Recent Advances in High-Content Imaging and Analysis in iPSC-Based Modelling of Neurodegenerative Diseases
Abstract
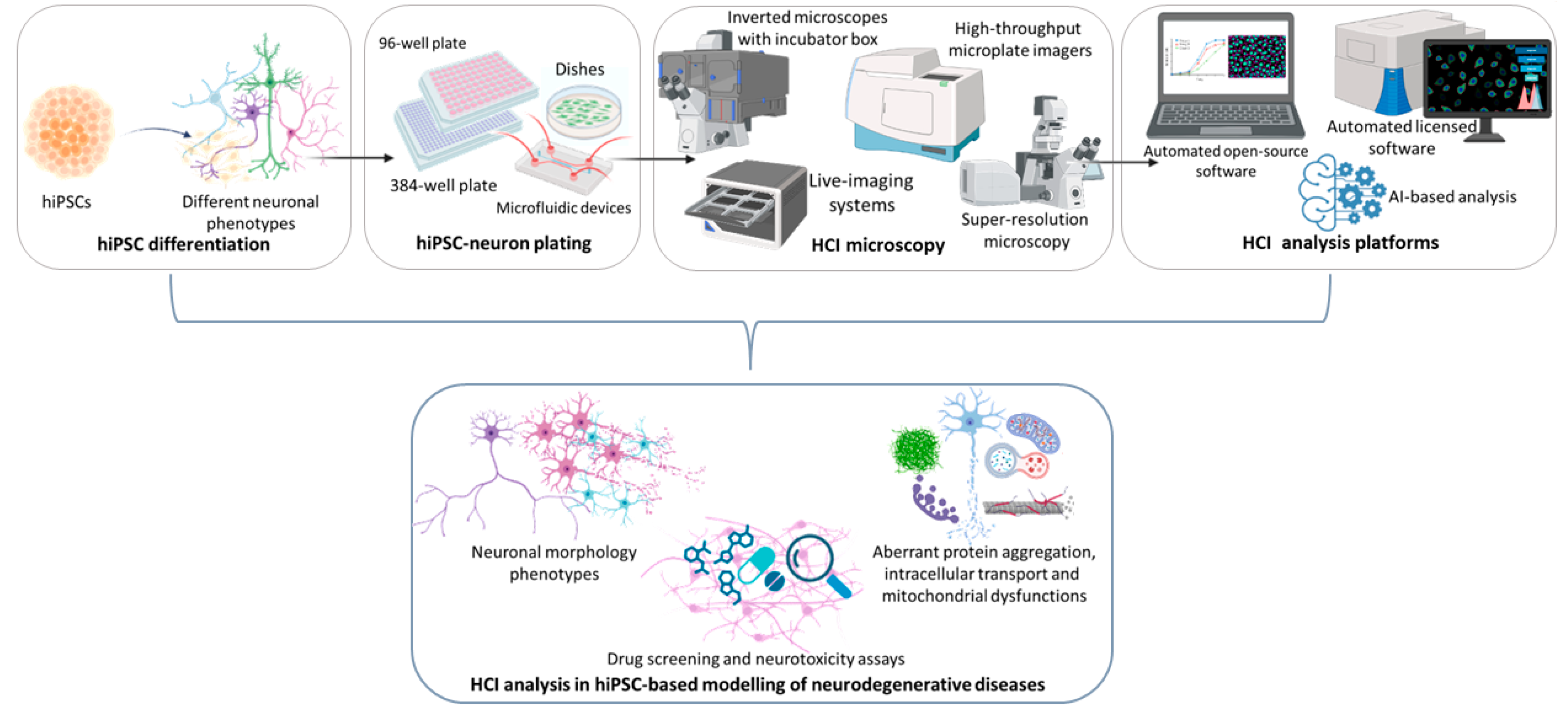
:1. Introduction
Most Common Microscope Settings and Platform Analysis in iPSC-Based Neuronal Models
|
Open-Source Analysis Software Advantages: Open-Source, Enabling Single-Cell Tracing and Measurements, Free Plug-Ins Download for Several Cellular Analysis Types, Feasible for Custom-Made Algorithm Analysis or Macros | ||||
|---|---|---|---|---|
| Analysis Platform Software | HCI Analysis | Plug-In and Tools | Microscopes (Used in the Mentioned References) | References |
| CellProfiler (automated) (https://cellprofiler.org/, accessed on 24 September 2023) | -neurite outgrowth | -Software analysis pipeline: https://doi.org/10.5281/zenodo.6642365 (accessed on 24 September 2023) | -Operetta CLS High-Content Analysis System—with Harmony software (PerkinElmer, Waltham, U.S.) | -CellProfiler software: [31]; -[32] |
| -mitochondrial fitness, neuronal toxicity quantification of neuronal branching complexity | -Software analysis pipeline: https://github.com/StemCellMetab/Mitochondrial-membranepotential (accessed on 24 September 2023) | -Operetta CLS High-Content Analysis System with Harmony software (PerkinElmer, Waltham, U.S.) | -[33,34] | |
| -mitochondrial function, morphology and cell viability | -Software analysis pipeline: automated synaptic imaging assay (ASIA), https://github.com/thayerlab/ASIA-pipelines scripts written (accessed on 24 September 2023) | -Opera High-Content Screening System, (live imaging) (PerkinElmer, Waltham, U.S.) | -[35] | |
| -discrimination in synaptic density changes | -Nikon, Tokyo, Japan A1 confocal microscope (Nikon, Tokyo, Japan) | -[36] | ||
| ImageJ (semi-automated) (https://imagej.nih.gov/ij/download.html, accessed on 24 September 2023) | -Neurite outgrowth, growth cone, axonal swellings | -ImageJ-NeuronJ plug-in -ImageJ-Neurite tracer macro | -Axioplan2 (Carl Zeiss AG, Oberkochen, Germany), LSM-710 (Carl Zeiss AG, Oberkochen, Germany), BZ9000 (Keyence, Itasca, U.S.), or IN Cell Analyzer 6000 (GE Healthcare, Chicago, U.S.) | -ImageJ software: [24,37]; -NeuronJ plug-in: [38]; -Neurite tracer macro: [39]; -[40] |
| -Neurite outgrowth, axon degeneration index; protein aggregates automated quantification | -ROI manager tool, Threshold function, analyze particles plug-in; cell counter plug-in -Image Mining: custom-made image processing and analysis application with an extendable “plug-in” infrastructure (based on data mining, AI, machine learning, image retrieval, image processing, computer vision and database) | -Opera High-Content Screening System (PerkinElmer, Waltham, U.S.) | -Axon degeneration index: [41,42,43]; -Image mining: [44]; -[26] | |
| -Motility of fluorescently labeled organelle and neurite number quantification | -Pairwise Stitching plug-in; Simple Neurite Tracer plug-in with Sholl Analysis; -segmented line and ROI manger tool; -Multiple Kymograph plug-in; -Custom MATLAB GUI (Kymograph Suite) (Manually tracing of individual organelles) | -UltraView Vox Spinning Disk Confocal system (PerkinElmer, Waltham, U.S.) with a Nikon Eclipse Ti inverted microscope (Nikon, Tokyo, Japan); inverted DMI6000B microscope (Leica Microsystems, Wetzlar, Germany) using LAS-X software (Leica Microsystems, Wetzlar, Germany). | -Pairwise Stitching: [45]; -Sholl Analysis: [46,47]; -[48] | |
| -Membrane trafficking | -Reslice function (Kymograph construction) (https://imagej.nih.gov/ij/plugins/radial-reslice/index.html, accessed on 24 September 2023) | -Incucyte SX1 live-cell analysis system (Sartorius, Göttingen, Germany); Nikon, Tokyo, Japan Eclipse Ti microscope (with optical autofocus system and a motorized piezo stage) spinning disk microscope (Nikon, Tokyo, Japan) (real-time quantitative live imaging); -Andor Ixon Ultra (EM-CCD) camera and the MetaMorph software imaging system (Molecular Devices, San Jose, U.S.); | -[49] | |
| -Neuronal local neuronal secretory system | -Custom-made macro intracellular for quantification of intracellular markers colocalization (%) | -Leica SP8 confocal microscope and a LASX imaging system (Leica Microsystems, Wetzlar, Germany). | -[50] | |
