Abstract
The productivity of plants is hindered by unfavorable conditions. To perceive stress signals and to transduce these signals to intracellular responses, plants rely on membrane-bound receptor-like kinases (RLKs). These play a pivotal role in signaling events governing growth, reproduction, hormone perception, and defense responses against biotic stresses; however, their involvement in abiotic stress responses is poorly documented. Plant RLKs harbor an N-terminal extracellular domain, a transmembrane domain, and a C-terminal intracellular kinase domain. The ectodomains of these RLKs are quite diverse, aiding their responses to various stimuli. We summarize here the sub-classes of RLKs based on their domain structure and discuss the available information on their specific role in abiotic stress adaptation. Furthermore, the current state of knowledge on RLKs and their significance in abiotic stress responses is highlighted in this review, shedding light on their role in influencing plant–environment interactions and opening up possibilities for novel approaches to engineer stress-tolerant crop varieties.
1. Introduction
Plants are continuously challenged by biotic and abiotic stresses. Being sessile, they cannot escape from these threats, and they must be able to perceive, process and respond in a timely and efficient manner. Abiotic stresses, such as low or high temperature, drought, high salinity, or the presence of heavy metals, affect their physiology, growth and development, thus hampering agricultural productivity. It is estimated that abiotic stresses lead to more than 50 percent of yield losses in most plant species [,]. Recognition of the stress signal initiates changes at the cellular and molecular levels leading to transcriptional reprogramming which is instrumental for their growth under diverse conditions. Thus, it becomes crucial to understand how different environmental stimuli are perceived and translated into signaling events [,]. Overall, stress response is orchestrated by a plethora of signaling molecules, such as ion channels, transcription factors, protein kinases and phosphatases [,], and secondary messengers such as calcium, cyclic nucleotides or reactive oxygen species (ROS) [,].
It has been well established that protein kinases such as mitogen-activated protein kinases (MAPKs), receptor-like kinases (RLKs), sucrose nonfermenting 1-related protein kinases, and calcium-dependent protein kinases, play a vital role in modulating plant growth and development during abiotic stress []. Eukaryotic protein kinases (ePKs) can phosphorylate serine, threonine or tyrosine residues of their substrates, thus changing their activities. [,]. ePKs are a highly complex superfamily consisting of 1.5–2.5% of all eukaryotic genes []. Kinome is the term used to describe the complete set of protein kinases encoded in a genome []. In plants, RLKs form one of the largest gene families that share structural similarity with animal receptor tyrosine kinases [], and their closest homologs are Drosophila melanogaster Pelle [] and mammalian interleukin receptor-associated kinases [,]. A typical RLK harbors a highly variable extracellular domain for ligand binding, a transmembrane domain and a cytoplasmic kinase domain for signal propagation [,]. RLKs play a pivotal role in sensing developmental cues, mediating plant growth, reproduction, stomatal patterning, pollen tube guidance, symbiosis, hormone signal transduction [,,], and adaptation to biotic and abiotic stresses [,,] (Figure 1). For instance, the leucine-rich repeat receptor-like protein kinase (LRR-RLK) brassinosteroid insensitive 1 (BRI1) regulates root elongation, seed germination, stomatal function, pathogen attack and senescence and is also involved in various abiotic stresses, such as abnormal temperatures, drought and high osmotic pressure [,,,,]. In Arabidopsis thaliana, overexpression of lectin-like protein kinase 1 (AtLPK1) results in enhanced resistance to infection by the necrotrophic fungus Botrytis cinerea as well as improved seed germination and cotyledon greening under high salt stress, indicating that it is vital for both biotic and abiotic stress responses [].
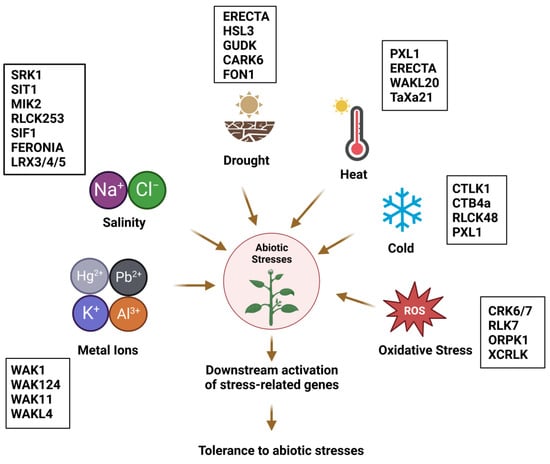
Figure 1.
Overview of the role of plant RLKs in diverse abiotic stress responses with representation of some examples. For details and abbreviations, cf. text.
Genome analyses have shown the presence of 610 RLKs in A. thaliana and over 1131 in rice []. Interestingly, an RLK subfamily that possesses only the cytoplasmic kinase domain has been designated as a receptor-like cytoplasmic kinase (RLCK) family. There are 200 RLCKs in Arabidopsis and 379 in rice [,]. RLCKs are attached to the plasma membrane through N-terminal putative myristoylation and/or palmitoylation motifs [,]. Studies have also reported their association with RLKs and their role as regulatory elements to relay intracellular signals via transphosphorylation [,,]. Recent advances suggest their role in hormone signaling pathways, plant immune responses, growth and development, embryonic patterning and organ abscission but their biological function in abiotic stress responses has not been well investigated [,,]. Boisson-Dernier et al. [] have reported that MARIS (MRI), a member of the RLCK-VIII subfamily, functions downstream of ANXUR1 and -2 which are RLKs of the Catharanthus roseus RLK1-like (CrRLK1L) subfamily. Activation of the ROS-generating NADPH oxidases regulate root hair growth and cell wall integrity of pollen tubes in A. thaliana. Botrytis-induced kinase 1 (BIK1), a RCLK VII member, forms a complex with both flagellin sensing 2 (FLS2) and brassinosteroid insensitive1-associated kinase 1 (BAK1) to activate microbe-associated molecular patterns (MAMP) triggered immunity [,,,]. However, there is also evidence that the FLS2 is involved in abiotic stress tolerance [].
Much research has been conducted that provides fundamental insights into mechanisms by which different RLKs activate defenses against microbes [,,,]. However, only a handful of studies have shed light on the role of RLKs in abiotic stress responses and the potential mechanisms underlying RLK-mediated abiotic stress tolerance [,]. A deeper understanding of kinase signaling cascades in responses to fluctuating environmental conditions such as drought, heat, cold, or salt is paramount to engineer stress-tolerant crops []. This review provides insights into the classification of RLKs and the biological functions of RLKs and RLCKs in coordinating the responses of different plant species against abiotic stresses such as salinity, drought and oxidative stress. Furthermore, this review also highlights arising avenues of future research to unravel novel targets of plant stress responses.
2. Classification of Arabidopsis RLKs
On the basis of signature motifs in their extracellular domains, RLKs are categorized into 14 classes: []: leucine-rich repeat (LRR), lectin (C-Lectin and L-Lectin), wall-associated kinase (WAK), extension-like, proline-rich extension-like (PERK), CrRLK, self-incompatibility domain (S-domain), CRINKLY4-like (CR-like), the domain of unknown function 26 (DUF26), lysin motif (LysM), thaumatin, leaf rust kinase-like (LRK), receptor-like kinase in flowers (RKF), and chitinase (glycoside hydrolase)-type domain proteins []. Such a diversity makes the RLK family one of the most versatile gene families and enables them to react to a variety of external stimuli by binding to proteins, polysaccharides, lipids and other ligands [,,].
2.1. Leucine-Rich Repeat-Receptor-like Kinases (LRR-RLKs)
LRR-RLKs form the largest RLK subfamily in A. thaliana, consisting of 239 genes and 15 subfamilies based on the amino acid relationships between their kinase domains [,]. The LRR is a tandem repeat of 24 amino acids with conserved leucines [,]. Genetic and biochemical experiments have shown that LRR-RLKs can recognize various ligands such as small molecules [,,], peptides and entire proteins [,]. These receptors often form heterodimers with other LRR units that act as co-receptors, such as BAK1 or suppressor of BAK1-interacting receptor-like kinase1 (BIR1)-1 (SOBIR) for the activation of signaling cascades [,,,,]. LRR-RLKs in Arabidopsis are involved in cell division, proliferation, differentiation, stem cell balance, pathogen resistance, hormone perception and stress adaptation [,,] (Table 1). Considering abiotic stress, Pitorre et al. [] uncovered a role of RLK7 in oxidative stress tolerance in A. thaliana. By generating knockout and overexpressing lines, Oryza sativa stress-induced protein kinase gene 1 (OsSIK1) was reported to play a positive role in salinity and drought stress by the upregulation of antioxidative enzyme activities. The leaves of OsSIK1-overexpressing plants accumulated less H2O2 than those of mutants and control plants []. Yang et al. [] identified a novel LRR-RLK from Glycine soja (GsLRPK) that is involved in cold signaling. Overexpression of GsLRPK in yeast and Arabidopsis improved cold tolerance by stimulating the expression of cold-inducible marker genes such as Kinase 1 (KIN1) and Cold regulated 15b (COR15b).
2.2. Lectin Domain-Containing Receptor-like Kinases (LecRLKs)
Lectins are widespread proteins that contain at least one noncatalytic domain with the capability to bind reversibly to a specific mono- or oligosaccharide []. A noxious protein called ricin was found in the seeds of the castor bean (Ricinus communis L.) in 1888 that was later identified as the first lectin []. The LecRLK family consists of 75 members in Arabidopsis and 173 members in rice []. Few lectins, such as the calnexins, calreticulins and malectins that are responsible for protein folding in the endoplasmic reticulum, are prevalent in plants, fungi, and animals [,,,,]. LecRLKs are classified into several groups based on their structural features and sequence similarities. L-type LecRLKs harbor a legume–lectin protein-like extracellular domain []. The extracellular domain of B-type lectins resembles the bulb–lectin proteins and were renamed as GNA-related (Galanthus nivalis agglutinin-related) or G lectins. Earlier, they were called S-domain RLKs as they contain an S-locus that participates in a self-incompatibility reaction []. There is the presence of an epidermal growth factor (EGF) domain and/or a PAN motif which is potentially crucial for protein–protein and protein–carbohydrate interactions [,,]. While C-type (calcium-dependent) lectin motifs are present in mammalian proteins that regulate innate immune responses [,], plant LecRKs function in pollen development, pathogen resistance [,,], wounding [,], regulation of stomatal immunity [], herbivory [,] and tolerance to abiotic stresses []. An Arabidopsis LecRK-b2 which is expressed during seed germination positively regulates abscisic acid (ABA), salt and osmotic stress responses []. Pohlia nutans lectin-like protein kinase 1 (PnLecRLK1) that localizes at the plasma membrane has been shown to enhance chilling stress tolerance and ABA sensitivity which allows Antarctic mosses to survive under extreme conditions. Moreover, its expression was induced by various abiotic stresses, including cold, salt, drought, ABA, and methyl jasmonic acid (MeJA) treatments. PnLecRLK1-overexpressing lines showed elevated transcription levels of genes in the C-repeat binding factor (CBF) signaling pathway, such as AtCBF1, AtCBF2, AtCBF3 and AtCOLD RESPONSIVE47 (AtCOR47) [].
2.3. Wall Associated Kinases (WAKs)
WAKs physically connect the extracellular matrix to the plasma membrane and allow communication between the two subcellular compartments [,]. These receptors can perceive the pectin breakdown products, oligogalacturonides []. Five WAK isoforms (WAK1–WAK5), clustered in 30 kilobases, share 40 to 64% similarity in their extracellular amino terminal and 86% in their cytoplasmic kinase domains []. WAKs encompass epidermal growth factor (EGF)-like repeats at the amino terminal side adjacent to the transmembrane domains []. In Arabidopsis, 26 WAK-like (WAKL) genes have been discovered due to their sequence similarity to WAKs [,]. Intriguingly, rice has 125 WAKs [], barley has 91 WAKs [], maize has more than 100 WAKs [] and Brachypodium distachyon has 115 WAKs []. WAK1, −2, −3, and −5 are expressed in green organs; WAK1 and −2 are expressed in flowers and siliques; and WAK2 is expressed in roots, while WAK4 is only expressed in siliques []. Besides their involvement in cell expansion, development, wounding and pathogen invasion, their expression is also stimulated by heavy metals, and ozone [,,,,]. Bot et al. [] investigated the role of AtWAKL10 in response to nitric oxide (NO) and its involvement in plant defense against pathogens and abiotic stresses. Using transcriptome analysis, the mRNA levels of WAKL10 changed in response to the NO donor S-nitroso-L-cysteine (CysNO). Both oxidative and nitrosative stresses had distinct effects on the growth of atwakl10 plants; CysNO treatment resulted in a greater growth rate but S-nitrosoglutathione and methyl-viologen treatment ceased its growth. AtWAKL10 positively regulates salt stress but has adverse effects under drought conditions. Analysis of the promoter region confirmed the presence of cis-regulatory elements that are crucial for abiotic stress tolerance. These results add to our understanding of the mechanisms by which plants can ameliorate stress based on the intricate interactions between NO signaling and AtWAKL10. Furthermore, AtWAKL10 could be an interesting protein to be investigated for the crosstalk between NO-induced responses to biotic and abiotic stress.
2.4. Lysin Motif Receptor-like Kinase Family (LysM-RLKs)
LysM, about 44–65 amino acids long, was first discovered in the antimicrobial protein, lysozyme, of Bacillus phage ϕ29 []. X-ray crystallography and homology modelling uncovered the three-dimensional βααβ structure of LysM that consists of two α-helices stacked on one side of a two-stranded antiparallel β-sheet [,,]. Their extracellular region is composed of three LysM modules separated by cysteine pair motifs (CxC) which are required for disulfide bridge formation and stabilization of the protein structure [,]. This kinase family is a second major class of plant pattern recognition receptors that can recognize proteinaceous microbial patterns such as short and long chitooligomers, lipochitooligomers, and peptidoglycans, mainly N-acetyl glucosamine [,,,,]. These RLKs can detect both symbiotic and pathogenic microorganisms. It has been reported that the A. thaliana genome contains five LysM-RLKs. LYK1/CERK1, a classical LysM member, is implicated in chitin recognition, which is a component of the fungal cell wall. Additionally, LYK4 and LYK5 are also necessary for the formation of heterodimers to activate chitin-mediated immune responses [,,]. In the past few years, substantial progress has been made in elucidating the signaling pathways and gene networks of LysM-RLKs in plant immune activation and symbiosis, but we are still far away from understanding their role in abiotic stresses.
