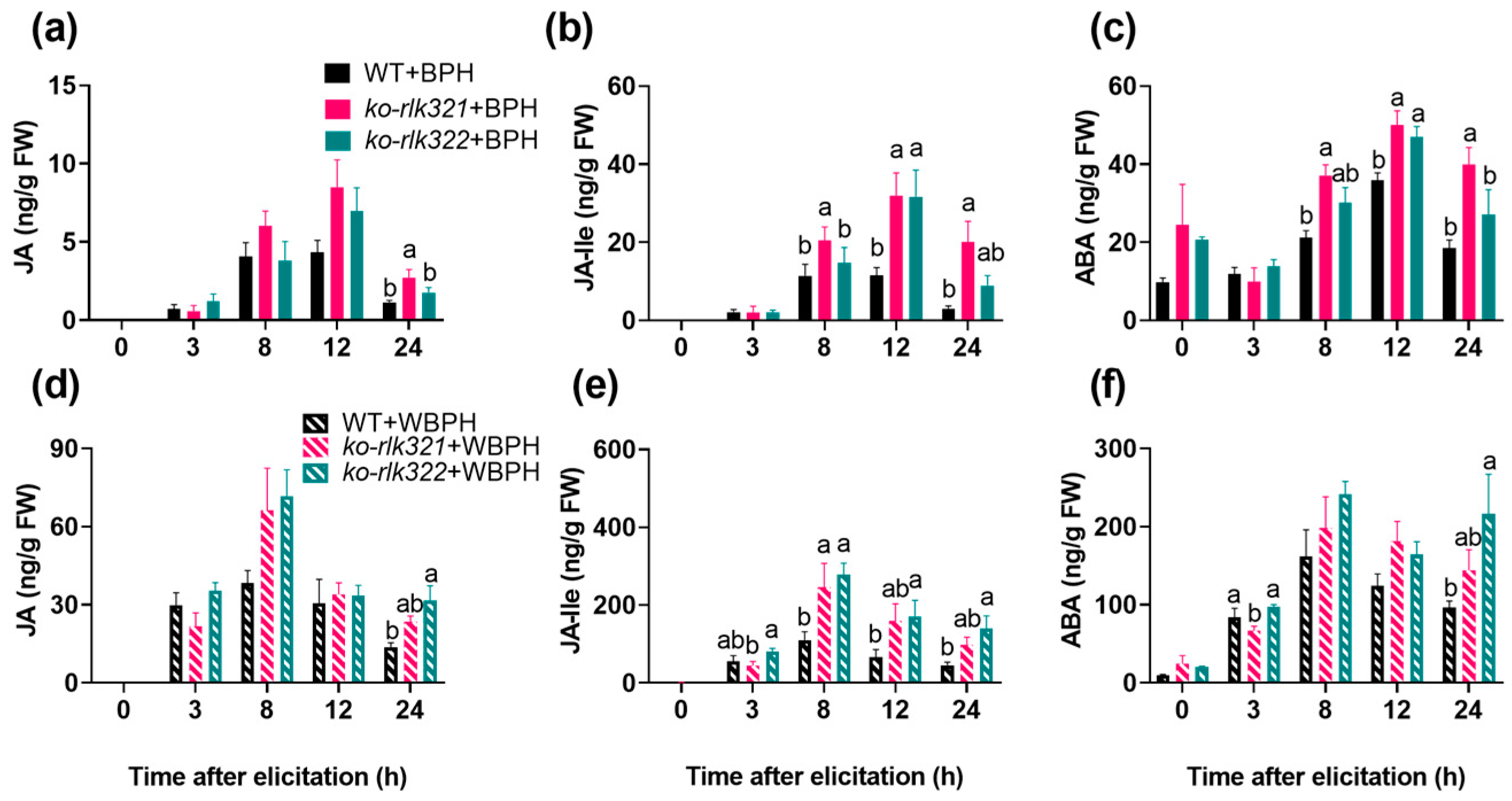
2.4. OsRLK7-1 Negatively Regulates the Biosynthesis of Induced JA, JA-Ile, and ABA
2.5. Knocking Out OsRLK7-1 Enhances the Phenolamide Synthesis of Planthopper-Induced Rice
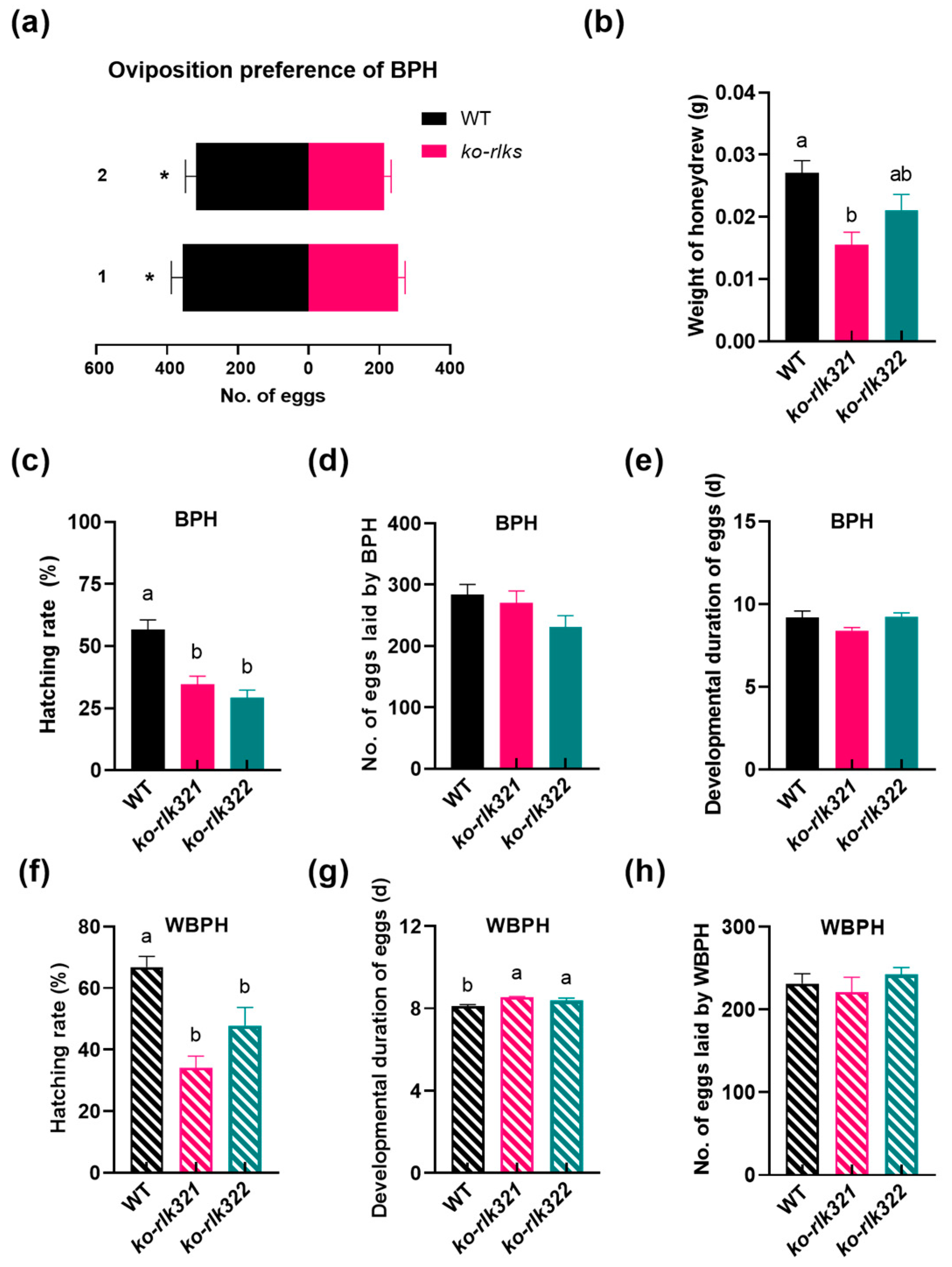
2.6. Knocking Out OsRLK7-1 Enhances the Resistance of Rice to BPH and WBPH
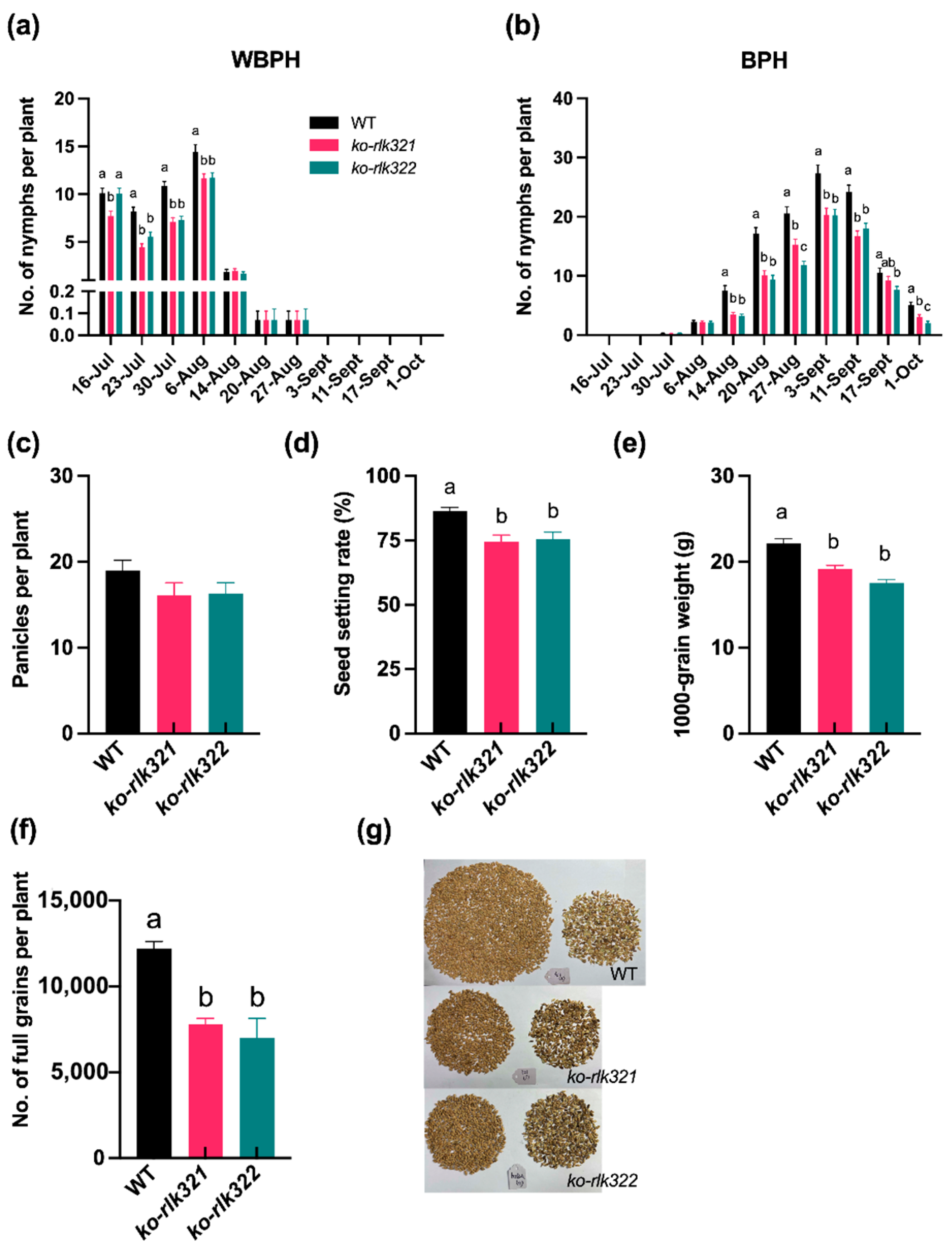
2.7. Knocking Out OsRLK7-1 Decreases the Population Density of BPH and WBPH and Rice Yield in the Field
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Insects
4.2. Plant Treatment
4.3. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
4.4. Cloning of OsRLK7-1 and Sequence Analysis
4.5. Subcellular Localization Assay
4.6. Generation of Rice Lines with OsRLK7-1 Knocked Out
4.7. BPH and WBPH Bioassays
4.8. Phytohormone Analysis
4.9. Phenolamides Measurements
4.10. Field Experiments
4.11. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dvorak, P.; Krasylenko, Y.; Zeiner, A.; Samaj, J.; Takac, T. Signaling toward reactive oxygen species-scavenging enzymes in plants. Front. Plant Sci. 2020, 11, 618835. [Google Scholar] [CrossRef]
- Lin, F.; Li, S.; Wang, K.; Tian, H.; Gao, J.; Zhao, Q.; Du, C. A leucine-rich repeat receptor-like kinase, OsSTLK, modulates salt tolerance in rice. Plant Sci. 2020, 296, 110465. [Google Scholar] [CrossRef]
- Li, C.; Xu, M.; Cai, X.; Han, Z.; Si, J.; Chen, D. Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs. Int. J. Mol. Sci. 2022, 23, 3945. [Google Scholar] [CrossRef]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional roles in stress tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef]
- Fischer, I.; Diévart, A.; Droc, G.; Dufayard, J.; Chantret, N. Evolutionary dynamics of the leucine-rich repeat receptor-like kinase (LRR-RLK) subfamily in Angiosperms. Plant Physiol. 2016, 170, 1595–1610. [Google Scholar] [CrossRef]
- Wu, Y.; Xun, Q.; Guo, Y.; Zhang, J.; Cheng, K.; Shi, T.; He, K.; Hou, S.; Gou, X.; Li, J. Genome-wide expression pattern analyses of the Arabidopsis leucine-rich repeat receptor-like kinases. Mol. Plant 2016, 9, 289–300. [Google Scholar] [CrossRef]