| -Discrimination in synaptic density changes | -Software analysis pipeline:automated synaptic imaging assay (ASIA), https://github.com/thayerlab/ASIA-pipelines scripts written (accessed on 24 September 2023) | -Nikon Eclipse Ti-E inverted confocal microscope and the NIS Element software (Nikon, Tokyo, Japan) + Carl Zeiss LSM 880 AiryScan confocal microscope and the Zen Black 2.3 software, within the AiryScan super-resolution mode (Carl Zeiss AG, Oberkochen, Germany) | -[36] | |
| -Axonal outgrowth and muscle maturation | -ImageJ macro for calculating pillar deflection: Method A: Supplementary Data 4 of [15]. Method B: Supplementary Data 5 of [15]. | -Nikon A1 confocal microscope controlled with the JOBS module of Nikon Elements software (Nikon, Tokyo, Japan) -Zeiss, Axiovert 200 (Phase-contrast) (Carl Zeiss AG, Oberkochen, Germany); -Olympus, model no. FV-1000 (Confocal laser microscope with motorized stage) (Olympus, Tokyo, Japan) Tokai Hit, INUG2F-ZM (Tokai Hit, Fujinomiya, Japan) (Phase-contrast and fluorescent microscope with a stage-top incubator) | -[15] | |
| -Autophagy LC3-based assay | -Customed R script (https://www.r-project.org/; accessed on 24 September 2023) version 3.5.2 and data processing with Bioconductor R package cellHTS2 (https://www.bioconductor.org/packages//2.7/bioc/html/cellHTS2.html, accessed on 24 September 2023) Coloc2 plug-in for Fiji (providing Pearson’s R correlation) (https://imagej.net/Coloc2; accessed on 24 September 2023); | -Opera Phenix High-Content screening System with Harmony software (PerkinElmer, Waltham, U.S.) | -[51] | |
| -Intracellular transport | -plusTipTracker software (for microtubule dynamics video quantification) | -Olympus Inverted FV1000 confocal microscope (Olympus, Tokyo, Japan); -STED imaging was performed on a custom built, dual color, beam scanning system; -Leica SP5 microscope equipped with a controlled environment chamber (Leica Microsystems, Wetzlar, Germany). | -plusTipTracker: [52]; -[53] | |
| Licensed Analysis Software Advantages: Allowed with Licensed Microscopes, Powerful Image Analysis Capabilities with Highly Flexible and Easy-to-Use Building Blocks to Analyze Simple and Complex Phenotypes of Cells, Automated Cell Tracking, Automated Multiple Segmentation and Co-Localization Analysis, Fast Automated Cell Analysis (Minutes) Enabling Multi-Threaded, Parallel Image Processing, Teachable Interface for Analysis Creation, and Batch Processing for Large Time-Lapse Image Datasets. | ||||
|---|---|---|---|---|
| Analysis Platform Software | HCI Analysis | Building Blocks for Analysis Segmentation and Tools | Required Microscopes | References |
|
-Harmony High-Content Imaging and Analysis Software(PerkinElmer, Waltham, U.S.) (https://www.perkinelmer.com/it/product/harmony-4-8-office-hh17000001, accessed on 24 September 2023) | -Neurite outgrowth and neuron maturation assessment |
-Find nuclei, Find neurites, Calculate Intensity Properties | -Opera PhenixPlus CLS High-Content screening System (PerkinElmer, Waltham, U.S.) with CSIRO Neurite analysis software (https://www.csiro.au/en/research/technology-space/data/neurite-analysis-software, accessed on 24 September 2023) | -[54] |
| -Intracellular protein aggregation | -Find Spot | -Operetta or Opera Phenix CLS High-Content Analysis System (PerkinElmer, Waltham, U.S.) | -[55] | |
| -neurite outgrowth | -nuclear parameters neurite parameters | -Opera CLS High-Content Analysis System (PerkinElmer, Waltham, U.S.) | -[56] | |
|
-Columbus (image data storage and analysis system allowed for connection with Harmony software) (PerkinElmer, Waltham, U.S.) (https://www.perkinelmer.com/it/product/harmony-4-8-office-hh17000001, accessed on 24 September 2023) | -Mitochondrial fitness and neuronal toxicity and quantification of neuronal branching complexity | -Software analysis pipeline: https://github.com/StemCellMetab/Mitochondrial-membrane-potential (accessed on 24 September 2023) | -Operetta CLS High-Content Analysis System (PerkinElmer, Waltham, U.S. | -[33,34] |
| -Autophagy LC3-based assay | -Opera Phenix CLS High-Content screening System (PerkinElmer, Waltham, U.S.) | -[51] | ||
| -MetaMorph Microscopy Automation and Image Analysis Software (Molecular Devices, San Jose, U.S.) Automated (https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy, accessed on 24 September 2023) | -Neurite outgrowth | -Software analysis: https://www.moleculardevices.com/applications/neurite-outgrowth (accessed on 24 September 2023) | -MetaMorph Microscopy Automation and Image Analysis Software (Molecular Devices, San Jose, U.S.) | -[57] |
| -Membrane trafficking | -MetaMorph Microscopy Automation and Image Analysis Software (Molecular Devices, San Jose, U.S.) | -[49] | ||