Espinoza et al. [] have unraveled the molecular mechanism that contributes to cross-tolerance between chitin and salt. Interestingly, CERK1 expression was upregulated under salt stress and the cerk1 mutant was more prone to salt stress but was not affected by osmotic stress. Transcriptome studies have shown similar expression profiles of chitin and salt treatments. It would be interesting to investigate the link between chitin-mediated plant defense responses and salt stress responses. Moreover, cerk1 plants have shown an aberrant rise in the levels of cytosolic calcium ([Ca2+]cyt) after salt application. Bimolecular fluorescence complementation and co-immunoprecipitation experiments have revealed the interaction between CERK1 and ANNEXIN1, a Ca2+-permeable channel that mediates the salt-elicited [Ca2+]cyt signal []. The above evidence helps us to comprehend the crosstalk between biotic and abiotic signaling pathways. Furthermore, the question as to which Ca2+-channels are involved in biotic and abiotic signaling and whether there is a crosstalk is still enigmatic.
2.5. Cysteine-Rich Repeat Domain-Containing Receptor-like Kinases (CRKs)
Most CRKs have two domain of unknown function 26 (DUF26) motifs in their extracellular region with three conserved cysteine (C) residues in a C-X8-C-X2-C configuration [,,]. These conserved C residues are postulated to be involved in the formation of disulfide bridges to stabilize the three-dimensional structure of these kinases or in ROS redox regulation [,,]. The DUF26 domain has an antifungal property which is why it is also called a stress–antifungal domain (PF01657) [,,] or GINKBILOBIN2 (GNK2), and its role in salt stress response has also been reported []. Multi-omics and molecular genetic analyses have led to the identification of 44 CRKs in A. thaliana and 1074 CRKs in 14 other crops. However, the role of only 63 members has been characterized and the function of the other members is still obscure [].
Functional characterization of CRKs has unveiled their involvement in development [,,], defense [,,,,,], cell death [,,], acclimation to various abiotic stresses such as salt [,,], osmotic stress [], UV light, oxidative [], heat [], cold stress [] and drought [,,]. In addition, existing evidence shows the upregulation of CRKs by O3 [,], salicylic acid (CRK4, CRK5, CRK6, CRK10, CRK11, CRK19, CRK20, CRK45), pathogens and their microbial patterns [,,,]. Bioinformatic studies have revealed the presence of W-Box elements (TTGAC) which are the binding sites for WRKY transcription factors in the promoters of AtCRKs, implying that WRKYs play a role in the regulation of these kinases [,,,,,].
The presence of conserved C residues points towards the role of CRKs in ROS signaling. Bourdais et al. [] analyzed the effect of chloroplast ROS inducers, methyl viologen (paraquat) and 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), on different crk mutants, and higher photoinhibition was observed in crk2, crk5, crk8, crk17, crk20, crk40, and crk45 mutants compared with the wild-type. Furthermore, crk2 and crk45 mutants also exhibited higher electrolyte leakage upon exposure to light stress. Another study demonstrated the role of CRK2 in improving germination, root length, and callose deposition under high salt stress, thereby suggesting that it is necessary for salt tolerance [].
2.6. CRINKLY4 (CR4) Family of Receptor-like Kinases
These kinases were first described in maize where the CR4 gene was isolated by mutator–transposon tagging. cr4 mutants were shorter, had crinkly leaves, and had defects in the differentiation of the leaf epidermis []. In addition, the epidermal cells of these mutants were aberrant in shape, cell wall thickness, cuticle formation, vesicle trafficking and showed tumor-like outgrowths [,]. The characteristic feature of this family is the presence of 7 crinkly repeats of 39 amino acids in the extracellular region and a domain homologous to tumor necrosis factor receptor (TNFR) cysteine-rich region [,]. Arabidopsis CR4 (ACR4), an ortholog of CR4, contains all of the features of maize CR4 and is strongly expressed in the protodermal cells of the embryo, in the L1 layer of the shoot apical meristem, in the epidermis of leaf primordia, in the small daughter cells after the first asymmetric pericycle cell division and in the root stem cell niche [,,,]. Localization studies in Nicotiana benthamiana leaf epidermal cells have revealed the accumulation of ACR4 at the plasmodesmata []. Mounting evidence supports the idea of a role for ACR4 in regulating asymmetric cell division in columella stem cells, embryonic development, and lateral root formation [,,,,,,,]. Furthermore, ACR4 and an LRR-RLK, CLAVATA1, can form homo- and heterodimers and participate in the maintenance of root meristem in response to the signaling peptide clavata 3/embryo surrounding region 40 (CLE40) [].
Interestingly, Zereen and Ingram [] have demonstrated that the acr4 knockout line is less susceptible to the necrotrophic fungal pathogen Botrytis cinerea, and the expression of ACR4 is significantly downregulated in response to this fungus. It is speculated that this might be due to an enhanced expression of LIPOXYGENASE2, which codes for an important enzyme involved in the biosynthesis of jasmonic acid []. This finding indicates that it is potentially involved in plant defense responses. While much is known about the role of these kinases in plant growth and development, their role in abiotic stress responses has not been explored. Knowledge of their regulatory networks and downstream signaling components warrants future investigations.
2.7. Proline-Rich Extensin like Kinases (PERKs)
The PERK family of kinases consist of an extensin-like extra-cellular domain rich in prolines followed by a typical transmembrane, serine/threonine kinase domain and these were first discovered in Brassica napus (BnPERK1; [,]). This family has fifteen members in Arabidopsis, eight in Oryza sativa, two in Hordeum vulgare, six in Solanum tuberosum, and nineteen in Brassica rapa [,]. AtPERK members have differential expression patterns, most of which are widely expressed while some are expressed in certain tissues. For instance, AtPERK1 is expressed predominantly in the vascular tissues of cotyledons, developing leaves, and roots; AtPERK2 is expressed in rosette leaf veins, stems, and pollen []; AtPERK8 and AtPERK13 in root hairs [,,]; and AtPERK5, AtPERK6, AtPERK7, AtPERK11, and AtPERK12 in pollen [,,].
AtPERKs play multifaceted roles in plants, including in the regulation of apical dominance, cell proliferation, and root or pollen tube growth [,,,]. Additionally, they might also be promising candidates for sensing cell wall integrity, as their extracellular domain resides inside the cell wall, similar to WAKs []. Their role in plant–pathogen interactions, abiotic stresses, and hormone signaling has emerged only recently [,]. Feng et al. [] have pinpointed the role of ZEBRA LEAF 15 (Z15), a single copy gene that encodes a PERK, in moderate low-temperature signaling in rice. Z15 mutation hampered the cell development and chloroplast structure, leading to transversely striped leaves with yellow-green or white-green sectors. Further, the expression of the downstream cold-response genes OsWRKY71 and OsMYB4 has also been affected in the z15 mutant upon exposure to moderate low temperature (18 °C). Recently, PERK13 was shown to participate in root hair growth under phosphate deficiency. Knocking out and overexpressing PERK13 extended the root hair elongation period and led to higher ROS generation in root hairs under phosphate-limiting conditions [].
To date, ligands for these receptor kinases have not been reported, owing to the redundancy of these kinase genes [,]. These studies link PERK13-controlled developmental programs to abiotic stress.
2.8. Catharanthus Roseus RLK1-like Kinases (CrRLK1Ls)
This family obtained its name after the first identified member, CrRLK1, from Madagascar periwinkle (C. roseus). Family members have been discovered in different species, including angiosperms (monocots, eudicots), gymnosperms, and early diverging lineages [,,,]. These receptors consist of two malectin-like domains (MLDs) in their extracellular region that are homologous to the carbohydrate-binding malectin protein from Xenopus laevis [,,]. CrRLK1s have diverse functions including responses to biotic and abiotic stresses [], plant growth [], morphogenesis [], reproduction [], immunity [], hormone signal transduction [], RNA metabolism [] and energy production []. Seventeen CrRLK1L members are present in A. thaliana, twenty-three in S. lycopersicum, twenty in O. sativa, and forty in Gossypium hirsutum [,,,].
Eight of the seventeen Arabidopsis CrRLK1Ls have been well investigated to date: FERONIA (FER), ANXUR1 (ANX1), ANXUR2 (ANX2), HERCULES1 (HERK1), HERCULES2 (HERK2), [Ca2+]cyt -associated protein kinase 1 (CAP1/ERULUS), THESEUS1 (THE1), and CURVY1 (CVY1) []. Among these, FER is probably the most extensively characterized CrRLK1L. FER is named after the Etruscan goddess of fertility []. It serves as a sensor for salt, osmotic, heat, and cold stresses [,,]. Kim et al. [] have identified a Feronia-temperature sensitive (fer-ts) mutant, with G41S substitution in its extracellular domain, that is unable to form root hairs at high temperatures (30 °C). FER controls RHO GTPase (RAC/ROP)-mediated NADPH oxidase-dependent ROS production in root hairs []. Kim et al. [] have observed a reduction in ROS levels in the fer-ts mutant at elevated temperatures. Yin et al. [] have examined the role of FER2 and -3 in heat stress tolerance. Interestingly, FER2 and -3 mRNA levels were enhanced under heat stress, and this was dependent on Brassinazole resistant 1 (BZR1), a transcription factor that regulates brassinosteroid (BR) signaling. In addition, chromatin immunoprecipitation real-time quantitative PCR has revealed that BZR1 was bound to the FER2 and FER3 promoters and is responsible for regulating their expression. The fer2/3 mutants showed a decrease in BR-dependent apoplastic H2O2 accumulation, ROS-detoxification ability, and heat stress tolerance. These results suggest that FER interferes with BZR1-dependent BR signaling and thus ROS production [].
Additionally, the role of cell wall sensors, FER/HERK1/THE1-4, was also investigated in salt stress responses. Plants impaired in a functional HERK1 and THE1 were severely sensitive to salt stress, comparable to the fer-4 loss of function mutants. fer-4 and herk1the1-4 mutants showed a higher activation of MAP protein kinase 6 and elevated transcript levels of salt-induced marker genes. Moreover, FER alone or with HERK1/THE1-4 demonstrates salt stress-induced pectin de-methyl esterification leading to the activation of downstream signaling events []. In conclusion, the few well-investigated CrRLK1Ls are clearly involved in abiotic stress responses.
2.9. Leucine-Rich Repeat Extensins (LRXs)
The plant cell wall is composed of a complex network of polysaccharides such as cellulose, hemicellulose, pectin, and also proteins that constitute 20% of the primary cell wall [,]. Cell wall proteins (CWPs) are vital for wall modifications, signaling and harbor highly repetitive sequence motifs [,,]. CWPs are categorized into three main classes: proline-rich-proteins (PRPs), glycine-rich-proteins (GRPs), and hydroxyproline-rich-glycoproteins (HRGPs) [,,]. Extensins (EXTs) are cell wall structural proteins that belong to the HRGPs family. They contain a repeating serine-(hydroxyproline)4 motif where the serine and hydroxyproline residues undergo glycosylation with often four to five oligoarabinosides becoming attached to hydroxyproline residues and galactose monosaccharides to serine by several glycosyltranferases [,,]. Extensins are basic in nature, with isoelectric points of ∼10, due to their high lysine content, and they form polyproline II helices [].
LRXs are chimeric, cell wall localized proteins with a conserved N-terminal LRR domain and a highly variable C-terminal extensin domain []. LRR domain plays a role in protein–protein interactions while the highly glycosylated extensin domain is probably important for anchoring to polysaccharides such as pectins, in the apoplast [,,]. Arabidopsis contains 11 LRX members that can be subdivided into 2 groups on the basis of their tissue-specific expression. LRX1–LRX7 are expressed in vegetative tissue while LRX8–LRX11 are specifically expressed in pollen. LRX1 and LRX2 are involved in cell wall development and defects in root hairs have been observed for atlrx1 and atlrx1/atlrx2 double mutants [,]. LRXs bind peptide hormones known as rapid alkalinization factors (RALFs; [,,,]). The lrx345 triple mutant, the fer-4 mutant, and RALF22 or RALF23 overexpressing lines exhibited similar phenotypic aspects such as reduced growth, and they were extremely sensitive to salt stress, indicating that LRX3/4/5 and FER potentially work together in the same pathway. In addition, biochemical data show that LRX3/4/5 is associated with several RALF peptides, including RALF22 and RALF23. This restricts binding of these peptides with FER and its internalization. These findings indicate that LRXs, RALFs, and FER form a signaling hub to coordinate cell wall integrity, plant growth, and salt stress responses [].
2.10. Thaumatin Domain-Containing Receptor-like Kinases
Pathogenesis-related (PR) proteins are produced as a result of the defense response in the host plant upon pathogen invasion [,]. These are classified into 17 families (PR1–PR17) based on their mode of action and sequence similarity. Thaumatin-like proteins (TLPs) are structurally similar to thaumatin, a sweet-tasting protein from a rainforest shrub, Thaumatococcus daniellii []. These proteins belong to the PR5 family and harbor the conserved signature motif G-X-[GF]-X-C-X-T-[GA]-D-C-X-(1,2)-G-X-(2,3)-C [,]. Based on their molecular weight, TLPs can be either long (L-type; 22 to 26 kDa) or small (S-type; 18 kDa or less). Their salient characteristic is the presence of 10 and 16 conserved cysteine residues in S-type and L-type TLPs, respectively []. These cysteine residues form disulfide bridges which are important for their stability under extreme conditions such as heat and extreme pHs []. TLPs can regulate plant responses to various biotic and abiotic stresses, such as pathogenic elicitors, osmotic stress, cold stress, wounding, and plant hormones [].