- Dufayard, J.F.; Bettembourg, M.; Fischer, I.; Droc, G.; Guiderdoni, E.; Perin, C.; Chantret, N.; Dievart, A. New insights on leucine-rich repeats receptor-like kinase orthologous relationships in Angiosperms. Front. Plant Sci. 2017, 8, 381. [Google Scholar] [CrossRef]
- Chen, T. Identification and characterization of the LRR repeats in plant LRR-RLKs. BMC Mol. Cell Biol. 2021, 22, 9. [Google Scholar] [CrossRef]
- Li, X.; Ahmad, S.; Guo, C.; Yu, J.; Cao, S.; Gao, X.; Li, W.; Li, H.; Guo, Y. Identification and characterization of LRR-RLK family genes in potato reveal their involvement in peptide signaling of cell fate decisions and biotic/abiotic stress responses. Cells 2018, 7, 120. [Google Scholar] [CrossRef]
- Chen, D.; Guo, H.; Chen, S.; Yue, Q.; Wang, P.; Chen, X. Receptor-like kinase HAESA-like 1 positively regulates seed longevity in Arabidopsis. Planta 2022, 256, 21. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.; Lease, K.; Doke, J.; Tax, F.; Walker, J. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liang, C.; Hou, S.; Wang, X.; Chen, D.; Shen, J.; Zhang, W.; Wang, M. The LRR-RLK protein HSL3 regulates stomatal closure and the drought stress response by modulating hydrogen peroxide homeostasis. Front. Plant Sci. 2020, 11, 548034. [Google Scholar] [CrossRef]
- Ouyang, S.Q.; Liu, Y.F.; Liu, P.; Lei, G.; He, S.J.; Ma, B.; Zhang, W.K.; Zhang, J.S.; Chen, S.Y. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010, 62, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Liu, X.; Wang, Q.; Chen, Y.; Liu, C.; Qiu, Y.; Zhang, W. OsRPK1, a novel leucine-rich repeat receptor-like kinase, negatively regulates polar auxin transport and root development in rice. Biochim. Biophys. Acta 2014, 1840, 1676–1685. [Google Scholar] [CrossRef] [PubMed]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tor, M.; de Vries, S.; Zipfel, C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to Hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar] [CrossRef]