|
-IN Cell Analyzer 6000 software (GE Healthcare, Chicago, U.S.) | -Neurite outgrowth |
-Segmentation of ROI (Dendrites, cell bodies, and axons.) | -IN Cell Analyzer 6000 high-performance and high-content automated laser-based confocal imaging platform and ImageXpress Micro Confocal High-Content Imaging System (GE Healthcare, Chicago, U.S.) | -[58] |
| -Intracellular protein aggregation | -IN Cell Analyzer 6000 IN Cell Developer Toolbox version 1.9 (GE Healthcare, Chicago, U.S.) | -[59] | ||
| -Cell population assays, fluorescence intensity analysis, neurite length analysis | -IN Cell Analyzer 6000 IN Cell Developer Toolbox version 1.9 (GE Healthcare, Chicago, U.S.) | -[60] | ||
|
-Neuronal classification and outgrowth convolutional neural network analysis (random forest classification using total neurite length, number of cells, and average size of neuronal soma as random classifiers) |
-* Keras/TensorFlow framework (v1.13.1)12 on GTX1080Ti by using CUDA 10.0. scikit-learn (v0.23.2), gradient-weighted class activation mapping (Grad-CAM) and guided Grad-CAM algorithm | -IN Cell Analyzer 6000 high-performance and high-content automated laser-based confocal imaging platform (GE Healthcare, Chicago, U.S.) | -[61,62]; -[27] | |
|
-Image segmentation in individual mitochondria (masking of somatic, axonal, and dendritic mitochondria) | -Cell bodies count and analysis of the number, area, median circularity, and length of mitochondria |
-IN Cell Analyzer 6000 confocal microscope (GE Healthcare, Chicago, U.S.) and GE Developer Toolbox (1.9.2, build 2415) software (GE Healthcare, Chicago, U.S.) | -[63,64] | |
|
-CL-Quant Automated Image Analysis Software (Nikon, Tokyo, Japan) (https://www.nikon.com/company/news/2019/1008_cl-quant_01.html, accessed on 24 September 2023) | -Cell population assays, fluorescence intensity analysis, neurite length analysis | -Nuclei and neurite tracing, fiber objects quantification (neurite lengths) | -BioStation CT (Nikon, Tokyo, Japan) | -[60] |
|
-Cellomics software (Thermo Fisher Scientific, Waltham, U.S.) (https://www.thermofisher.com/it/en/home/brands/thermo-scientific/cellomics.html, accessed on 24 September 2023) | -Cell population assays, fluorescence intensity analysis, neurite length analysis | -Nuclei and neurite tracing, fiber objects quantification (neurite lengths) | -ArrayScan high-content system (Thermo Fisher Scientific, Waltham, U.S.) | -[65] |
| -Imaris (Bitplane, Belfast, UK) (Not requiring specific HCI microscope) (https://www.oxinst.com/search-results?search=IMARIS&businesses=bitplane, accessed on 24 September 2023) | -Membrane trafficking | -Surface function 3D cellular structures reconstruction from different image dataset |
-Andor Ixon Ultra (EM-CCD) camera and the MetaMorph software (Molecular Devices, San Jose, U.S.) imaging system Leica SP8 confocal microscope and a LASX imaging system (Leica Microsystems, Wetzlar, Germany) | -[49] |
| -Axonal outgrowth and muscle maturation | -Zeiss, Axiovert 200 (Phase-contrast) (Carl Zeiss AG, Ober-kochen, Germany); Olympus, model no. FV-1000 (Confocal laser microscope with motorized stage) (Olympus, Tokyo, Japan) with a stage-top incubator (Tokai Hit, INUG2F-ZM, Tokai Hit, Fujinomiya, Japan) (Phase-contrast and fluorescent microscope) | -[15] | ||
2. HCI Analysis of Neuronal Dysmorphogenesis in iPSC-Based Neurodegenerative Diseases Modelling
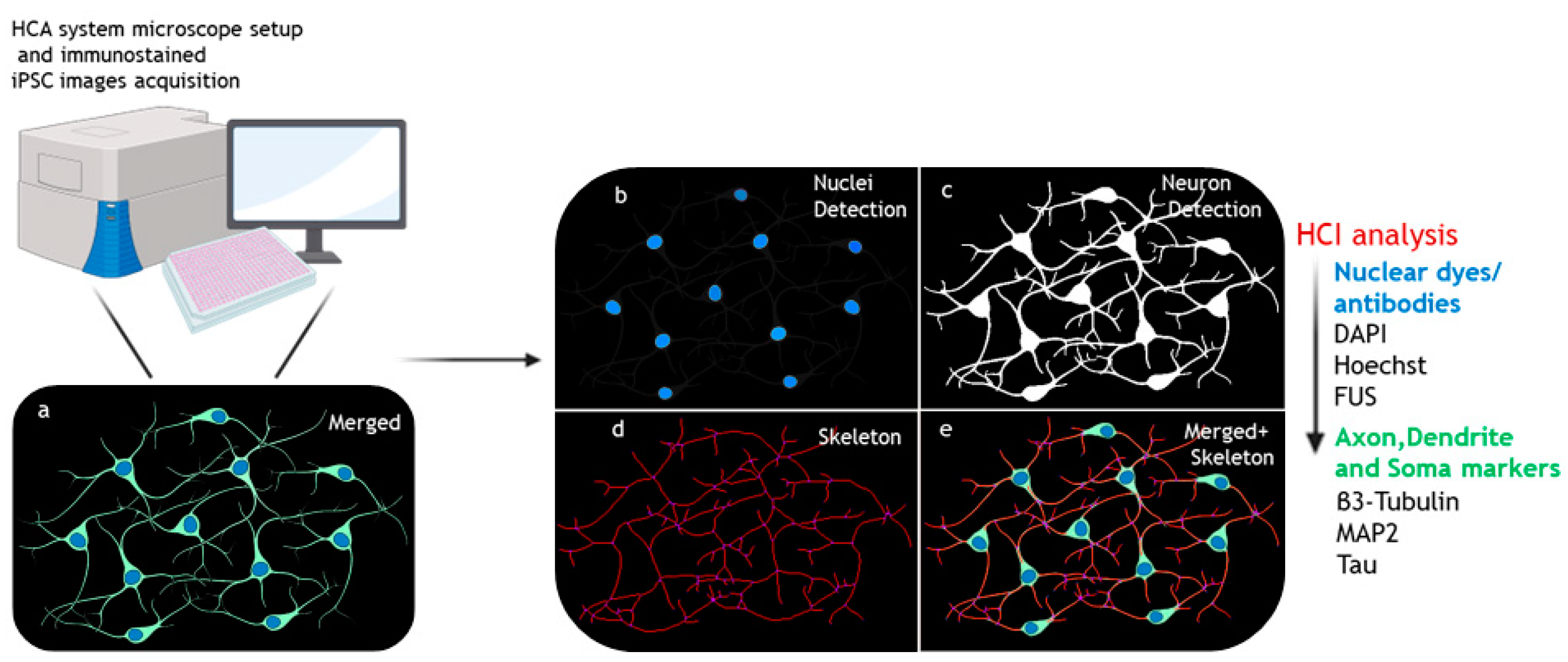
2.1. Experimental Design and Analysis Setting for hiPSC-Based HCI for Neuronal Morphology Phenotypes
2.2. HCI Analysis of Neuronal Morphology Phenotypes in Patient iPSC-Based Neurodegeneration Modelling
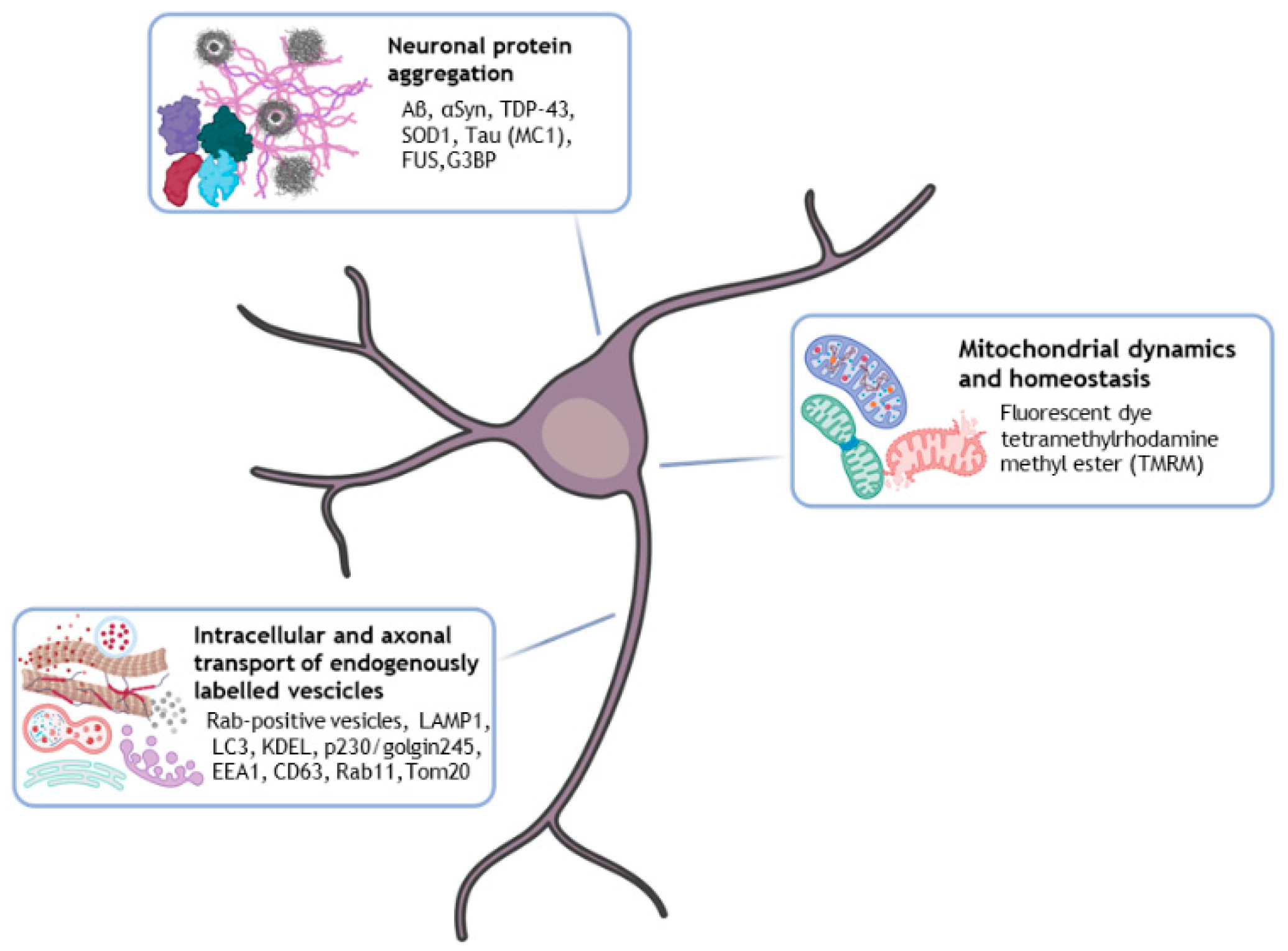
3. HCI Analysis for Aberrant Neuronal Protein Aggregation and Intracellular Transport in iPSC-Based Neurodegenerative Disease Models
3.1. HCI Analysis for Aberrant Neuronal Protein Aggregation in iPSC-Based Neurodegenerative Disease Models
3.2. HCI Analysis of Axonal Transport of Endogenously Labelled Vesicles in Neurodegenerative Patient iPSC-Derived Neurons
3.3. iPSC-Derived Neurons HCI Analysis of Mitochondrial Dynamics and Homeostasis
4. HCI Analysis for Drug Screening and Neurotoxicity Assays in hiPSC-Based Neurodegenerative Disease Modelling
4.1. Experimental Design and Analysis Settings
4.2. Neuromuscular Diseases
4.3. Parkinson’s Disease
4.4. Alzheimer’s Disease
4.5. Neurotoxicity Assays