Thaumatin-like protein kinases (TLPKs) have a typical RLK structure, with an extracellular thaumatin-like domain that has an antifungal and chitinase activity []. The pathogenesis-related 5 RLK (PR5K) from A. thaliana was the first characterized TLPK that is majorly expressed in inflorescence stems and roots. It is speculated that it might be a receptor for pathogen-derived elicitors []. Three PR5K genes are present in Arabidopsis while one gene exists in rice []. Their expression can be triggered by various hormones and pathogens [,]. Recently, the involvement of AtPR5Ks in ABA-mediated drought stress responses was delineated. Among the three atpr5k1-1, atpr5k2-1, and atpr5k3-1 mutants, a drought tolerance phenotype was detected only for atpr5k2-1. On the contrary, AtPR5K2-overexpressing plants were found to be hypersensitive to drought stress. Furthermore, AtPR5K2 interacts with the core components involved in ABA signaling, type 2C protein phosphatases ABA-insensitive 1 (ABI1) and ABI2 and the SNF1-related protein kinase 2 (SnRK2.6) proteins []. Because this association is abolished in the presence of exogenously applied ABA, AtPR5K2 may function as a negative regulator of ABA signaling under drought conditions [].
2.11. Chitinase (Glycoside Hydrolase)-Type Domain Containing Receptor-like Kinases
Chitinase-related RLK1 (CHRK1) was first isolated from tobacco. The kinase contained an extracellular domain that is closely related to the class V chitinase of tobacco and to bacterial chitinases, though it does not have chitinase activity itself due to the lack of a glutamic acid residue. It is proposed that CHRK1 might take part in defense against pathogens as its activity was induced upon invasion by fungal pathogen and tobacco mosaic virus []. By generating a GFP construct and transfecting it into animal cells, the fusion protein resides in the plasma membrane. There are more than 600 RLKs in the Arabidopsis genome, but none of them have been seen to have a chitinase-like sequence []. Lee et al. [] showed that CHRK1 functions in plant development and also regulates cytokinin homeostasis in tobacco. In comparison with other RLK classes, these kinases have received less research attention, and not much is known about their functions in biotic and abiotic stress responses.
2.12. Leaf Rust Kinase 10-like (LRK 10-like)
A novel extracellular recognition domain is present in the LRK10 protein. The LRK10 gene was initially discovered in wheat (TaLRK10) using a homology-based approach based on disease resistance against the leaf-rust-causing fungal pathogen Puccinia recondite []. The most closely associated Arabidopsis homolog of TaLRK10, A. thaliana leaf rust 10 disease-resistance locus receptor-like protein kinase 1 (AtLRK10L1), uses distinct promoters to generate two transcripts, AtLRK10L1.1 and AtLRK10L1.2. Microarray data have disclosed the contribution of LRK10Ls in plant growth and stress responses, specifically AtLRK10L1.2, in controlling the time of flowering and defense responses against pathogens []. Moreover, LRK10L1.2 participates in ABA signaling at the seedling stage and its localization at the plasma membrane is required for this process as transgenic plants expressing its splicing variant generated a protein that relocated to the endoplasmic reticulum and were seen to be insensitive to high levels of ABA. In addition, LRK10L1.2 is involved in drought tolerance by facilitating stomatal closure, which was revealed by higher water loss in lrk10l1-2 mutants under drought conditions. Physiological and molecular functions of the LRK10-like N-terminal domains in abiotic and biotic stress responses have not yet been explored in detail [].

Table 1.
Functions of RLKs in different abiotic stress responses.
Table 1.
Functions of RLKs in different abiotic stress responses.
| Genes | Species | Type of RLK | Function | References |
|---|---|---|---|---|
| Salt Stress | ||||
| OsSRK1 | Oryza sativa | S-receptor protein kinases | Controls leaf development and provides adaptation against salinity | [] |
| PsLecRLK | Pisum sativum | Lectin | Mitigates salt stress by lowering oxidative damage and increasing the expression of stress-responsive genes thus, retaining ion homeostasis | [] |
| SIT1 | Oryza sativa | Lectin | Negatively regulates salt stress by inducing ethylene and ROS that suppresses plant growth and causes plant death | [] |
| MIK2 | A. thaliana | LRR | Controls the direction of root growth, alters the cell wall structure in the root tip and provides adaptation to salt stress | [] |
| OsRLCK253 | Oryza sativa | RLCK | Interacts with OsSAP11 and prevents yield losses during salt and drought | [] |
| GhSIF1 | Gossypium hirsutum | LRR-RLK | Negative regulator of salt stress responses | [] |
| FERONIA | A. thaliana | CrRLK1L | Required for restoration of root growth, cell wall stiffness after salt exposure | [] |
| LRX 3/4/5 | A. thaliana | LRX | Forms a signaling network with RALF 22/23 and FER which is pivotal for plant development and adaptation to salt stress | [] |
| TaSR | Triticum aestivum | LRR–RLK | Participates in salt tolerance by increasing Na+ efflux | [] |
| PnRLK-1 | Pohlia nutans | RLCK | Regulates plant sensitivity to ABA and adaptation to salt and oxidative stress | [] |
| GsSRK | Glycine soja | G-type lectin | Vital for plant response to salt stress | [] |
| RLK 7 | A. thaliana | LRR-RLK | Associates with PAMP-INDUCED SECRETED PEPTIDE 3, activates MPK3/6 and ultimately increases salt stress resistance through maintenance of ionic homeostasis | [] |
| STRK1 | Oryza sativa | RLCK | Confers tolerance against salt stress by activating and phosphorylating Catalase C that maintains H2O2 balance. Boosts grain yield under salt stress | [] |
| PaLectinL16 | Prunus avium | Lectin | Provides protection against salt stress by increasing the activities of antioxidant enzymes | [] |
| RPK1 | A. thaliana and Oryza sativa | Leucine-rich repeat RLK | Negatively regulates salt stress responses, reduces proline synthesis, and inhibits the expression of SALT OVERLY SENSITIVE 3 | [] |
| AtLPK1 | A. thaliana | Lectin | Functions in salt stress responses by increasing seed germination and cotyledon greening, also participates in pathogen resistance, thus acting as a mediator between abiotic and biotic stress responses | [] |
| OsRLCK 311 | Oryza sativa | RLCK | Regulates stomatal responses under salt stress and binds to aquaporin protein, PIP2;1 | [] |
| Drought Stress | ||||
| HSL3 | A. thaliana | LRR-RLK | Negatively impacts plant response to moisture deficit conditions through ABA-mediated stomatal closure induced by the generation of H2O2 in the guard cells | [] |
| GUDK | Oryza sativa | RLCK | Provides protection against drought stress by activating APETALA2/ETHYLENE RESPONSE FACTOR OsAP37 which triggers the transcription of stress-regulated genes resulting in high yield | [] |
| GbRLK | Gossypium barbadense | Probable G-type lectin | Crucial for drought and salinity stress tolerance and activation of ABA-dependent signaling events | [] |
| CARK6 | A. thaliana | RLCK | Participates in ABA-mediated drought tolerance | [] |
| FON1 | Oryza sativa | LRR-RLK | Involved in drought stress tolerance in rice by regulating the expression of ABA-responsive genes | [] |
| LP2 | Oryza sativa | LRR-RLK | Acts as a negative regulator in drought response. Interacts with drought-responsive aquaporins and is transcriptionally regulated by C2H2 zinc finger transcriptional factor DROUGHT AND SALT TOLERANCE | [] |
| AtLRK10L1.2 | A. thaliana | LRK 10-like | Takes part in ABA signaling and provides tolerance against drought stress by enhancing stomatal closure | [] |
| OsSIK2 | Oryza sativa | S-RLKs | Reduces the accumulation of H2O2 under salt stress, participates in dark-induced leaf senescence and plays a vital role under drought conditions | [] |
| CRK45 | A. thaliana | Cysteine-rich RLK | Imparts tolerance against drought stress and controls expression of ABA responsive genes | [] |
| AtPR5K2 | A. thaliana | Thaumatin-like RLK | Plays a negative role in ABA signaling during drought stress by phosphorylating ABI1 and ABI2 | [] |
| LRK2 | Oryza sativa | LRR-RLK | Positive regulator of the drought stress response and tiller size in rice | [] |
| OsESG1 | Oryza sativa | S-domain RLK | Participates in drought tolerance by enhancing the activities of antioxidants and expression of stress-regulated genes | [] |
| OsSIK1 | Oryza sativa | LRR-RLK | Inhibits stomatal development in rice leaves which reduces water loss and thereby, providing tolerance against drought stress. Confers adaptation to salt stress by activation of antioxidant enzymes | [] |
| ScRIPK | Saccharum spp. Hybrids | RLCK | Positively regulates drought tolerance and is a negative regulator of plant defense | [] |
| OsRLCK241 | Oryza sativa | RLCK | Confers tolerance against drought and salt stress by enhancing ROS detoxification, osmolyte production and upregulating the expression of stress-responsive genes | [] |
| Oxidative Stress | ||||
| ORPK1/ZAR1 | A. thaliana | LRR-RLK | Positively controls oxidative stress responses and promotes lateral root formation | [] |
| XCRLK | Oryza sativa | RLCK | Fine tunes ROS levels by detoxifying H2O2, thus protecting rice plants against oxidative stress | [] |
| CRK7 | A. thaliana | Cysteine-rich RLK | Important for the coordinated response to extracellular but not chloroplastic ROS | [] |
| Heavy metal stress | ||||
| WAK1 | A. thaliana | WAK | Involved in tolerance against aluminum toxicity | [] |
| WAKL4 | A. thaliana | WAKL | Plays a vital role in root mineral nutrient responses such as Na+, K+, Cu2+, and Zn2+ | [] |
| OsWAK124 | Oryza sativa | WAK-RLP | Functions in environmental (heavy) metal stress responses such as Cd2+, Cu2+, and Al3+ | [] |
| OsWAK11 | Oryza sativa | WAK | Regulates plant response to metal stress and wounding | [] |
| Cold Stress | ||||
| GsLRPK | Glycine soja | LRR-RLK | Functions in cold tolerance by inducing the expression of cold-inducible marker genes | [] |
| CTLK1 | Medicago truncatula | LRR-RLK | Improves cold tolerance by modulating the expression of antioxidant genes, enzyme activities and proline accumulation | [] |
| NDW | Solanum lycopersicum | Unknown | Participates in plant growth regulation, cold adaptation and disease resistance against Botrytis cinerea | [] |
| CTB4a | Oryza sativa | LRR-RLK | Confers cold tolerance at the booting stage and improves seed set by regulating pollen fertility and interacts with a beta subunit of ATP synthase, AtpB | [] |
| OsRLCK48 | Oryza sativa | RLCK | Its expression is downregulated under cold stress | [] |
| Heat stress | ||||
| TMS10 | Oryza sativa | LRR-RLK | Plays a role in tapetal degeneration and male fertility under high temperatures | [] |
| ERECTA | A. thaliana | LRR-RLK | Introduction of ERECTA gene in Pinellia ernate disrupted the summer dormancy. It is crucial for preventing plant cells from cellular damage caused by high heat and positively regulates transpiration efficiency in rice and tomato | [,] |
| AtPXL1 | A. thaliana | LRR-RLK | Interacts with histidine-rich dehydrin1, light-harvesting protein complex I and is involved in signaling under cold and heat stress | [] |
| FER | A. thaliana | CrRLK1L | Required for root hair development under elevated temperatures | [] |
| CaHSL1 | Capsicum annuum | LRR-RLK | Provides thermotolerance against high temperature and high humidity | [] |
| CaWAKL20 | Capsicum annuum | WAKL | Negative regulator of plant thermotolerance as it suppresses the expression of ABA-responsive genes | [] |
| TaXa21 | Triticum aestivum | LRR-RLK | Positively mediates high temperature plant resistance to P. striiformis f. sp. Tritici by interacting with TaWRKY76 and TaWRKY62 | [] |
3. Biological Functions of RLKs in Abiotic Stress Responses
Direct sensing of abiotic stress by RLKs may occur via their extracellular domains, which transduce the information to the cytoplasmic compartment to initiate appropriate downstream responses []. Proper signaling of the RLKs requires an intact plasma membrane around the stress-exposed cell. In order to identify candidate RLKs involved in stress sensing, (a) the expression levels of an RLK gene might respond to a stress treatment a, (b) manipulation of the RLK mRNA or RLK protein level in a plant could alter stress-induced responses which occur in the un-manipulated plants, and (c) manipulation of the RLK levels alters the stress resistance phenotype of the plant in comparison with unmanipulated plants. In all three cases, the correlation between input and output can be positive or negative, depending on whether the RLK is an activator or repressor of the stress response. Basically, these three criteria were used to identify RLKs involved in abiotic stress resistance. We describe the well-investigated RLK from different plant species that are involved in salt, drought, oxidative, temperature and metal stress.