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.M.; Chai, J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013, 342, 624–628. [Google Scholar] [CrossRef]
- Peng, H.C.; Kaloshian, I. The tomato leucine-rich repeat receptor-like kinases SlSERK3A and SlSERK3B have overlapping functions in bacterial and nematode innate immunity. PLoS ONE 2014, 9, e93302. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L.; Yang, Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta 2005, 222, 107–117. [Google Scholar] [CrossRef]
- Poretsky, E.; Ruiz, M.; Ahmadian, N.; Steinbrenner, A.D.; Dressano, K.; Schmelz, E.A.; Huffaker, A. Comparative analyses of responses to exogenous and endogenous antiherbivore elicitors enable a forward genetics approach to identify maize gene candidates mediating sensitivity to herbivore-associated molecular patterns. Plant J. 2021, 108, 1295–1316. [Google Scholar] [CrossRef]
- Cheng, X.; Zhu, L.; He, G. Towards understanding of molecular interactions between rice and the brown planthopper. Mol. Plant 2013, 6, 621–634. [Google Scholar] [CrossRef]
- Muduli, L.; Radhan, S.K.P.; Mishra, A.; Bastia, D.N.; Samal, K.C.; Agrawal, P.K.; Dash, M. Understanding brown planthopper resistance in rice: Genetics, biochemical and molecular breeding approaches. Rice Sci. 2021, 28, 532–546. [Google Scholar] [CrossRef]
- Ye, M.; Kuai, P.; Hu, L.; Ye, M.; Sun, H.; Erb, M.; Lou, Y. Suppression of a leucine-rich repeat receptor-like kinase enhances host plant resistance to a specialist herbivore. Plant Cell Environ. 2020, 43, 2571–2585. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xing, F.; He, Q.; Tahir ul Qamar, M.; Chen, L.-L.; Xing, Y. Conserved imprinted genes between intra-subspecies and inter-subspecies are involved in energy metabolism and seed development in rice. Int. J. Mol. Sci. 2020, 21, 9618. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Han, S.; Zhou, X.; Zhao, C.; Guo, L.; Zhang, J.; Liu, F.; Huo, Q.; Zhao, W.; Guo, Z.; et al. Phosphorylation and ubiquitination of OsWRKY31 are integral to OsMKK10-2-mediated defense responses in rice. Plant Cell 2023, 35, 2391–2412. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Glauser, G.; Lou, Y.; Erb, M.; Hu, L. Molecular dissection of early defense signaling underlying volatile-mediated defense regulation and herbivore resistance in rice. Plant Cell 2019, 31, 687–698. [Google Scholar] [CrossRef]
- Zheng, X.; Fang, A.; Qiu, S.; Zhao, G.; Wang, J.; Wang, S.; Wei, J.; Gao, H.; Yang, J.; Mou, B.; et al. Ustilaginoidea virens secretes a family of phosphatases that stabilize the negative immune regulator OsMPK6 and suppress plant immunity. Plant Cell 2022, 34, 3088–3109. [Google Scholar] [CrossRef]
- Wang, L.; Chen, J.; Zhao, Y.; Wang, S.; Yuan, M. OsMAPK6 phosphorylates a zinc finger protein OsLIC to promote downstream OsWRKY30 for rice resistance to bacterial blight and leaf streak. J. Integr. Plant Biol. 2022, 64, 1116–1130. [Google Scholar] [CrossRef]
- Koo, S.C.; Moon, B.C.; Kim, J.K.; Kim, C.Y.; Sung, S.J.; Kim, M.C.; Cho, M.J.; Cheong, Y.H. OsBWMK1 mediates SA-dependent defense responses by activating the transcription factor OsWRKY33. Biochem. Biophys. Res. Commun. 2009, 387, 365–370. [Google Scholar] [CrossRef]
- Huangfu, J.; Li, J.; Li, R.; Ye, M.; Kuai, P.; Zhang, T.; Lou, Y. The transcription factor OsWRKY45 negatively modulates the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 2016, 17, 697. [Google Scholar] [CrossRef]
- Ye, M.; Song, Y.; Baerson, S.; Long, J.; Wang, J.; Pan, Z.; Lin, W.; Zeng, R. Ratoon rice generated from primed parent plants exhibit enhanced herbivore resistance. Plant Cell Environ. 2017, 40, 779–787. [Google Scholar] [CrossRef]
- Shi, J.; Sun, Z.; Hu, X.; Jin, H.; Foba, C.; Liu, H.; Wang, C.; Liu, L.; Li, F.; Wang, M. Rice defense responses are induced upon leaf rolling by an insect herbivore. BMC Plant Biol. 2019, 19, 514. [Google Scholar] [CrossRef] [PubMed]