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bonaventura, G.; Iemmolo, R.; Attaguile, G.A.; La Cognata, V.; Pistone, B.S.; Raudino, G.; D’agata, V.; Cantarella, G.; Barcellona, M.L.; Cavallaro, S. Ipscs: A preclinical drug research tool for neurological disorders. Int. J. Mol. Sci. 2021, 22, 4596. [Google Scholar] [CrossRef]
- Li, L.; Chao, J.; Shi, Y. Modeling neurological diseases using iPSC-derived neural cells: IPSC modeling of neurological diseases. Cell Tissue Res. 2018, 371, 143–151. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, D.; Shang, Y.; Qi, X. BBA—Molecular Basis of Disease Using induced pluripotent stem cell neuronal models to study neurodegenerative diseases. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165431. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Rivetti di Val Cervo, P.; Besusso, D.; Conforti, P.; Cattaneo, E. hiPSCs for predictive modelling of neurodegenerative diseases: Dreaming the possible. Nat. Rev. Neurol. 2021, 17, 381–392. [Google Scholar] [CrossRef]
- Allsopp, T.; Ebneth, A.; Cabrera-Socorro, A. Adapting hPSCs cells to develop therapies for CNS disorders: Potential, facts and challenges. Stem Cell Res. 2019, 41, 101581. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Ting, H.-C.; Liu, C.-A.; Su, H.-L.; Chiou, T.-W.; Lin, S.-Z.; Harn, H.-J.; Ho, T.-J. Induced Pluripotent Stem Cell (iPSC)-Based Neurodegenerative Disease Models for Phenotype Recapitulation and Drug Screening. Molecules 2020, 25, 2000. [Google Scholar] [CrossRef]
- Chory, E.J.; Gretton, D.W.; DeBenedictis, E.A.; Esvelt, K.M. Enabling high-throughput biology with flexible open-source automation. Mol. Syst. Biol. 2021, 17, e9942. [Google Scholar] [CrossRef]
- Dettinger, P.; Kull, T.; Arekatla, G.; Ahmed, N.; Zhang, Y.; Schneiter, F.; Wehling, A.; Schirmacher, D.; Kawamura, S.; Loeffler, D.; et al. Open-source personal pipetting robots with live-cell incubation and microscopy compatibility. Nat. Commun. 2022, 13, 2999. [Google Scholar] [CrossRef]
- Li, L.; Zhou, Q.; Voss, T.C.; Quick, K.L.; LaBarbera, D.V. High-throughput imaging: Focusing in on drug discovery in 3D. Methods 2016, 96, 97–102. [Google Scholar] [CrossRef]
- Xia, X.; Wong, S.T. Concise review: A high-content screening approach to stem cell research and drug discovery. Stem Cells 2012, 30, 1800–1807. [Google Scholar] [CrossRef]
- Kepiro, M.; Varkuti, B.H.; Davis, R.L. High Content, Phenotypic Assays and Screens for Compounds Modulating Cellular Processes in Primary Neurons. Methods Enzymol. 2018, 610, 219–250. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, S.N.; Ceulemans, H.; Boyd, J.D.; Carpenter, A.E. Image-based profiling for drug discovery: Due for a machine-learning upgrade? Nat. Rev. Drug Discov. 2021, 20, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Schikora, J.; Kiwatrowski, N.; Förster, N.; Selbach, L.; Ostendorf, F.; Pallapies, F.; Hasse, B.; Metzdorf, J.; Gold, R.; Mosig, A.; et al. A propagated skeleton approach to high throughput screening of neurite outgrowth for in vitro parkinson’s disease modelling. Cells 2021, 10, 931. [Google Scholar] [CrossRef] [PubMed]
- Osaki, T.; Uzel, S.G.M.; Kamm, R.D. On-chip 3D neuromuscular model for drug screening and precision medicine in neuromuscular disease. Nat. Protoc. 2020, 15, 421–449. [Google Scholar] [CrossRef]
- Trask, O.J. Guidelines for Microplate Selection in High Content Imaging. Methods Mol. Biol. 2018, 1683, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Overland, A.C.; Rauch, J.N.; Oupicka, L.; Rock, D.M.; Appledorn, D.M. Quantitative live-cell analysis for optimization of culture conditions and evaluation of cell health in human induced pluripotent stem cell-derived neurons. IncuCyte 2017, 1–8. Available online: https://www.sartorius.com/en/products/live-cell-imaging-analysis/live-cell-analysis-resources/quantitative-live-cell-analysis-for-optimization-of-culture-conditions-and-evaluation-of-cell-health-in-human-induced-pluripotent-stem-cell-derived-neurons-application-note (accessed on 24 September 2023).
- Hosny, N.A.; Song, M.; Connelly, J.T.; Ameer-Beg, S.; Knight, M.M.; Wheeler, A.P. Super-resolution imaging strategies for cell biologists using a spinning disk microscope. PLoS ONE 2013, 8, e74604. [Google Scholar] [CrossRef]
- Hsiao, Y.-T.; Wu, T.-Y.; Wu, B.-K.; Chu, S.-W.; Hsieh, C.-L. Spinning disk interferometric scattering confocal microscopy captures millisecond timescale dynamics of living cells. Opt. Express 2022, 30, 45233–45245. [Google Scholar] [CrossRef]