3.1. Salt Stress
Salt stress is a major environmental constraint that affects 20% of irrigated land leading to germination failure, reduced growth, and yield losses [,,]. Excess salt causes ionic (mainly due to Na+, Cl−, and SO42−) and osmotic stresses, as well as secondary stresses such as oxidative stress and nutritional deficiencies []. Plants are classified as halophytes or glycophytes depending upon their ability to tolerate salinity. Halophytes have the capacity to tolerate high salt concentrations (400 mM NaCl), while glycophytes are sensitive to high salt conditions []. In order to maintain their ion homeostasis, plants must sense excess salt in the apoplast (cf. Table 1). OsSRK1 is an atypical S-receptor-like kinase whose expression is enhanced upon ABA, salt, and polyethylene glycol (PEG) 4000 treatment. OsSRK1-overexpression (OsSRK1-OX) plants have been shown to be sensitive to ABA but, interestingly, were found to be highly tolerant to salt stress in comparison with the wild type. The mechanism underlying OsSRK1-mediated salt tolerance is unclear but can be attributed to the induction of a set of genes involved in dehydration, such as DEHYDRATION RESPONSIVE ELEMENT-BINDING, OsDREB1A. This makes SRK1 an interesting candidate in rice agriculture []. LecRLKs have been extensively studied for their role in environmental stress conditions. Vaid et al. [] have investigated the role of PsLecRLK, an L-type LecRLK from Pisum sativum, under salinity stress conditions. When the expression pattern of PsLecRLK was analyzed in response to salinity, temperature stress, wounding and hormone applications, an 80-fold induction was observed under salt stress. Overexpression of PsLecRLK in transgenic tobacco plants resulted in a salt-tolerant phenotype which was evident from higher biomass, germination rate, growth, and photosynthetic pigment content. Moreover, estimation of malondialdehyde, 3,3′-diaminobenzidine and nitroblue tetrazolium staining indicated lesser ROS levels and lower membrane damage in these plants. PsLecRLK overexpression resulted in a lesser accumulation of Na+ ions and a higher expression of water channels and transporters involved in ion homeostasis under salt stress. This indicates that PsLecRLK stabilizes the membranes and ion homeostasis under salt stress by stimulating transport processes across the plasma membrane. In another study, Salt Intolerance 1 (SIT1) from rice was shown to negatively regulate salt tolerance. This is expressed in root epidermal cells and becomes activated under salt stress. SIT1 activates MAPK3 and MAPK6 leading to ethylene generation. Finally, SIT1 enhances the ROS generation that inhibits growth and causes plant death under salinity, which in turn is dependent on MAPK3/6 and ethylene signaling in A. thaliana []. Furthermore, maintenance of cell wall integrity is crucial for plants in order to acclimatize to saline conditions []. MDIS1-INTERACTING RECEPTOR LIKE KINASE2 (MIK2) of the sub-family XIIb of LRR-RLKs is vital for regulating the responses activated by the suppression of cellulose biosynthesis. In addition, MIK2’s role in salt stress tolerance is dependent on THE1, though it does not require THE1 in order to provide resistance to Fusarium oxysporum in roots (Figure 2) []. Apparently, this is an interesting starting point from which to experimentally dissect the signaling leading to abiotic and biotic stress responses. 20⁄AN1 zinc-finger domain-containing stress-associated proteins (SAPs) are central for imparting tolerance against abiotic stresses []. Giri et al. [] have shown that OsSAP11 interacts with OsRLCK253 at the nuclear membrane and only weakly at the plasma membrane. By generating overexpressor lines, it was observed that OsSAP11 and OsRLCK253 enhanced plant survival and reduced yield losses under salt and drought stress. Transcript profiles indicated the upregulation of genes involved in the biosynthesis of antioxidant compounds such as anthocyanin and carotenoids, which might be a reason for the protection of these plants against different stresses. Stress-induced factor genes (SIF1–SIF4) belong to a multifunctional kinase family which is involved in biotic and abiotic stress responses in Arabidopsis. Phylogenetic analysis revealed six LRR-RLKs in cotton that are homologous to Arabidopsis SIFs and, among these, GhSIF1 showed a 46–47% amino acid sequence similarity. Yuan et al. [] have characterized the role of GhSIF1 in salt tolerance by knocking it out using a virus-induced gene silencing system in cotton. The plants were better protected against salt stress in the transient assay system and they exhibited enhanced growth, lower electrolyte leakage, and higher chlorophyll content than the control plants. These results demonstrate that GhSIF1 negatively regulates salt tolerance. Apparently, the participation of RLKs in salt stress tolerance ranges from the activation of general responses to abiotic stress to more specific responses stabilizing the ion homeostasis. This includes alterations in gene expression and enzyme activity.
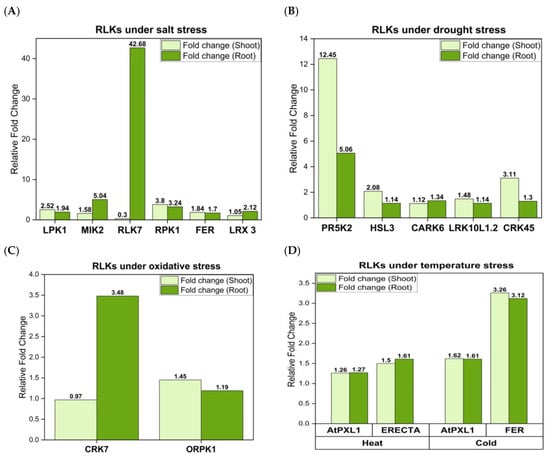
Figure 2.
Fold change of RLK mRNA levels in shoots and roots under different stress treatments in Arabidopsis. (A) Salt stress, (B) drought stress, (C) oxidative stress, and (D) temperature stress (heat and cold). The above graphs represent maximum relative fold change values at a particular time point after stress exposure. Expression of these RLKs under different stress conditions was analyzed by Arabidopsis eFP Browser at BAR website (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi, accessed on 18 August 2023).
K+, the most prevalent cation in plants, is crucial for the maintenance of cell turgor, carbohydrate metabolism, starch synthesis, osmoregulation, and enzyme activation [,]. Moreover, K+ uptake and transport play a vital role in providing tolerance against various abiotic stresses, such as drought and salinity. K+ enhances the activity of antioxidant enzymes, thereby decreasing the accumulation of ROS and also regulates stomatal opening under water deficient conditions []. Salinity tolerance is acquired by establishing K+ homeostasis and a low Na+/K+ ratio in the plant [,]. When plants are exposed to salt stress, Na+ competes with K+ for uptake sites at the plasma membrane []. As a result, membrane integrity is disrupted due to depolarization that leads to the efflux of K+ via K+ channels (depolarization-activated outward-rectifying K+ channels) [].
Salt stress causes an increase in cytosolic free calcium (Ca2+) concentration [] which is sensed by a salt overlay sensitive (SOS) pathway that consists of SOS3, SOS2, and SOS1 []. SOS3, a myristoylated Ca2+ binding protein senses the elevation in Ca2+ levels and binds with Ca2+ which activates SOS2 or calcineurin-B-like protein (CBL)-interacting protein kinase (CIPK24), a serine/threonine protein kinase. The SOS3–SOS2 complex activates SOS1, a Na+/H+ antiporter, which is responsible for the extrusion of Na+ from the cytosol [,]. CBLs interact with plant Ser/Thr kinases, namely the CIPKs, which are homologous to yeast and animal sucrose non-fermenting (SNF) protein kinases []. The CBL–CIPK network has been extensively studied under K+-limiting conditions in Arabidopsis. Exposure of plants to long term salt stress decreases K+ content and stimulates K+ transporters. CBL1/CBL9 associates with CIPK23 and targets it to the plasma membrane of root cells to phosphorylate a voltage-gated high-affinity K+ channel, Arabidopsis K+ transporter 1 (AKT1). Its activation enhances K+ uptake [].
3.2. Drought Stress
Drought is an inevitable environmental factor that impacts 64% of land area worldwide and impedes photosynthesis, stomatal movement, ion uptake, metabolism, and can even cause plant death []. It is well documented that plants have developed phenotypic plasticity through intricate signaling networks involving RLKs []. Several RLKs have been linked to drought stress responses [] (Figure 3). ABA is an essential stress hormone that can regulate the expression of osmotic stress-responsive genes and facilitates stomatal closure under drought stress []. HAESA-LIKE3 (HSL3) is an LRR-RLK that is induced by ABA, H2O2 and water deficiency. In the hsl3 mutant, over-accumulation of H2O2 and a higher net flow of anions in the guard cells was observed, resulting in the closure of stomata and thereby providing tolerance against drought stress. HSL3 acts as a negative regulator of drought stress by tweaking the levels of H2O2 in the guard cells []. ERECTA (ER), another LRR-RLK from Arabidopsis has been implicated in development and disease protection. However, its role in water deficiency has not been explored. Recently, two ER genes from Sorghum bicolor L., SbER1–1 and SbER2–1 were cloned, and their expression was evaluated under moderate and severe drought stress. SbER2 transcript levels were greatly enhanced in response to drought, and a 35S::SbER2–1-eGFP fusion protein was localized on the plasma membrane and in chloroplasts. This suggests a role for SbER2-1 in photosynthesis. Overexpression of SbER2–1 in Arabidopsis and maize resulted in higher drought tolerance of the shoot. Differentially expressed gene analysis of maize lines overexpressing SbER2–1 depicted genes enriched in glutathione metabolism in leaves and phenylpropanoid biosynthesis in stems. Moreover, these plants also showed higher lignin content as well as water use efficiency, indicating that SbER2–1 is a vital target for genetic engineering in order to produce plants that are resistant to drought stress []. An RLCK, GROWTH UNDER DROUGHT KINASE (GUDK) is responsible for imparting drought tolerance under both vegetative and reproductive stages in rice. This was demonstrated by the use of gudk loss-of-function mutant lines as they are sensitive to salinity, osmotic stress, and ABA at the seedling stage. The grain yield was reduced in these lines under well-watered conditions and drought stress. Furthermore, phosphoproteomics unraveled the transcription factor APETALA2/ETHYLENE RESPONSE FACTOR, OsAP37 as a target of GUDK which, in turn, activates genes involved in photosynthesis, carbon metabolism, and drought tolerance []. Another study by Zhao et al. [] provides insights into the mechanism by which an RLK gene from cotton, GbRLK, imparts tolerance against various abiotic stresses including drought. The GbRLK promoter was fused to the β-glucuronidase (GUS) gene and histochemical staining indicated induction of GUS levels upon exposure to ABA, PEG, salt, and Verticillium dahlia infection. Overexpression of GbRLK in Arabidopsis reduced water loss and these transgenic plants are more tolerant towards drought. Furthermore, these lines showed hypersensitivity to ABA, indicating that GbRLK is involved in ABA-mediated signal transduction. Additionally, the upregulation of various stress-responsive genes, RESPONSIVE TO DESICCATION (AtRD20, AtRD22, and AtRD26) and antioxidant genes such as CATALASE 1 (AtCAT1), COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE (AtCCS), and COPPER/ZINC SUPEROXIDE DISMUTASE 2 (AtCSD2) was also detected, which can mitigate the adverse effect of these stresses by detoxification of ROS. Another RLCK VIII subfamily member in Arabidopsis, cytosolic ABA receptor kinase 6 (CARK6), participates in the regulation of germination and root growth and stimulates drought resistance by interacting with ABA receptors. Thus, further studies are needed to unravel the molecular mechanism underlying its role in ABA signaling and to manipulate it for crop improvement []. These examples demonstrate that those RLKs which are involved in drought tolerance, either crosstalk with ABA signaling or—in the case of CARK6—interact directly with ABA receptors.
3.3. Oxidative Stress
Multiple lines of evidence support the dual role of ROS as both toxic by-products of aerobic metabolism as well as vital signaling molecules for inter- and intracellular communication in plants [,]. ROS generation is stimulated by various environmental stresses that include excessive light, wounding, ozone, drought, UV irradiations, pathogen invasion, low and high temperatures, and heavy metals []. Oxidative stress occurs as a result of the overproduction and accumulation of ROS in various organelles. It can cause cellular damage to biomolecules such as DNA, proteins, and lipids that may even lead to cell death [,]. RLK signaling often results in apoplastic ROS generation, indicating an intricate connection between RLKs and ROS burst []. Because ROS production is a general response to many abiotic and biotic stresses, it is not surprising that elevated ROS levels have been described for many of the activated RLKs described here. Recently, the role of a previously uncharacterized LRR-RLK, Oxidative-stress Related Protein Kinase 1 (ORPK1) from Arabidopsis was dissected by Yang and Jiang []. AtORPK1 mutants were shown to be extremely sensitive to oxidative stress while the overexpression of AtORPK1 enhanced the oxidative stress tolerance of transgenic plants. The expression of antioxidant enzyme genes such as IRON SUPEROXIDE DISMUTASE (FeSOD), CATALASE1 (CAT1), and ASCORBATE PEROXIDASE1 (APX1) was downregulated in orpk1 mutants and upregulated in overexpression lines. Examining how this regulation occurs at the molecular level is an interesting task for future studies. Fluorescent resonance energy transfer (FRET) analyses have demonstrated that AtORPK1 dimerizes at the plasma membrane upon ligand binding. This results in the autophosphorylation of the complex and endocytosis. In the endosome/prevascular compartment, the C-terminal kinase domain of AtORPK1 interacts directly with AtKAPP. Trafficking and binding to AtKAPP is essential for the activation of downstream antioxidant genes []. Taken together, these results suggest that AtORPK1 positively regulates oxidative stress signaling in plants. Analysis of rice proteomes revealed a set of differentially expressed proteins upon infection with Xanthomonas oryzae pv. Oryzicola (Xoc), with the Xoc-associated receptor-like kinase (XCRLK) a candidate among these, one whose role in biotic and abiotic stress responses has been investigated. mRNA levels of XCRLK were seen to be significantly upregulated by ABA, indole acetic acid, and H2O2 application, implying that it might participate in the response to several stresses and phytohormones. Overexpression of XCRLK has resulted in enhanced resistance towards Xoc and these plants accumulated lesser H2O2 compared with wild-type seedlings. Additionally, the expression of the resistance genes, PHENYLALANINE AMMONIA-LYASE 1 (PAL1) and PR5, and the oxidation-related genes, WRKY77 and WRKY13, was seen to be higher in XCRLK-overexpressing transgenic plants, indicating that XCRLK is essential for providing resistance against Xoc and enhancing the antioxidant ability in rice []. In summary, the broad spectrum of stress responses with ROS participation makes it difficult to define a specific role of RLKs in abiotic tolerance responses. In many cases, ROS production is far downstream of stress perception. However, AtORPK1 provides an example for a mechanism of how an RLK directly controls oxidative stress resistance.