- Chavan, S.N.; De Kesel, J.; Desmedt, W.; Degroote, E.; Singh, R.R.; Nguyen, G.T.; Demeestere, K.; De Meyer, T.; Kyndt, T. Dehydroascorbate induces plant resistance in rice against root-knot nematode Meloidogyne graminicola. Mol. Plant Pathol. 2022, 23, 1303–1319. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Sun, Y.; Yan, H.; Li, C.; Ge, F. O3-Induced priming defense associated with the abscisic acid signaling pathway enhances plant resistance to Bemisia tabaci. Front. Plant Sci. 2020, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Gao, Y.; Chen, W.; Wang, W.; Gong, L.; Liu, X.; Luo, J. Spatiotemporal distribution of phenolamides and the genetics of natural variation of hydroxycinnamoyl spermidine in rice. Mol. Plant 2015, 8, 111–121. [Google Scholar] [CrossRef]
- Erb, M.; Reymond, P. Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 2019, 70, 527–557. [Google Scholar] [CrossRef]
- Lyapina, I.; Filippova, A.; Fesenko, I. The role of peptide signals hidden in the structure of functional proteins in plant immune responses. Int. J. Mol. Sci. 2019, 20, 4343. [Google Scholar] [CrossRef]
- Hu, L.; Ye, M.; Kuai, P.; Ye, M.; Erb, M.; Lou, Y. OsLRR-RLK1, an early responsive leucine-rich repeat receptor-like kinase, initiates rice defense responses against a chewing herbivore. New Phytol. 2018, 219, 1097–1111. [Google Scholar] [CrossRef]
- Hou, S.; Wang, X.; Chen, D.; Yang, X.; Wang, M.; Turra, D.; Di Pietro, A.; Zhang, W. The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog. 2014, 10, e1004331. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Boudsocq, M.; Neubauer, J.; Frei, D.F.N.; Leonhardt, N.; Pateyron, S.; Gwinner, F.; Tamby, J.P.; Ortiz-Masia, D.; et al. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 2015, 82, 232–244. [Google Scholar] [CrossRef]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, X.; He, Y.; Sang, T.; Wang, P.; Dai, S.; Zhang, S.; Meng, X. Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis. Plant Cell 2020, 32, 2621–2638. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.; Xu, L.; Wang, Q.; Lou, Y. Expressing OsMPK4 impairs plant growth but enhances the resistance of rice to the striped stem borer Chilo suppressalis. Int. J. Mol. Sci. 2018, 19, 1182. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, C.; Zhang, Y.; Meng, X.; Zhou, X.; Chu, C.; Wang, X. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 2012, 80, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Hu, Y.; Tang, X.; Zhou, P.; Deng, X.; Wang, H.; Guo, Z. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 2012, 236, 1485–1498. [Google Scholar] [CrossRef]
- Hu, L.; Ye, M.; Li, R.; Zhang, T.; Zhou, G.; Wang, Q.; Lu, J.; Lou, Y. The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of mitogen-activated protein kinase activity. Plant Physiol. 2015, 169, 2907–2921. [Google Scholar] [CrossRef]
- Zeng, J.; Zhang, T.; Huangfu, J.; Li, R.; Lou, Y. Both allene oxide synthases genes are involved in the biosynthesis of herbivore-induced jasmonic acid and herbivore resistance in rice. Plants 2021, 10, 442. [Google Scholar] [CrossRef]
- Wang, W.; Yu, Z.; Meng, J.; Zhou, P.; Luo, T.; Zhang, J.; Wu, J.; Lou, Y. Rice phenolamindes reduce the survival of female adults of the white-backed planthopper Sogatella furcifera. Sci. Rep. 2020, 10, 5778. [Google Scholar] [CrossRef]
- Ding, X.; Huang, X.; Sun, L.; Wu, J.; Liu, J. Influence of abscisic acid-biosynthesis inhibitor fluridone on the feeding behavior and fecundity of Nilaparvata lugens. Insects 2019, 10, 57. [Google Scholar] [CrossRef]
- Liu, J.; Du, H.; Ding, X.; Zhou, Y.; Xie, P.; Wu, J. Mechanisms of callose deposition in rice regulated by exogenous abscisic acid and its involvement in rice resistance to Nilaparvata lugens Stål (Hemiptera: Delphacidae). Pest Manag. Sci. 2017, 73, 2559–2568. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, D.; Gao, D.; Zhao, W.; Du, H.; Qiu, Z.; Huang, J.; Wen, P.; Wang, Y.; Li, Q.; et al. Cytokinin confers brown planthopper resistance by elevating jasmonic acid pathway in rice. Int. J. Mol. Sci. 2022, 23, 5946. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, L.; Ding, X.; Fan, W.; Liu, J. Transcriptome analysis reveals crosstalk between the abscisic acid and jasmonic acid signaling pathways in rice-mediated defense against Nilaparvata lugens. Int. J. Mol. Sci. 2022, 23, 6319. [Google Scholar] [CrossRef] [PubMed]
- Alamgir, K.; Hojo, Y.; Christeller, J.; Fukumoto, K.; Isshiki, R.; Shinya, T.; Baldwin, I.; Galis, I. Systematic analysis of rice (Oryza sativa) metabolic responses to herbivory. Plant Cell Environ. 2016, 39, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, J.; Li, J.; Zhou, G.; Wang, Q.; Bian, W.; Erb, M.; Lou, Y. Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. eLife 2015, 4, e4805. [Google Scholar] [CrossRef]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y. CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 2017, 10, 1246–1249. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in Monocot and Dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.; Shen, Z.; Gao, Q.; Jin, N.; Lou, Y. Knocking Out OsRLK7-1 Impairs Rice Growth and Development but Enhances Its Resistance to Planthoppers. Int. J. Mol. Sci. 2023, 24, 14569. https://doi.org/10.3390/ijms241914569
Han S, Shen Z, Gao Q, Jin N, Lou Y. Knocking Out OsRLK7-1 Impairs Rice Growth and Development but Enhances Its Resistance to Planthoppers. International Journal of Molecular Sciences. 2023; 24(19):14569. https://doi.org/10.3390/ijms241914569
Chicago/Turabian StyleHan, Shanjie, Zhifan Shen, Qing Gao, Nuo Jin, and Yonggen Lou. 2023. "Knocking Out OsRLK7-1 Impairs Rice Growth and Development but Enhances Its Resistance to Planthoppers" International Journal of Molecular Sciences 24, no. 19: 14569. https://doi.org/10.3390/ijms241914569
APA StyleHan, S., Shen, Z., Gao, Q., Jin, N., & Lou, Y. (2023). Knocking Out OsRLK7-1 Impairs Rice Growth and Development but Enhances Its Resistance to Planthoppers. International Journal of Molecular Sciences, 24(19), 14569. https://doi.org/10.3390/ijms241914569