- Qin, S.; Isbaner, S.; Gregor, I.; Enderlein, J. Doubling the resolution of a confocal spinning-disk microscope using image scanning microscopy. Nat. Protoc. 2021, 16, 164–181. [Google Scholar] [CrossRef]
- Hamilton, N.A. Open source tools for fluorescent imaging. Methods Enzymol. 2012, 504, 393–417. [Google Scholar] [CrossRef]
- Guiet, R.; Burri, O.; Seitz, A. Open Source Tools for Biological Image Analysis. Methods Mol. Biol. 2019, 2040, 23–37. [Google Scholar] [CrossRef]
- Lucas, A.M.; Ryder, P.V.; Li, B.; Cimini, B.A.; Eliceiri, K.W.; Carpenter, A.E. Open-source deep-learning software for bioimage segmentation. Mol. Biol. Cell 2021, 32, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.B.; Dobson, E.T.A.; Rueden, C.T.; Tomancak, P.; Jug, F.; Eliceiri, K.W. The ImageJ ecosystem: Open-source software for image visualization, processing, and analysis. Protein Sci. 2021, 30, 234–249. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, D.; Yuasa, S.; Fukuda, K. Induced Pluripotent Stem Cell-Based Drug Screening by Use of Artificial Intelligence. Pharmaceuticals 2022, 15, 562. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, N.; Prodromidou, K.; Kouroupi, G.; Boumpoureka, I.; Samiotaki, M.; Panayotou, G.; Xilouri, M.; Kloukina, I.; Stefanis, L.; Grailhe, R.; et al. High content screening and proteomic analysis identify a kinase inhibitor that rescues pathological phenotypes in a patient-derived model of Parkinson’s disease. NPJ Park. Dis. 2022, 8, 15. [Google Scholar] [CrossRef]
- Imamura, K.; Yada, Y.; Izumi, Y.; Morita, M.; Kawata, A.; Arisato, T.; Nagahashi, A.; Enami, T.; Tsukita, K.; Kawakami, H.; et al. Prediction Model of Amyotrophic Lateral Sclerosis by Deep Learning with Patient Induced Pluripotent Stem Cells. Ann. Neurol. 2021, 89, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Botté, A.; Lainé, J.; Xicota, L.; Heiligenstein, X.; Fontaine, G.; Kasri, A.; Rivals, I.; Goh, P.; Faklaris, O.; Cossec, J.C.; et al. Ultrastructural and dynamic studies of the endosomal compartment in down syndrome. Acta Neuropathol. Commun. 2020, 8, 89. [Google Scholar] [CrossRef]
- Hong, S.; Wilton, D.K.; Stevens, B.; Richardson, D.S. Structured Illumination Microscopy for the Investigation of Synaptic Structure and Function. Methods Mol. Biol. 2017, 1538, 155–167. [Google Scholar] [CrossRef]
- Paul, T.C.; Johnson, K.A.; Hagen, G.M. Super-resolution imaging of neuronal structure with structured illumination microscopy. bioRxiv 2023. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [PubMed]
- Lickfett, S.; Menacho, C.; Zink, A.; Telugu, N.S.; Beller, M.; Diecke, S.; Cambridge, S.; Prigione, A. High-content analysis of neuronal morphology in human iPSC-derived neurons. STAR Protoc. 2022, 3, 101567. [Google Scholar] [CrossRef] [PubMed]
- Zink, A.; Conrad, J.; Telugu, N.S.; Diecke, S.; Heinz, A.; Wanker, E.; Priller, J.; Prigione, A. Assessment of Ethanol-Induced Toxicity on iPSC-Derived Human Neurons Using a Novel High-Throughput Mitochondrial Neuronal Health (MNH) Assay. Front. Cell Dev. Biol. 2020, 8, 590540. [Google Scholar] [CrossRef] [PubMed]
- Zink, A.; Haferkamp, U.; Wittich, A.; Beller, M.; Pless, O.; Prigione, A. High-content screening of mitochondrial polarization in neural cells derived from human pluripotent stem cells. STAR Protoc. 2022, 3, 101602. [Google Scholar] [CrossRef] [PubMed]
- Little, D.; Luft, C.; Mosaku, O.; Lorvellec, M.; Yao, Z.; Paillusson, S.; Kriston-Vizi, J.; Gandhi, S.; Abramov, A.Y.; Ketteler, R.; et al. A single cell high content assay detects mitochondrial dysfunction in iPSC-derived neurons with mutations in SNCA. Sci. Rep. 2018, 8, 9033. [Google Scholar] [CrossRef] [PubMed]
- Green, M.V.; Pengo, T.; Raybuck, J.D.; Naqvi, T.; McMullan, H.M.; Hawkinson, J.E.; Marron Fernandez de Velasco, E.; Muntean, B.S.; Martemyanov, K.A.; Satterfield, R.; et al. Automated Live-Cell Imaging of Synapses in Rat and Human Neuronal Cultures. Front. Cell. Neurosci. 2019, 13, 467. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Popko, J.; Fernandes, A.; Brites, D.; Lanier, L.M. Automated analysis of NeuronJ tracing data. Cytom. Part A J. Int. Soc. Anal. Cytol. 2009, 75, 371–376. [Google Scholar] [CrossRef]
- Pool, M.; Thiemann, J.; Bar-or, A.; Fournier, A.E. NeuriteTracer: A novel ImageJ plugin for automated quantification of neurite outgrowth. J. Neurosci. Methods 2008, 168, 134–139. [Google Scholar] [CrossRef]