3.4. Temperature Stress
The earliest physiological change that occurs in response to low temperature is the reduction in membrane fluidity that results from alterations in lipid composition []. Moreover, it can also lead to the rearrangement of the cytoskeleton, dehydration of cells and tissues, electrolyte leakage, Ca2+ influx, ultrastructural modifications in photosynthetic apparatus, and electron transport []. In recent years, studies on the identification of plant sensors and transcriptional networks involved in low-temperature sensing have been gaining momentum, as this knowledge will aid in the development of crops with improved winter resilience []. An increasing number of studies have suggested a role for RLKs in the regulation of cold adaptation. CBFs or DREBs regulate the expression of COR genes and play a pivotal role in cold acclimation []. Transcription levels of Cold tolerance LRR-RLK1 (CTLK1) in Medicago truncatula and M. falcate were upregulated by cold treatment and the loss of MtCTLK1 resulted in lower survival rates, while its overexpression enhanced the survival rate and lowered the 50% ion leakage after freezing stress. Furthermore, MtCTLK1 overexpressors had higher antioxidant enzyme activities and proline levels along with higher mRNA levels of associated genes. Additionally, the induction of CBFs and CBF-dependent cold-responsive genes was lower in the mtctlk1 mutants but higher in the MtCTLK1-overexpressor plants, indicating that MtCTLK1 protects M. truncatula from cold stress. The above evidence suggests a positive role for MtCTLK1 in providing tolerance against cold stress through regulation of the CBF pathway, antioxidant defense system, and proline accumulation []. Using forward genetic screens, an LRR-RLK, CTB4a (cold tolerance at booting stage), was cloned and was found to play a role in seed setting by increasing the pollen fertility during cold stress in rice. Biochemical assays revealed the interaction between CTB4a and AtpB, the beta subunit of ATP synthases, demonstrating that this association is crucial for the synthesis of ATP. The ATP might provide energy for improving seed setting, and thus yield, under cold stress conditions [].
The involvement of RLKs in plant thermotolerance and their underlying mechanisms is poorly documented, although a few studies have elucidated how RLKs buffer the detrimental effects of high temperatures. The LRR-RLKs Thermo-Sensitive Genic Male Sterile 10 (TMS10) and its close homolog TMS10-Like (TMS10L), have redundant functions in regulating male fertility under variable temperatures. tms10 mutant plants did not produce viable pollen grains and showed male sterility under high temperatures (>24 °C) because of abnormalities in tapetal degeneration and pollen wall formation. However, it is partially fertile under low temperatures (23–24 °C). Interestingly, tms10 tms10l-1 and tms10 tms10l-2 double mutants displayed male sterility under both low and high temperatures. These observations suggest that TMS10L and TMS10 function together to safeguard the effect of changing temperatures on male fertility in rice []. Previous studies have documented the importance of multi-functional ER genes in plant development and in response to environmental changes []. To break summer dormancy and to provide heat tolerance, Juneidi et al. [] introduced a heat responsive ER gene into the Chinese medicinal plant, Pinellia ternata. The transgenic lines were better adapted to heat stress. Moreover, higher leaf stomatal conductance, water-use efficiency, and carbon assimilation were detected in ER overexpressor genotypes on exposure to heat stress than in the controls. These results were confirmed under field conditions, where the plants were exposed to 35 °C for 90 days, and highlight the thermo-tolerant ability of ER in P. ternata. In conclusion, although there is sufficient evidence for the involvement of RLKs in cold and heat stress adaptation, the underlying molecular mechanisms are poorly investigated. The signaling pathway for plant adaptation to cold was quite intensively studied and contains well defined signaling components, while adaptation to heat is more complex and diversified. Therefore, the interference of RLKs that leads to cold adaptation is probably easier to investigate than that which leads to heat adaptation.
3.5. Metal Stress
Although the metals and metalloids collectively known as heavy metals (HMs) play significant roles in diverse biological processes, such as metabolic reactions and antioxidant defense systems, extensive exposure to certain metals can be lethal to microorganisms and plants []. These are taken up by the plants, which results in the inhibition of germination, root elongation, reduced crop yields and the generation of ROS []. Plants have acquired various strategies to tolerate and detoxify metals and heavy metals and these are mediated by a nexus of signaling pathways, including RLKs. WAK1 has been reported to be a key gene responsible for imparting tolerance against aluminum (Al3+) toxicity in Arabidopsis. Al3+ exposure results in the upregulation of WAK1 mRNA and protein levels after 3 and 6 hours, respectively. Moreover, overexpression of WAK1 leads to a 3-fold increase in root growth as compared with wild-type plants in the presence of Al3+ []. Analysis of the WAKL4 promoter fused with the GUS reporter gene indicated that it was strongly activated by various mineral nutrients, including Na+, K+, copper (Cu2+), nickel (Ni2+), and zinc (Zn2+). A T-DNA introduced into the promoter region of the WAKL4 gene inhibited its expression by Na+, K+, Cu2+, Zn2+ but, interestingly, not by Ni2+. Wakl4–1 mutants are hypersensitive to Na+, K+, Cu2+, and Zn2+, as indicated by the reduction in root lengths, while they are tolerant towards toxic levels of Ni2+. In addition, WAKL4 is essential for the expression of Zn2+ transporter genes under Zn2+ limiting conditions. Overall, these results highlight the importance of WAKL4 in plant mineral responses []. Yin and Hou [] investigated the expression patterns of OsWAK124 under numerous stress environments, including NaCl, AlCl3, CuSO4, and CdSO4. GUS staining revealed that, under normal conditions, OsWAK124 primarily expresses at the shoot–root junction. However, on exposure to the above-mentioned stresses, its promoter activity is also induced in non-meristematic tissues, such as leaf, stem, and root. Furthermore, overexpression of OsWAK124 rendered rice plants more resistant to Al3+, Cu2+ and Cd2+, indicating that it participates in (heavy) metal stress responses. Additionally, OsWAK11 contributes to Cu2+ detoxification when plants are exposed to excess amounts of Cu2+. By knocking out of OsWAK11, the plants became hypersensitive to Cu2+ due to the higher uptake of Cu2+ by OsWAK11-RNAi seedlings into the cytoplasm. Intriguingly, there was also a significant decrease in Cu2+ levels in the pectin and hemicellulose fractions, suggesting that OsWAK11 controls the Cu2+ binding ability in the cell wall. It has been reported that the degree of pectin methyl esterification (PME) is negatively related to PME activity in Arabidopsis []. OsWAK11-RNAi exhibited greater cell wall methyl esterification than wild-type plants, resulting in fewer transcription levels of the PME encoding gene OsPME14 under normal and Cu2+-stressed conditions. Moreover, disruption of OsPME14 resembled the phenotypes of OsWAK11-RNAi lines under Cu2+ stress, indicating the possibility of OsPME14 as a downstream component of OsWAK11. In conclusion, under Cu2+ toxicity, plants activate OsWAK11 to modify the properties of the cell wall so that the immobilization of Cu2+ in the cell wall is enhanced [].
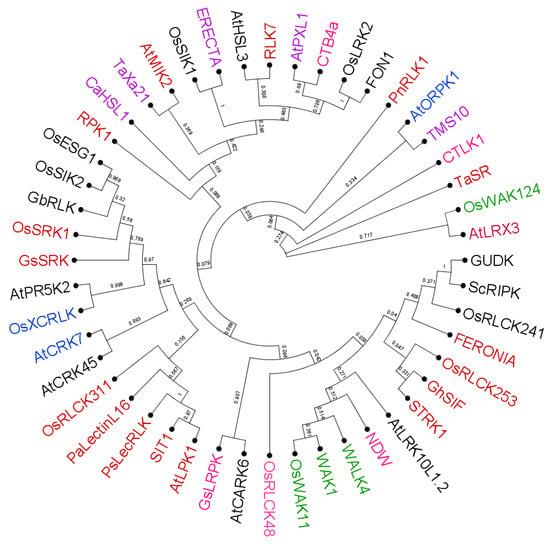
Figure 3.
Phylogenetic tree for the RLKs involved in various abiotic stresses in different plant species. Different colors represent various stress conditions in which the RLKs mentioned in Table 1 are involved. The RLKs colored in red are involved in salt stress; green in (heavy) metal stress; blue in oxidative stress; violet in heat stress; pink in cold stress; and black in drought stress. Multiple sequence alignment of their amino acid sequences was performed by MUSCLE program. Phylogenetic tree was constructed by the neighbor-joining method, with 1000 bootstrap replications using the MEGA 11.0.13 software (https://www.megasoftware.net, accessed on 21 August 2023/; Kumar et al. []).
4. Conclusions and Perspectives
RLKs act as molecular hubs for communication between cells and the extracellular environment, which is essential for the coordinated development and growth of a plant under stress conditions. Great progress has been made in characterizing RLKs but our knowledge about their specificity and downstream components in abiotic stress responses is still fragmentary. Although great attention has been given to studying the role of LRR-RLKs in biotic stress responses, there is a pressing need to integrate these less-studied kinases into abiotic stress research as this will reveal novel stress-responsive components which can be leveraged for engineering crops with broad-spectrum resistance.
The involvement of RLKs in various biotic stress responses is well documented, while much less is known about their involvement in abiotic stress responses. This raises the question as to whether this is due to the lack of experiments or whether RLKs are more involved in biotic stress responses. Therefore, it is important to identify the specific ligands that activate the RLK-induced signaling pathways. Besides protein–protein interactions in or at the plasma membrane after receptor activation, some of the downstream signaling molecules might be involved in multiple biotic and abiotic stress responses. This includes Ca2+, ROS and MAPKs, although different stimuli establish, e.g., different Ca2+ signatures or produce ROS in different compartments. Therefore, it might be important to understand which of the downstream components are involved in establishing the crosstalk between the signaling pathways without losing the specificity of the response.
The phylogenetic tree (Figure 3) demonstrates that there is little relationship between the evolution of the RLKs and the abiotic stress by which it they can be activated. Closer inspection of the tree might help to identify novel RLKs in different plant species involved in a specific stress response, or vice versa.
Most of the studies have examined a plant’s response to one stress at a time, but under natural conditions, plants face multiple abiotic and biotic stresses simultaneously, thus, it becomes imperative to understand how RLKs integrate signals from diverse stimuli to fine-tune their defense strategies and to allocate resources effectively. Powerful molecular tools, such as single-cell-omics, cellular and subcellular imaging, metabolomics, and phenomics analyses, will be indispensable in order to gain an in-depth understanding of kinase-mediated signal transduction pathways under abiotic stresses []. Genetic studies have revealed only a handful of ligand–receptor pairs and most of the RLKs are still orphans. Ligand identification is a challenging task due to high redundancy, but is nonetheless pivotal if we are to unravel their functions and the intricate signaling web they regulate. Although research on model plants, Arabidopsis and rice have contributed significantly to our understanding of the molecular mechanisms underpinning responses to various stressors, identification of RLKs in other crops will help us to design strategies for crop improvement specifically tailored to their genetic backgrounds. Furthermore, dissection of the interaction partners of RLKs when responding to biotic and abiotic stresses through phosphoproteomics may be fruitful for targeted genetic manipulations and the design of crops with enhanced resilience to multiple stresses.
Author Contributions
A.G. conceptualized the study, designed the figures and wrote the manuscript. R.O. proofread and edited the final draft. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (CRC1127, project ID: 239748522 to R.O.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Akanksha Gandhi is supported by the International Max-Planck-Research School (Max-Planck Institute for Chemical Ecology).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef]
- Bray, E.A.; Bailey-Serres, J.; Weretilnyk, E. Biochemistry and Molecular Biology of Plants; American Society of Plant Physiologists: Rockville, Md, USA, 2000; pp. 1158–1203. [Google Scholar]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Pereira, A. Plant abiotic stress challenges from the changing environment. Front. Plant Sci. 2016, 7, 1123. [Google Scholar] [CrossRef]
- Tuteja, N.; Sopory, S.K. Chemical signaling under abiotic stress environment in plants. Plant Signal. Behav. 2008, 3, 525–536. [Google Scholar] [CrossRef]
- Sierla, M.; Waszczak, C.; Vahisalu, T.; Kangasjärvi, J. Reactive oxygen species in the regulation of stomatal movements. Plant Physiol. 2016, 171, 1569–1580. [Google Scholar] [CrossRef]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef]
- Wankhede, D.P.; Misra, M.; Singh, P.; Sinha, A.K. Rice mitogen activated protein kinase kinase and mitogen activated protein kinase interaction network revealed by in-silico docking and yeast two-hybrid approaches. PLoS ONE 2013, 8, e65011. [Google Scholar] [CrossRef]
- Hanks, S.K.; Quinn, A.M.; Hunter, T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 1988, 241, 42–52. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Shiu, S.-H. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 2619–2639. [Google Scholar] [CrossRef]
- Manning, G.; Plowman, G.D.; Hunter, T.; Sudarsanam, S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002, 27, 514–520. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef]
- Belvin, M.P.; Anderson, K.V. A conserved signaling pathway: The Drosophila toll-dorsal pathway. Annu. Rev. Cell Dev. Biol. 1996, 12, 393–416. [Google Scholar] [CrossRef]
- Cao, Z.; Henzel, W.J.; Gao, X. IRAK: A kinase associated with the interleukin-1 receptor. Science 1996, 271, 1128–1131. [Google Scholar] [CrossRef] [PubMed]
- Flannery, S.; Bowie, A.G. The interleukin-1 receptor-associated kinases: Critical regulators of innate immune signalling. Biochem. Pharmacol. 2010, 80, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.-H.; Mayer, K.F.; Li, W.-H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Wang, G.; Zhou, J.-M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef]
- Diévart, A.; Clark, S.E. LRR-containing receptors regulating plant development and defense. Development 2004, 131, 251–261. [Google Scholar] [CrossRef]
- Lindner, H.; Müller, L.M.; Boisson-Dernier, A.; Grossniklaus, U. CrRLK1L receptor-like kinases: Not just another brick in the wall. Curr. Opin. Plant Biol. 2012, 15, 659–669. [Google Scholar] [CrossRef]
- Stone, J.M.; Trotochaud, A.E.; Walker, J.C.; Clark, S.E. Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 1998, 117, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Zhang, X.-C.; Neece, D.; Ramonell, K.M.; Clough, S.; Kim, S.-y.; Stacey, M.G.; Stacey, G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008, 20, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Breiden, M.; Simon, R. Q&A: How does peptide signaling direct plant development? BMC Biol. 2016, 14, 58. [Google Scholar]
- Gudesblat, G.E.; Schneider-Pizoń, J.; Betti, C.; Mayerhofer, J.; Vanhoutte, I.; Van Dongen, W.; Boeren, S.; Zhiponova, M.; De Vries, S.; Jonak, C. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 2012, 14, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [PubMed]
- Krishna, P. Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 2003, 22, 289–297. [Google Scholar] [CrossRef]
- Zhu, J.-Y.; Sae-Seaw, J.; Wang, Z.-Y. Brassinosteroid signalling. Development 2013, 140, 1615–1620. [Google Scholar] [CrossRef]
- Huang, P.; Ju, H.-W.; Min, J.-H.; Zhang, X.; Kim, S.-H.; Yang, K.-Y.; Kim, C.S. Overexpression of L-type lectin-like protein kinase 1 confers pathogen resistance and regulates salinity response in Arabidopsis thaliana. Plant Sci. 2013, 203, 98–106. [Google Scholar] [CrossRef]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress. Mol. plant 2008, 1, 732–750. [Google Scholar] [CrossRef]
- Veronese, P.; Nakagami, H.; Bluhm, B.; AbuQamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.A.; Hirt, H.; Mengiste, T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 2006, 18, 257–273. [Google Scholar] [CrossRef]
- Tang, W.; Kim, T.-W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.-Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.-J.D.; Liebrand, T.W.; Yadeta, K.A.; Coaker, G. PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol. 2015, 169, 2950–2962. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luo, X.; Wu, W.; Liang, Y.; Xu, N.; Wang, Z.; Zou, H.; Liu, J. Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J. 2020, 39, e102856. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhou, J.-M. Receptor-like cytoplasmic kinases: Central players in plant receptor kinase–mediated signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef]
- Lin, W.; Ma, X.; Shan, L.; He, P. Big roles of small kinases: The complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J. Integr. Plant Biol. 2013, 55, 1188–1197. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, J. Regulatory role of receptor-like cytoplasmic kinases in early immune signaling events in plants. FEMS Microbiol. Rev. 2020, 44, 845–856. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Kessler, S.A.; Grossniklaus, U. The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 2011, 62, 1581–1591. [Google Scholar] [CrossRef]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef]
- Laluk, K.; Luo, H.; Chai, M.; Dhawan, R.; Lai, Z.; Mengiste, T. Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell 2011, 23, 2831–2849. [Google Scholar] [CrossRef]
- Yu, X.; Liu, Z.; Qin, A.; Zhou, Y.; Zhao, Z.; Yang, J.; Hu, M.; Liu, H.; Liu, Y.; Sun, S. FLS2-RBOHD module regulates changes in the metabolome of Arabidopsis in response to abiotic stress. Plant-Environ. Interact. 2023, 4, 36–54. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, J.M. Receptor-L ike Kinases in Plant Innate Immunity. J. Integr. Plant Biol. 2013, 55, 1271–1286. [Google Scholar] [CrossRef] [PubMed]
- Böhm, H.; Albert, I.; Fan, L.; Reinhard, A.; Nürnberger, T. Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 2014, 20, 47–54. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. cell 2014, 54, 263–272. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ding, Y.; Jiang, Q.; Wang, F.; Sun, J.; Zhu, C. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 2017, 36, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Soltabayeva, A.; Dauletova, N.; Serik, S.; Sandybek, M.; Omondi, J.O.; Kurmanbayeva, A.; Srivastava, S. Receptor-like Kinases (LRR-RLKs) in Response of Plants to Biotic and Abiotic Stresses. Plants 2022, 11, 2660. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.L.; Daniell, H. Plastid biotechnology for crop production: Present status and future perspectives. Plant Mol. Biol. 2011, 76, 211–220. [Google Scholar] [CrossRef]
- Jose, J.; Ghantasala, S.; Roy Choudhury, S. Arabidopsis transmembrane receptor-like kinases (RLKs): A bridge between extracellular signal and intracellular regulatory machinery. Int. J. Mol. Sci. 2020, 21, 4000. [Google Scholar] [CrossRef]
- Haffani, Y.Z.; Silva, N.F.; Goring, D.R. Receptor kinase signalling in plants. Can. J. Bot. 2004, 82, 1–15. [Google Scholar] [CrossRef][Green Version]
- Shiu, S.-H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci.’s STKE 2001, 2001, re22. [Google Scholar] [CrossRef] [PubMed]
- Lehti-Shiu, M.D.; Zou, C.; Hanada, K.; Shiu, S.-H. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009, 150, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Dunne, A.; O’Neill, L.A. The interleukin-1 receptor/Toll-like receptor superfamily: Signal transduction during inflammation and host defense. Sci.’s STKE 2003, 2003, re3. [Google Scholar] [CrossRef] [PubMed]
- Torii, K.U. Leucine-rich repeat receptor kinases in plants: Structure, function, and signal transduction pathways. Int. Rev. Cytol. 2004, 234, 1–46. [Google Scholar] [PubMed]
- Wang, Z.-Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Shinohara, H.; Sakagami, Y.; Matsubayashi, Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 2008, 319, 294. [Google Scholar] [CrossRef]
- Yang, S.-L.; Xie, L.-F.; Mao, H.-Z.; Puah, C.S.; Yang, W.-C.; Jiang, L.; Sundaresan, V.; Ye, D. Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 2003, 15, 2792–2804. [Google Scholar] [CrossRef]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Gust, A.A.; Felix, G. Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 2014, 21, 104–111. [Google Scholar] [CrossRef]
- Hohmann, U.; Lau, K.; Hothorn, M. The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 2017, 68, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, U.; Santiago, J.; Nicolet, J.; Olsson, V.; Spiga, F.M.; Hothorn, L.A.; Butenko, M.A.; Hothorn, M. Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc. Natl. Acad. Sci. USA 2018, 115, 3488–3493. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Han, Z.; Wang, J.; Lin, G.; Chai, J. Structural insights into ligand recognition and activation of plant receptor kinases. Curr. Opin. Struct. Biol. 2017, 43, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Wu, X.N.; Gilbert, M.; Schulze, W.X. Classification and interactions of LRR receptors and co-receptors within the Arabidopsis plasma membrane–an overview. Front. Plant Sci. 2019, 10, 472. [Google Scholar] [CrossRef] [PubMed]
- Morillo, S.A.; Tax, F.E. Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 2006, 9, 460–469. [Google Scholar] [CrossRef]
- Su, Y.; Peng, X.; Shen, S. Identification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in paper mulberry and their potential roles in response to cold stress. Comput. Biol. Chem. 2022, 97, 107622. [Google Scholar] [CrossRef]
- Pitorre, D.; Llauro, C.; Jobet, E.; Guilleminot, J.; Brizard, J.-P.; Delseny, M.; Lasserre, E. RLK7, a leucine-rich repeat receptor-like kinase, is required for proper germination speed and tolerance to oxidative stress in Arabidopsis thaliana. Planta 2010, 232, 1339–1353. [Google Scholar] [CrossRef]
- Ouyang, S.Q.; Liu, Y.F.; Liu, P.; Lei, G.; He, S.J.; Ma, B.; Zhang, W.K.; Zhang, J.S.; Chen, S.Y. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010, 62, 316–329. [Google Scholar] [CrossRef]
- Yang, L.; Wu, K.; Gao, P.; Liu, X.; Li, G.; Wu, Z. GsLRPK, a novel cold-activated leucine-rich repeat receptor-like protein kinase from Glycine soja, is a positive regulator to cold stress tolerance. Plant Sci. 2014, 215, 19–28. [Google Scholar] [CrossRef]
- Peumans, W.J.; Van Damme, E. Lectins as plant defense proteins. Plant Physiol. 1995, 109, 347. [Google Scholar] [CrossRef]
- Van Damme, E.J.; Lannoo, N.; Peumans, W.J. Plant lectins. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2008; Volume 48, pp. 107–209. [Google Scholar]
- Vaid, N.; Pandey, P.K.; Tuteja, N. Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice. Plant Mol. Biol. 2012, 80, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Van Holle, S.; Van Damme, E.J. Messages from the past: New insights in plant lectin evolution. Front. Plant Sci. 2019, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Opas, M.; Tharin, S.; Milner, R.; Michalak, M. Identification and localization of calreticulin in plant cells. Protoplasma 1996, 191, 164–171. [Google Scholar] [CrossRef]
- Powers-Fletcher, M.V.; Jambunathan, K.; Brewer, J.L.; Krishnan, K.; Feng, X.; Galande, A.K.; Askew, D.S. Impact of the lectin chaperone calnexin on the stress response, virulence and proteolytic secretome of the fungal pathogen Aspergillus fumigatus. PLoS ONE 2011, 6, e28865. [Google Scholar] [CrossRef]
- Schallus, T.; Jaeckh, C.; Fehér, K.; Palma, A.S.; Liu, Y.; Simpson, J.C.; Mackeen, M.; Stier, G.; Gibson, T.J.; Feizi, T. Malectin: A novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 2008, 19, 3404–3414. [Google Scholar] [CrossRef]
- Franck, C.M.; Westermann, J.; Boisson-Dernier, A. Plant malectin-like receptor kinases: From cell wall integrity to immunity and beyond. Annu. Rev. Plant Biol. 2018, 69, 301–328. [Google Scholar] [CrossRef]
- Vaid, N.; Macovei, A.; Tuteja, N. Knights in action: Lectin receptor-like kinases in plant development and stress responses. Mol. Plant 2013, 6, 1405–1418. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Loaiza-Figueroa, F. Gametophytic self-incompatibility is controlled by a single major locus on chromosome 1 in Lycopersicon peruvianum. Proc. Natl. Acad. Sci. USA 1985, 82, 5093–5096. [Google Scholar] [CrossRef]
- Tordai, H.; Bányai, L.; Patthy, L. The PAN module: The N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999, 461, 63–67. [Google Scholar] [CrossRef]
- Loris, R. Principles of structures of animal and plant lectins. Biochim. Biophys. Acta Gen. Subj. 2002, 1572, 198–208. [Google Scholar] [CrossRef]
- Naithani, S.; Chookajorn, T.; Ripoll, D.R.; Nasrallah, J.B. Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc. Natl. Acad. Sci. USA 2007, 104, 12211–12216. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.; Eichbaum, Q.; Sheriff, S.; Ezekowitz, R.A.B. The collectins in innate immunity. Curr. Opin. Immunol. 1996, 8, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Hawgood, S.; Akiyama, J.; Brown, C.; Allen, L.; Li, G.; Poulain, F.R. GM-CSF mediates alveolar macrophage proliferation and type II cell hypertrophy in SP-D gene-targeted mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001, 280, L1148–L1156. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Patel, A.; Mathieu, M.; Kim, S.-Y.; Xu, D.; Stacey, G. A lectin receptor-like kinase is required for pollen development in Arabidopsis. Plant Mol. Biol. 2008, 67, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; de Sain, M.; Weide, R.; Gouget, A.; Klamer, S.; Canut, H.; Govers, F. The lectin receptor kinase LecRK-I. 9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011, 7, e1001327. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kuo, Y.-C.; Mishra, S.; Tsai, C.-H.; Chien, C.-C.; Chen, C.-W.; Desclos-Theveniau, M.; Chu, P.-W.; Schulze, B.; Chinchilla, D. The lectin receptor kinase-VI. 2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 2012, 24, 1256–1270. [Google Scholar] [CrossRef]