- Rehbach, K.; Kesavan, J.; Hauser, S.; Ritzenhofen, S.; Jungverdorben, J.; Schüle, R.; Schöls, L.; Peitz, M.; Brüstle, O. Multiparametric rapid screening of neuronal process pathology for drug target identification in HSP patient-specific neurons. Sci. Rep. 2019, 9, 9615. [Google Scholar] [CrossRef]
- Kouroupi, G.; Taoufik, E.; Vlachos, I.S.; Tsioras, K.; Antoniou, N.; Papastefanaki, F.; Chroni-Tzartou, D.; Wrasidlo, W.; Bohl, D.; Stellas, D.; et al. Defective synaptic connectivity and axonal neuropathology in a human iPSC-based model of familial Parkinson’ s disease. Proc. Natl. Acad. Sci. USA 2017, 114, E3679–E3688. [Google Scholar] [CrossRef] [PubMed]
- Kneynsberg, A.; Collier, T.J.; Manfredsson, F.P.; Kanaan, N.M. Quantitative and semi-quantitative measurements of axonal degeneration in tissue and primary neuron cultures. J. Neurosci. Methods 2016, 266, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Zygogianni, O.; Antoniou, N.; Kalomoiri, M.; Kouroupi, G.; Taoufik, E.; Matsas, R. In Vivo Phenotyping of Familial Parkinson’s Disease with Human Induced Pluripotent Stem Cells: A Proof-of-Concept Study. Neurochem. Res. 2019, 44, 1475–1493. [Google Scholar] [CrossRef] [PubMed]
- Dorval, T.; Ogier, A.; Genovesio, A.; Lim, H.K.; Kwon, D.Y.; Lee, J.; Worman, H.J.; Dauer, W.; Grailhe, R. Contextual Automated 3D Analysis of Subcellular Organelles Adapted to High-Content Screening. J. Biomol. Screen. 2010, 15, 847–857. [Google Scholar] [CrossRef]
- Preibisch, S.; Saalfeld, S.; Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 2009, 25, 1463–1465. [Google Scholar] [CrossRef]
- Ferreira, T.A.; Blackman, A.V.; Oyrer, J.; Jayabal, S.; Chung, A.J.; Watt, A.J.; Sjöström, P.J.; van Meyel, D.J. Neuronal morphometry directly from bitmap images. Nat. Methods 2014, 11, 982–984. [Google Scholar] [CrossRef]
- Longair, M.H.; Baker, D.A.; Armstrong, J.D. Simple Neurite Tracer: Open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 2011, 27, 2453–2454. [Google Scholar] [CrossRef]
- Boecker, C.A.; Olenick, M.A.; Gallagher, E.R.; Ward, M.E.; Holzbaur, E.L.F. ToolBox: Live Imaging of intracellular organelle transport in induced pluripotent stem cell-derived neurons. Traffic 2020, 21, 138–155. [Google Scholar] [CrossRef]
- Wang, J.; Daniszewski, M.; Hao, M.M.; Hernández, D.; Pébay, A.; Gleeson, P.A.; Fourriere, L. Organelle mapping in dendrites of human iPSC-derived neurons reveals dynamic functional dendritic Golgi structures. Cell Rep. 2023, 42, 112709. [Google Scholar] [CrossRef]
- Roselli, S.; Satir, T.M.; Camacho, R.; Fruhwürth, S.; Bergström, P.; Zetterberg, H.; Agholme, L. APP-BACE1 Interaction and Intracellular Localization Regulate Aβ Production in iPSC-Derived Cortical Neurons. Cell. Mol. Neurobiol. 2023, 43, 3653–3668. [Google Scholar] [CrossRef]
- Papandreou, A.; Luft, C.; Barral, S.; Kriston-Vizi, J.; Kurian, M.A.; Ketteler, R. Automated high-content imaging in iPSC-derived neuronal progenitors. SLAS Discov. 2023, 28, 42–51. [Google Scholar] [CrossRef]
- Applegate, K.T.; Besson, S.; Matov, A.; Bagonis, M.H.; Jaqaman, K.; Danuser, G. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 2011, 176, 168–184. [Google Scholar] [CrossRef] [PubMed]
- Paonessa, F.; Evans, L.D.; Solanki, R.; Larrieu, D.; Wray, S.; Hardy, J.; Jackson, S.P.; Livesey, F.J. Microtubules Deform the Nuclear Membrane and Disrupt Nucleocytoplasmic Transport in Tau-Mediated Frontotemporal Dementia. Cell Rep. 2019, 26, 582–593.e5. [Google Scholar] [CrossRef] [PubMed]
- Wali, G.; Li, Y.; Abu-Bonsrah, D.; Kirik, D.; Parish, C.L.; Sue, C.M. Generation of human-induced pluripotent-stem-cell-derived cortical neurons for high-throughput imaging of neurite morphology and neuron maturation. STAR Protoc. 2023, 4, 102325. [Google Scholar] [CrossRef] [PubMed]
- Manos, J.D.; Preiss, C.N.; Venkat, N.; Tamm, J.; Reinhardt, P.; Kwon, T.; Wu, J.; Winter, A.D.; Jahn, T.R.; Yanamandra, K.; et al. Uncovering specificity of endogenous TAU aggregation in a human iPSC-neuron TAU seeding model. iScience 2022, 25, 103658. [Google Scholar] [CrossRef]
- Sherman, S.P.; Bang, A.G. High-throughput screen for compounds that modulate neurite growth of human induced pluripotent stem cell-derived neurons. DMM Dis. Model. Mech. 2018, 11, dmm031906. [Google Scholar] [CrossRef]
- Chang, K.H.; Lee-Chen, G.J.; Huang, C.C.; Lin, J.L.; Chen, Y.J.; Wei, P.C.; Lo, Y.S.; Yao, C.F.; Kuo, M.W.; Chen, C.M. Modeling Alzheimer’s Disease by Induced Pluripotent Stem Cells Carrying APP D678H Mutation. Mol. Neurobiol. 2019, 56, 3972–3983. [Google Scholar] [CrossRef]
- Bassil, R.; Shields, K.; Granger, K.; Zein, I.; Ng, S.; Chih, B. Improved modeling of human AD with an automated culturing platform for iPSC neurons, astrocytes and microglia. Nat. Commun. 2021, 12, 5220. [Google Scholar] [CrossRef]
- Kondo, T.; Ebinuma, I.; Tanaka, H.; Nishikawa, Y.; Komiya, T.; Ishikawa, M.; Okano, H. Rapid and Robust Multi-Phenotypic Assay System for ALS Using Human iPS Cells with Mutations in Causative Genes. Int. J. Mol. Sci. 2023, 24, 6987. [Google Scholar] [CrossRef]
- Fujimori, K.; Ishikawa, M.; Otomo, A.; Atsuta, N.; Nakamura, R.; Akiyama, T.; Hadano, S.; Aoki, M.; Saya, H.; Sobue, G.; et al. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat. Med. 2018, 24, 1579–1589. [Google Scholar] [CrossRef]
- Philbrick, K.A.; Kline, T.L.; Weston, A.D.; Erickson, B.J.; Ka, P.; Yoshida, K.; Inoue, D. What Does Deep Learning See? Insights From a Classifier Trained. Am. J. Roentgenol. 2018, 211, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Rampasek, L.; Goldenberg, A. TensorFlow: Biology’s Gateway to Deep Learning? Cell Syst. 2016, 2, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Varkuti, B.H.; Kepiro, M.; Liu, Z.; Vick, K.; Avchalumov, Y.; Pacifico, R.; MacMullen, C.M.; Kamenecka, T.M.; Puthanveettil, S.V.; Davis, R.L. Neuron-based high-content assay and screen for CNS active mitotherapeutics. Sci. Adv. 2020, 6, eaaw8702. [Google Scholar] [CrossRef] [PubMed]
- MacMullen, C.; Davis, R.L. High-Throughput Phenotypic Assay for Compounds That Influence Mitochondrial Health Using iPSC-Derived Human Neurons. SLAS Discov. 2021, 26, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ward, M.E.; Chen, R.; Liu, K.; Tracy, T.E.; Chen, X.; Xie, M.; Sohn, P.D.; Ludwig, C.; Meyer-Franke, A.; et al. Scalable Production of iPSC-Derived Human Neurons to Identify Tau-Lowering Compounds by High-Content Screening. Stem Cell Rep. 2017, 9, 1221–1233. [Google Scholar] [CrossRef]
- Okano, H.; Morimoto, S. iPSC-based disease modeling and drug discovery in cardinal neurodegenerative disorders. Cell Stem Cell 2022, 29, 189–208. [Google Scholar] [CrossRef]
- Ross, C.A.; Akimov, S.S. Human-induced pluripotent stem cells: Potential for neurodegenerative diseases. Hum. Mol. Genet. 2014, 23, R17–R26. [Google Scholar] [CrossRef]
- Li, J.; Fraenkel, E. Phenotyping Neurodegeneration in Human iPSCs. Annu. Rev. Biomed. Data Sci. 2021, 4, 83–100. [Google Scholar] [CrossRef]
- Lin, H.-C.; He, Z.; Ebert, S.; Schörnig, M.; Santel, M.; Nikolova, M.T.; Weigert, A.; Hevers, W.; Kasri, N.N.; Taverna, E.; et al. NGN2 induces diverse neuron types from human pluripotency. Stem Cell Rep. 2021, 16, 2118–2127. [Google Scholar] [CrossRef]
- Kusena, J.W.T.; Shariatzadeh, M.; Thomas, R.J.; Wilson, S.L. Understanding cell culture dynamics: A tool for defining protocol parameters for improved processes and efficient manufacturing using human embryonic stem cells. Bioengineered 2021, 12, 979–996. [Google Scholar] [CrossRef]
- Connolly, N.M.C.; Theurey, P.; Adam-Vizi, V.; Bazan, N.G.; Bernardi, P.; Bolaños, J.P.; Culmsee, C.; Dawson, V.L.; Deshmukh, M.; Duchen, M.R.; et al. Guidelines on experimental methods to assess mitochondrial dysfunction in cellular models of neurodegenerative diseases. Cell Death Differ. 2018, 25, 542–572. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Wirthensohn, D.C.; Flagmeier, P.; Hughes, C.; Aprile, F.A.; Ruggeri, F.S.; Whiten, D.R.; Emin, D.; Xia, Z.; Varela, J.A.; et al. Different soluble aggregates of Aβ42 can give rise to cellular toxicity through different mechanisms. Nat. Commun. 2019, 10, 1541. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.M.; Cookson, M.R.; Van Den Bosch, L.; Zetterberg, H.; Holtzman, D.M.; Dewachter, I. Hallmarks of neurodegenerative diseases. Cell 2023, 186, 693–714. [Google Scholar] [CrossRef] [PubMed]
- Vuidel, A.; Cousin, L.; Weykopf, B.; Haupt, S.; Hanifehlou, Z.; Wiest-Daesslé, N.; Segschneider, M.; Lee, J.; Kwon, Y.J.; Peitz, M.; et al. High-content phenotyping of Parkinson’s disease patient stem cell-derived midbrain dopaminergic neurons using machine learning classification. Stem Cell Rep. 2022, 17, 2349–2364. [Google Scholar] [CrossRef]
- Trist, B.G.; Fifita, J.A.; Hogan, A.; Grima, N.; Smith, B.; Troakes, C.; Vance, C.; Shaw, C.; Al-Sarraj, S.; Blair, I.P.; et al. Co-deposition of SOD1, TDP-43 and p62 proteinopathies in ALS: Evidence for multifaceted pathways underlying neurodegeneration. Acta Neuropathol. Commun. 2022, 10, 122. [Google Scholar] [CrossRef]