- Riou, C.; Hervé, C.; Pacquit, V.; Dabos, P.; Lescure, B. Expression of an Arabidopsis lectin kinase receptor gene, lecRK-a1, is induced during senescence, wounding and in response to oligogalacturonic acids. Plant Physiol. Biochem. 2002, 40, 431–438. [Google Scholar] [CrossRef]
- Nishiguchi, M.; Yoshida, K.; Sumizono, T.; Tazaki, K. A receptor-like protein kinase with a lectin-like domain from lombardy poplar: Gene expression in response to wounding and characterization of phosphorylation activity. Mol. Genet. Genom. 2002, 267, 506–514. [Google Scholar] [CrossRef]
- Desclos-Theveniau, M.; Arnaud, D.; Huang, T.-Y.; Lin, G.J.-C.; Chen, W.-Y.; Lin, Y.-C.; Zimmerli, L. The Arabidopsis lectin receptor kinase LecRK-V. 5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 2012, 8, e1002513. [Google Scholar] [CrossRef]
- Gilardoni, P.A.; Hettenhausen, C.; Baldwin, I.T.; Bonaventure, G. Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell 2011, 23, 3512–3532. [Google Scholar] [CrossRef]
- Bonaventure, G. The Nicotiana attenuata LECTIN RECEPTOR KINASE 1 is involved in the perception of insect feeding. Plant Signal. Behav. 2011, 6, 2060–2063. [Google Scholar] [CrossRef]
- Joshi, A.; Dang, H.Q.; Vaid, N.; Tuteja, N. Pea lectin receptor-like kinase promotes high salinity stress tolerance in bacteria and expresses in response to stress in planta. Glycoconj. J. 2010, 27, 133–150. [Google Scholar] [CrossRef]
- Deng, K.; Wang, Q.; Zeng, J.; Guo, X.; Zhao, X.; Tang, D.; Liu, X. A lectin receptor kinase positively regulates ABA response during seed germination and is involved in salt and osmotic stress response. J. Plant Biol. 2009, 52, 493–500. [Google Scholar] [CrossRef]
- Liu, S.; Wang, J.; Chen, K.; Zhang, Z.; Zhang, P. The L-type lectin receptor-like kinase (PnLecRLK1) from the Antarctic moss Pohlia nutans enhances chilling-stress tolerance and abscisic acid sensitivity in Arabidopsis. Plant Growth Regul. 2017, 81, 409–418. [Google Scholar] [CrossRef]
- He, Z.-H.; Fujiki, M.; Kohorn, B.D. A cell wall-associated, receptor-like protein kinase. J. Biol. Chem. 1996, 271, 19789–19793. [Google Scholar] [CrossRef] [PubMed]
- He, Z.-H.; Cheeseman, I.; He, D.; Kohorn, B.D. A cluster of five cell wall-associated receptor kinase genes, Wak1–5, are expressed in specific organs of Arabidopsis. Plant Mol. Biol. 1999, 39, 1189–1196. [Google Scholar] [CrossRef]
- Brutus, A.; Sicilia, F.; Macone, A.; Cervone, F.; De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 2010, 107, 9452–9457. [Google Scholar] [CrossRef]
- Wagner, T.A.; Kohorn, B.D. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 2001, 13, 303–318. [Google Scholar] [CrossRef]
- Kohorn, B.; He, Z.; Fujiki, M. Elusin: A receptor-like kinase with an EGF domain in the cell wall. In Proceedings of the Phytochemical Society of Europe; Oxford University Press Inc.: Oxford, UK, 1996; pp. 297–304. [Google Scholar]
- Verica, J.A.; Chae, L.; Tong, H.; Ingmire, P.; He, Z.-H. Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol. 2003, 133, 1732–1746. [Google Scholar] [CrossRef] [PubMed]
- Verica, J.A.; He, Z.-H. The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 2002, 129, 455–459. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, C.; Li, L.; Meng, L.; Singh, J.; Jiang, N.; Deng, X.-W.; He, Z.-H.; Lemaux, P.G. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005, 139, 1107–1124. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Aguirre, J.A.; Singh, J. Genome-wide analysis of wall associated kinase (WAK) gene family in barley. Genomics 2021, 113, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Chao, Q.; Zhang, N.; Ye, J.; Tan, G.; Li, B.; Xing, Y.; Zhang, B.; Liu, H.; Fengler, K.A. A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 2015, 47, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Bacic, A.; Johnson, K.L.; Humphries, J. The role of Brachypodium distachyon wall-associated kinases (WAKs) in cell expansion and stress responses. Cells 2020, 9, 2478. [Google Scholar] [CrossRef] [PubMed]
- Decreux, A.; Messiaen, J. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 2005, 46, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.M.; Wagner, T.A.; Perret, M.; He, Z.-H.; He, D.; Kohorn, B.D. WAKs: Cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Cell Walls 2001, 47, 197–206. [Google Scholar]
- Kohorn, B.D. WAKs; cell wall associated kinases. Curr. Opin. Cell Biol. 2001, 13, 529–533. [Google Scholar] [CrossRef]
- Sivaguru, M.; Ezaki, B.; He, Z.-H.; Tong, H.; Osawa, H.; Baluška, F.E.; Volkmann, D.; Matsumoto, H. Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 2003, 132, 2256–2266. [Google Scholar] [CrossRef]
- Bot, P.; Mun, B.-G.; Imran, Q.M.; Hussain, A.; Lee, S.-U.; Loake, G.; Yun, B.-W. Differential expression of AtWAKL10 in response to nitric oxide suggests a putative role in biotic and abiotic stress responses. PeerJ 2019, 7, e7383. [Google Scholar] [CrossRef]
- Garvey, K.J.; Saedi, M.S.; Ito, J. Nucleotide sequence of Bacillus phage Ø29 genes 14 and 15: Homology of gene 15 with other phage lysozymes. Nucleic Acids Res. 1986, 14, 10001–10008. [Google Scholar] [CrossRef]
- Bateman, A.; Bycroft, M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 2000, 299, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Bielnicki, J.; Devedjiev, Y.; Derewenda, U.; Dauter, Z.; Joachimiak, A.; Derewenda, Z.S. B. subtilis ykuD protein at 2.0 Å resolution: Insights into the structure and function of a novel, ubiquitous family of bacterial enzymes. Prot. Struct. Funct. Bioinform. 2006, 62, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Mulder, L.; Lefebvre, B.; Cullimore, J.; Imberty, A. LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology 2006, 16, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.L. Energetics of protein folding. J. Mol. Biol. 2007, 371, 283–301. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, B.; Klaus-Heisen, D.; Pietraszewska-Bogiel, A.; Hervé, C.; Camut, S.; Auriac, M.-C.; Gasciolli, V.; Nurisso, A.; Gadella, T.W.; Cullimore, J. Role of N-glycosylation sites and CXC motifs in trafficking of Medicago truncatula Nod factor perception protein to plasma membrane. J. Biol. Chem. 2012, 287, 10812–10823. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Biswas, R.; Lenz, H.D.; Rauhut, T.; Ranf, S.; Kemmerling, B.; Götz, F.; Glawischnig, E.; Lee, J.; Felix, G. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 2007, 282, 32338–32348. [Google Scholar] [CrossRef]
- Gust, A.A.; Willmann, R.; Desaki, Y.; Grabherr, H.M.; Nürnberger, T. Plant LysM proteins: Modules mediating symbiosis and immunity. Trends Plant Sci. 2012, 17, 495–502. [Google Scholar] [CrossRef]
- Gough, C.; Cullimore, J. Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol. Plant-Microbe Interact. 2011, 24, 867–878. [Google Scholar] [CrossRef]
- Newman, M.-A.; Sundelin, T.; Nielsen, J.T.; Erbs, G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 2013, 4, 139. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, X.-C.; Stacey, G. Chitin signaling and plant disease resistance. Plant Signal. Behav. 2008, 3, 831–833. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Tanaka, K.; Zhang, X.-C.; Son, G.H.; Brechenmacher, L.; Nguyen, T.H.N.; Stacey, G. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012, 160, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. elife 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, C.; Liang, Y.; Stacey, G. Chitin receptor CERK 1 links salt stress and chitin-triggered innate immunity in Arabidopsis. Plant J. 2017, 89, 984–995. [Google Scholar] [CrossRef]
- Laohavisit, A.; Richards, S.L.; Shabala, L.; Chen, C.; Colaço, R.D.; Swarbreck, S.M.; Shaw, E.; Dark, A.; Shabala, S.; Shang, Z. Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol. 2013, 163, 253–262. [Google Scholar] [CrossRef]
- Bourdais, G.; Burdiak, P.; Gauthier, A.; Nitsch, L.; Salojärvi, J.; Rayapuram, C.; Idänheimo, N.; Hunter, K.; Kimura, S.; Merilo, E. Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 2015, 11, e1005373. [Google Scholar] [CrossRef]
- Chen, Z. A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol. 2001, 126, 473–476. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Brosché, M.; Salojärvi, J.; Kangasjärvi, S.; Idänheimo, N.; Mersmann, S.; Robatzek, S.; Karpiński, S.; Karpińska, B.; Kangasjärvi, J. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010, 10, 95. [Google Scholar] [CrossRef]
- Idänheimo, N.; Gauthier, A.; Salojärvi, J.; Siligato, R.; Brosché, M.; Kollist, H.; Mähönen, A.P.; Kangasjärvi, J.; Wrzaczek, M. The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 2014, 445, 457–462. [Google Scholar] [CrossRef]
- Miyakawa, T.; Miyazono, K.i.; Sawano, Y.; Hatano, K.i.; Tanokura, M. Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases. Proteins Struct. Funct. Bioinform. 2009, 77, 247–251. [Google Scholar] [CrossRef]
- Miyakawa, T.; Hatano, K.-i.; Miyauchi, Y.; Suwa, Y.-i.; Sawano, Y.; Tanokura, M. A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol. 2014, 166, 766–778. [Google Scholar] [CrossRef]
- Sawano, Y.; Miyakawa, T.; Yamazaki, H.; Tanokura, M.; Hatano, K.-I. Purification, characterization, and molecular gene cloning of an antifungal protein from Ginkgo biloba seeds. Biol. Chem. 2007, 388, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, L.-H.; Zhao, J.-F.; Song, Y.; Zhang, C.-J.; Guo, Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tian, H.; Chen, D.; Zhang, H.; Sun, M.; Chen, S.; Qin, Z.; Ding, Z.; Dai, S. Cysteine-rich receptor-like protein kinases: Emerging regulators of plant stress responses. Trends Plant Sci. 2023, 28, 776–794. [Google Scholar] [CrossRef] [PubMed]
- Pelagio-Flores, R.; Muñoz-Parra, E.; Barrera-Ortiz, S.; Ortiz-Castro, R.; Saenz-Mata, J.; Ortega-Amaro, M.A.; Jiménez-Bremont, J.F.; López-Bucio, J. The cysteine-rich receptor-like protein kinase CRK28 modulates Arabidopsis growth and development and influences abscisic acid responses. Planta 2020, 251, 2. [Google Scholar] [CrossRef]
- Burdiak, P.; Rusaczonek, A.; Witoń, D.; Głów, D.; Karpiński, S. Cysteine-rich receptor-like kinase CRK5 as a regulator of growth, development, and ultraviolet radiation responses in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 3325–3337. [Google Scholar] [CrossRef]
- Tyagi, S.; Sharma, A.; Singh, K.; Upadhyay, S.K. Genomic dissection and transcriptional profiling of Cysteine-rich receptor-like kinases in five cereals and functional characterization of TaCRK68-A. Int. J. Biol. Macromol. 2019, 134, 316–329. [Google Scholar]
- Wu, T.; Guo, F.; Xu, G.; Yu, J.; Zhang, L.; Wei, X.; Zhu, X.; Zhang, Z. The receptor-like kinase TaCRK-7A inhibits Fusarium pseudograminearum growth and mediates resistance to Fusarium crown rot in wheat. Biology 2021, 10, 1122. [Google Scholar] [CrossRef]
- Lee, D.S.; Kim, Y.C.; Kwon, S.J.; Ryu, C.-M.; Park, O.K. The Arabidopsis cysteine-rich receptor-like kinase CRK36 regulates immunity through interaction with the cytoplasmic kinase BIK1. Front. Plant Sci. 2017, 8, 1856. [Google Scholar] [CrossRef]
- Ohtake, Y.; Takahashi, T.; Komeda, Y. Salicylic acid induces the expression of a number of receptor-like kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 1038–1044. [Google Scholar] [CrossRef]
- Chern, M.; Xu, Q.; Bart, R.S.; Bai, W.; Ruan, D.; Sze-To, W.H.; Canlas, P.E.; Jain, R.; Chen, X.; Ronald, P.C. A genetic screen identifies a requirement for cysteine-rich–receptor-like kinases in rice NH1 (OsNPR1)-mediated immunity. PLoS Genet. 2016, 12, e1006049. [Google Scholar]
- Mou, S.; Meng, Q.; Gao, F.; Zhang, T.; He, W.; Guan, D.; He, S. A cysteine-rich receptor-like protein kinase CaCKR5 modulates immune response against Ralstonia solanacearum infection in pepper. BMC Plant Biol. 2021, 21, 382. [Google Scholar] [CrossRef] [PubMed]
- Ederli, L.; Madeo, L.; Calderini, O.; Gehring, C.; Moretti, C.; Buonaurio, R.; Paolocci, F.; Pasqualini, S. The Arabidopsis thaliana cysteine-rich receptor-like kinase CRK20 modulates host responses to Pseudomonas syringae pv. tomato DC3000 infection. J. Plant Physiol. 2011, 168, 1784–1794. [Google Scholar] [CrossRef]
- Chen, K.; Du, L.; Chen, Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 2003, 53, 61–74. [Google Scholar] [CrossRef]
- Chen, K.; Fan, B.; Du, L.; Chen, Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 2004, 56, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Acharya, B.R.; Raina, S.; Maqbool, S.B.; Jagadeeswaran, G.; Mosher, S.L.; Appel, H.M.; Schultz, J.C.; Klessig, D.F.; Raina, R. Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 2007, 50, 488–499. [Google Scholar] [CrossRef]
- Hunter, K.; Kimura, S.; Rokka, A.; Tran, H.C.; Toyota, M.; Kukkonen, J.P.; Wrzaczek, M. CRK2 enhances salt tolerance by regulating callose deposition in connection with PLD α 1. Plant Physiol. 2019, 180, 2004–2021. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, J.; Zhang, Y.; Qiu, J.; Li, Y.; Zheng, B.; Hu, F.; Dai, S.; Huang, X. A high-quality genome sequence of alkaligrass provides insights into halophyte stress tolerance. Sci. China Life Sci. 2020, 63, 1269–1282. [Google Scholar] [CrossRef]
- Almutairi, Z.M. Effect of nano-silicon application on the expression of salt tolerance genes in germinating tomato (‘Solanum lycopersicum’L.) seedlings under salt stress. Plant Omics 2016, 9, 106–114. [Google Scholar]
- Tanaka, H.; Osakabe, Y.; Katsura, S.; Mizuno, S.; Maruyama, K.; Kusakabe, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 2012, 70, 599–613. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, Z.; Zhu, W.; Liu, J.; Zhang, Y. Genome-wide identification and characterization of cysteine-rich receptor-like protein kinase genes in tomato and their expression profile in response to heat stress. Diversity 2021, 13, 258. [Google Scholar] [CrossRef]