- Miller, S.J.; Wray, S.; Sattler, R.; Zhang, C. Editorial: Mechanisms of Action in Neurodegenerative Proteinopathies. Front. Neurosci. 2022, 16, 968994. [Google Scholar] [CrossRef]
- Ciccocioppo, F.; Bologna, G.; Ercolino, E.; Pierdomenico, L.; Simeone, P.; Lanuti, P.; Pieragostino, D.; Del Boccio, P.; Marchisio, M.; Miscia, S. Neurodegenerative diseases as proteinopathies-driven immune disorders. Neural Regen. Res. 2020, 15, 850–856. [Google Scholar] [CrossRef]
- Guo, W.; Stoklund Dittlau, K.; Van Den Bosch, L. Axonal transport defects and neurodegeneration: Molecular mechanisms and therapeutic implications. Semin. Cell Dev. Biol. 2020, 99, 133–150. [Google Scholar] [CrossRef]
- Mignogna, M.L.; D’Adamo, P. Critical importance of RAB proteins for synaptic function. Small GTPases 2018, 9, 145–157. [Google Scholar] [CrossRef]
- Kaech, S.; Banker, G. Culturing hippocampal neurons. Nat. Protoc. 2006, 1, 2406–2415. [Google Scholar] [CrossRef]
- Chen, M.; Xu, L.; Wu, Y.; Soba, P.; Hu, C. The organization and function of the Golgi apparatus in dendrite development and neurological disorders. Genes Dis. 2023, 10, 2425–2442. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S. Proximity Ligation Assay (PLA). Methods Mol. Biol. 2022, 2422, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Antón-Fernández, A.; Vallés-Saiz, L.; Avila, J.; Hernández, F. Neuronal nuclear tau and neurodegeneration. Neuroscience 2023, 518, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Stanga, S.; Caretto, A.; Boido, M.; Vercelli, A. Mitochondrial Dysfunctions: A Red Thread across Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 3719. [Google Scholar] [CrossRef] [PubMed]
- Menduti, G.; Rasà, D.M.; Stanga, S.; Boido, M. Drug Screening and Drug Repositioning as Promising Therapeutic Approaches for Spinal Muscular Atrophy Treatment. Front. Pharmacol. 2020, 11, 592234. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.; Arnhold, S.; Addicks, K.; Doerfler, W. Staurosporine is a potent activator of neuronal, glial, and “CNS stem cell-like” neurosphere differentiation in murine embryonic stem cells. Mol. Cell. Neurosci. 2003, 23, 669–680. [Google Scholar] [CrossRef]
- Paul, K.C.; Krolewski, R.C.; Lucumi Moreno, E.; Blank, J.; Holton, K.M.; Ahfeldt, T.; Furlong, M.; Yu, Y.; Cockburn, M.; Thompson, L.K.; et al. A pesticide and iPSC dopaminergic neuron screen identifies and classifies Parkinson-relevant pesticides. Nat. Commun. 2023, 14, 2803. [Google Scholar] [CrossRef]
- Lukonin, I.; Zinner, M.; Liberali, P. Organoids in image-based phenotypic chemical screens. Exp. Mol. Med. 2021, 53, 1495–1502. [Google Scholar] [CrossRef]
- Lampart, F.L.; Iber, D.; Doumpas, N. Organoids in high-throughput and high-content screenings. Front. Chem. Eng. 2023, 5, 1120348. [Google Scholar] [CrossRef]
- Ramm, S.; Vary, R.; Gulati, T.; Luu, J.; Cowley, K.J.; Janes, M.S.; Radio, N.; Simpson, K.J. High-Throughput Live and Fixed Cell Imaging Method to Screen Matrigel-Embedded Organoids. Organoids 2023, 2, 1–19. [Google Scholar] [CrossRef]
- Gabriel, E.; Albanna, W.; Pasquini, G.; Ramani, A.; Josipovic, N.; Mariappan, A.; Riparbelli, M.G.; Callaini, G.; Karch, C.M.; Goureau, O.; et al. Generation of iPSC-derived human forebrain organoids assembling bilateral eye primordia. Nat. Protoc. 2023, 18, 1893–1929. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menduti, G.; Boido, M. Recent Advances in High-Content Imaging and Analysis in iPSC-Based Modelling of Neurodegenerative Diseases. Int. J. Mol. Sci. 2023, 24, 14689. https://doi.org/10.3390/ijms241914689
Menduti G, Boido M. Recent Advances in High-Content Imaging and Analysis in iPSC-Based Modelling of Neurodegenerative Diseases. International Journal of Molecular Sciences. 2023; 24(19):14689. https://doi.org/10.3390/ijms241914689
Chicago/Turabian StyleMenduti, Giovanna, and Marina Boido. 2023. "Recent Advances in High-Content Imaging and Analysis in iPSC-Based Modelling of Neurodegenerative Diseases" International Journal of Molecular Sciences 24, no. 19: 14689. https://doi.org/10.3390/ijms241914689
APA StyleMenduti, G., & Boido, M. (2023). Recent Advances in High-Content Imaging and Analysis in iPSC-Based Modelling of Neurodegenerative Diseases. International Journal of Molecular Sciences, 24(19), 14689. https://doi.org/10.3390/ijms241914689