- Campos Mantello, C.; Boatwright, L.; da Silva, C.C.; Scaloppi, E.J.; de Souza Goncalves, P.; Barbazuk, W.B.; Pereira de Souza, A. Deep expression analysis reveals distinct cold-response strategies in rubber tree (Hevea brasiliensis). BMC Genom. 2019, 20, 455. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Liang, S.; Wu, Z.; Bi, C.; Yu, Y.-T.; Wang, X.-F.; Zhang, D.-P. Overexpression of an Arabidopsis cysteine-rich receptor-like protein kinase, CRK5, enhances abscisic acid sensitivity and confers drought tolerance. J. Exp. Bot. 2016, 67, 5009–5027. [Google Scholar] [CrossRef] [PubMed]
- Marshall, A.; Aalen, R.B.; Audenaert, D.; Beeckman, T.; Broadley, M.R.; Butenko, M.A.; Caño-Delgado, A.I.; de Vries, S.; Dresselhaus, T.; Felix, G. Tackling drought stress: Receptor-like kinases present new approaches. Plant Cell 2012, 24, 2262–2278. [Google Scholar] [CrossRef] [PubMed]
- Tosti, N.; Pasqualini, S.; Borgogni, A.; Ederli, L.; Falistocco, E.; Crispi, S.; Paolocci, F. Gene expression profiles of O3-treated Arabidopsis plants. Plant Cell Environ. 2006, 29, 1686–1702. [Google Scholar] [CrossRef]
- Czernic, P.; Visser, B.; Sun, W.; Savouré, A.; Deslandes, L.; Marco, Y.; Van Montagu, M.; Verbruggen, N. Characterization of an Arabidopsis thaliana receptor-like protein kinase gene activated by oxidative stress and pathogen attack. Plant J. 1999, 18, 321–327. [Google Scholar] [CrossRef]
- Du, L.; Chen, Z. Identification of genes encoding receptor-like protein kinases as possible targets of pathogen-and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 2000, 24, 837–847. [Google Scholar] [CrossRef]
- Zeiner, A.; Colina, F.J.; Citterico, M.; Wrzaczek, M. CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASES–their evolution, structure and roles in stress response and development. J. Exp. Bot. 2023, 74, 4910–4927. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Becraft, P.W.; Stinard, P.S.; McCarty, D.R. CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 1996, 273, 1406–1409. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Guo, T.; Becraft, P.W. The maize CR4 receptor-like kinase mediates a growth factor-like differentiation response. Genesis 2000, 27, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Gifford, M.L.; Robertson, F.C.; Soares, D.C.; Ingram, G.C. ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 2005, 17, 1154–1166. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Watanabe, M.; Watanabe, D.; Tanaka, T.; Machida, C.; Machida, Y. ACR4, a putative receptor kinase gene of Arabidopsis thaliana, that is expressed in the outer cell layers of embryos and plants, is involved in proper embryogenesis. Plant Cell Physiol. 2002, 43, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Gifford, M.L.; Dean, S.; Ingram, G.C. The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development 2003, 130, 4249–4258. [Google Scholar] [CrossRef]
- Watanabe, M.; Tanaka, H.; Watanabe, D.; Machida, C.; Machida, Y. The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 2004, 39, 298–308. [Google Scholar] [CrossRef]
- De Smet, I.; Vassileva, V.; De Rybel, B.; Levesque, M.P.; Grunewald, W.; Van Damme, D.; Van Noorden, G.; Naudts, M.; Van Isterdael, G.; De Clercq, R. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 2008, 322, 594–597. [Google Scholar] [CrossRef]
- Stahl, Y.; Grabowski, S.; Bleckmann, A.; Kühnemuth, R.; Weidtkamp-Peters, S.; Pinto, K.G.; Kirschner, G.K.; Schmid, J.B.; Wink, R.H.; Hülsewede, A. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 2013, 23, 362–371. [Google Scholar] [CrossRef]
- Becraft, P.W.; Asuncion-Crabb, Y. Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 2000, 127, 4039–4048. [Google Scholar] [CrossRef]
- Cao, X.; Li, K.; Suh, S.-G.; Guo, T.; Becraft, P.W. Molecular analysis of the CRINKLY4 gene family in Arabidopsis thaliana. Planta 2005, 220, 645–657. [Google Scholar] [CrossRef]
- Johnson, K.L.; Degnan, K.A.; Ross Walker, J.; Ingram, G.C. AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J. 2005, 44, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Olsen, L.; Sun, B.; Lid, S.E.; Brown, R.C.; Lemmon, B.E.; Fosnes, K.; Gruis, D.; Opsahl-Sorteberg, H.-G.; Otegui, M.S. Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1. Plant Cell 2007, 19, 3127–3145. [Google Scholar] [CrossRef]
- Zereen, J.; Ingram, G. A possible involvement of ACR4, a receptor like kinase, in plant defense mechanism. Bangladesh Pharm. J. 2012, 15, 127–130. [Google Scholar] [CrossRef]
- Bell, E.; Creelman, R.A.; Mullet, J.E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 1995, 92, 8675–8679. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.F.; Goring, D.R. The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol. 2002, 50, 667–685. [Google Scholar] [CrossRef]
- Nakhamchik, A.; Zhao, Z.; Provart, N.J.; Shiu, S.-H.; Keatley, S.K.; Cameron, R.K.; Goring, D.R. A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant Cell Physiol. 2004, 45, 1875–1881. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.; Östlund, G. InParanoid 8: Orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015, 43, D234–D239. [Google Scholar] [CrossRef]
- Haffani, Y.; Silva-Gagliardi, N.; Sewter, S.; Aldea, M.G.; Zhao, Z.; Nakhamchik, A.; Cameron, R.; Goring, D. Altered expression of PERK receptor kinases in Arabidopsis leads to changes in growth and floral organ formation. Plant Signal. Behav. 2006, 1, 251–260. [Google Scholar] [CrossRef]
- Won, S.-K.; Lee, Y.-J.; Lee, H.-Y.; Heo, Y.-K.; Cho, M.; Cho, H.-T. Cis-element-and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 2009, 150, 1459–1473. [Google Scholar] [CrossRef]
- Humphrey, T.V.; Haasen, K.E.; Aldea-Brydges, M.G.; Sun, H.; Zayed, Y.; Indriolo, E.; Goring, D.R. PERK–KIPK–KCBP signalling negatively regulates root growth in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 71–83. [Google Scholar] [CrossRef]
- Hwang, I.; Kim, S.Y.; Kim, C.S.; Park, Y.; Tripathi, G.R.; Kim, S.-K.; Cheong, H. Over-expression of the IGI1 leading to altered shoot-branching development related to MAX pathway in Arabidopsis. Plant Mol. Biol. 2010, 73, 629–641. [Google Scholar] [CrossRef][Green Version]
- Borassi, C.; Sede, A.R.; Mecchia, M.A.; Salgado Salter, J.D.; Marzol, E.; Muschietti, J.P.; Estevez, J.M. An update on cell surface proteins containing extensin-motifs. J. Exp. Bot. 2016, 67, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhang, G.; Zhou, Y.; Zhang, Z.; Wang, W.; Du, Y.; Wu, Z.; Song, C.P. Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca2+ signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 2009, 60, 314–327. [Google Scholar] [CrossRef]
- Ruprecht, C.; Mendrinna, A.; Tohge, T.; Sampathkumar, A.; Klie, S.; Fernie, A.R.; Nikoloski, Z.; Persson, S.; Mutwil, M. FamNet: A framework to identify multiplied modules driving pathway expansion in plants. Plant Physiol. 2016, 170, 1878–1894. [Google Scholar] [CrossRef] [PubMed]
- Qanmber, G.; Liu, J.; Yu, D.; Liu, Z.; Lu, L.; Mo, H.; Ma, S.; Wang, Z.; Yang, Z. Genome-wide identification and characterization of the PERK gene family in Gossypium hirsutum reveals gene duplication and functional divergence. Int. J. Mol. Sci. 2019, 20, 1750. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Shi, J.; Zhang, T.; Zhong, Y.; Zhang, L.; Yu, G.; Zhang, T.; Zhu, X.; Xing, Y.; Yin, W. Zebra leaf 15, a receptor-like protein kinase involved in moderate low temperature signaling pathway in rice. Rice 2019, 12, 83. [Google Scholar] [CrossRef]
- Xue, C.; Li, W.; Shen, R.; Lan, P. PERK13 modulates phosphate deficiency-induced root hair elongation in Arabidopsis. Plant Sci. 2021, 312, 111060. [Google Scholar] [CrossRef]
- Sharma, I.; Russinova, E. Probing plant receptor kinase functions with labeled ligands. Plant Cell Physiol. 2018, 59, 1520–1527. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Muth, P.; Irmler, S.; Schröder, G.; Schröder, J. Novel type of receptor-like protein kinase from a higher plant (Catharanthus roseus): cDNA, gene, intramolecular autophosphorylation, and identification of a threonine important for auto-and substrate phosphorylation. J. Biol. Chem. 1996, 271, 26684–26689. [Google Scholar] [CrossRef] [PubMed]
- Franck, C.M.; Westermann, J.; Bürssner, S.; Lentz, R.; Lituiev, D.S.; Boisson-Dernier, A. The protein phosphatases ATUNIS1 and ATUNIS2 regulate cell wall integrity in tip-growing cells. Plant Cell 2018, 30, 1906–1923. [Google Scholar] [CrossRef]
- Dievart, A.; Gottin, C.; Périn, C.; Ranwez, V.; Chantret, N. Origin and diversity of plant receptor-like kinases. Annu. Rev. Plant Biol. 2020, 71, 131–156. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef]
- Gachomo, E.W.; Jno Baptiste, L.; Kefela, T.; Saidel, W.M.; Kotchoni, S.O. The Arabidopsis CURVY1 (CVY1) gene encoding a novel receptor-like protein kinase regulates cell morphogenesis, flowering time and seed production. BMC Plant Biol. 2014, 14, 221. [Google Scholar] [CrossRef] [PubMed]
- Nibau, C.; Cheung, A. New insights into the functional roles of CrRLKs in the control of plant cell growth and development. Plant Signal. Behav. 2011, 6, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef]
- Guo, H.; Li, L.; Ye, H.; Yu, X.; Algreen, A.; Yin, Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 7648–7653. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, T.; Wang, B.; Lin, Q.; Zhu, S.; Li, C.; Ma, Y.; Tang, J.; Xing, J.; Li, X. RALF1-FERONIA complex affects splicing dynamics to modulate stress responses and growth in plants. Sci. Adv. 2020, 6, eaaz1622. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Wang, L.; Li, C.; Liu, Y.; Zhu, S.; Qi, Y.; Liu, X.; Lin, Q.; Luan, S.; Yu, F. Receptor protein kinase FERONIA controls leaf starch accumulation by interacting with glyceraldehyde-3-phosphate dehydrogenase. Biochem. Biophys. Res. Commun. 2015, 465, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Deguchi, M.; Brustolini, O.J.; Santos, A.A.; Silva, F.F.; Fontes, E.P. The tomato RLK superfamily: Phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense. BMC Plant Biol. 2012, 12, 229. [Google Scholar] [CrossRef]
- Yong-Feng, H.; Qian, Y.; Sheng-Wei, Z.; Da-Ye, S.; Ying, S. Receptor-like kinase CrRLK1-L subfamily: Novel motifs in extracellular domain and biological functions in plants. Prog. Biochem. Biophys. 2011, 38, 891–899. [Google Scholar]
- Pu, C.-X.; Han, Y.-F.; Zhu, S.; Song, F.-Y.; Zhao, Y.; Wang, C.-Y.; Zhang, Y.-C.; Yang, Q.; Wang, J.; Bu, S.-L. The rice receptor-like kinases DWARF AND RUNTISH SPIKELET1 and 2 repress cell death and affect sugar utilization during reproductive development. Plant Cell 2017, 29, 70–89. [Google Scholar] [CrossRef]
- Niu, E.; Cai, C.; Zheng, Y.; Shang, X.; Fang, L.; Guo, W. Genome-wide analysis of CrRLK1L gene family in Gossypium and identification of candidate CrRLK1L genes related to fiber development. Mol. Genet. Genom. 2016, 291, 1137–1154. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Restrepo, J.-M.; Huck, N.; Kessler, S.; Gagliardini, V.; Gheyselinck, J.; Yang, W.-C.; Grossniklaus, U. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 2007, 317, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yu, F.; Liu, Y.; Du, C.; Li, X.; Zhu, S.; Wang, X.; Lan, W.; Rodriguez, P.L.; Liu, X. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E5519–E5527. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.-C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 2018, 28, 666–675.e5. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Yang, J.; Gu, F.; Park, S.; Combs, J.; Adams, A.; Mayes, H.B.; Jeon, S.J.; Bahk, J.D.; Nielsen, E. A temperature-sensitive FERONIA mutant allele that alters root hair growth. Plant Physiol. 2021, 185, 405–423. [Google Scholar] [CrossRef]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.-M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 2018, 59, 2239–2254. [Google Scholar] [CrossRef]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.-Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef]
- Gigli-Bisceglia, N.; Van Zelm, E.; Huo, W.; Lamers, J.; Testerink, C. Salinity stress-induced modification of pectin activates stress signaling pathways and requires HERK/THE and FER to attenuate the response. bioRxiv 2020. [CrossRef]
- Burke, D.; Kaufman, P.; McNeil, M.; Albersheim, P. The structure of plant cell walls: VI. A survey of the walls of suspension-cultured monocots. Plant Physiol. 1974, 54, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Kieliszewski, M.; de Zacks, R.; Leykam, J.F.; Lamport, D.T. A repetitive proline-rich protein from the gymnosperm Douglas fir is a hydroxyproline-rich glycoprotein. Plant Physiol. 1992, 98, 919–926. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Showalter, A.M. Structure and function of plant cell wall proteins. Plant Cell 1993, 5, 9. [Google Scholar] [PubMed]
- Wu, H.; De Graaf, B.; Mariani, C.; Cheung, A. Hydroxyproline-rich glycoproteins in plant reproductive tissues: Structure, functions and regulation. Cell. Mol. Life Sci. CMLS 2001, 58, 1418–1429. [Google Scholar] [CrossRef]
- Ringli, C.; Keller, B.; Ryser, U. Glycine-rich proteins as structural components of plant cell walls. Cell. Mol. Life Sci. CMLS 2001, 58, 1430–1441. [Google Scholar] [CrossRef]
- Keller, B. Structural cell wall proteins. Plant Physiol. 1993, 101, 1127. [Google Scholar] [CrossRef]
- Cassab, G.I. Plant cell wall proteins. Annu. Rev. plant Biol. 1998, 49, 281–309. [Google Scholar] [CrossRef]
- Kieliszewski, M.J.; Lamport, D.T. Extensin: Repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 1994, 5, 157–172. [Google Scholar] [CrossRef]
- Wilson, L.; Fry, J. Extensin—A major cell wall glycoprotein. Plant Cell Environ. 1986, 9, 239–260. [Google Scholar]
- Liu, X.; Wolfe, R.; Welch, L.R.; Domozych, D.S.; Popper, Z.A.; Showalter, A.M. Bioinformatic identification and analysis of extensins in the plant kingdom. PLoS ONE 2016, 11, e0150177. [Google Scholar] [CrossRef]
- Baumberger, N.; Doesseger, B.; Guyot, R.; Diet, A.; Parsons, R.L.; Clark, M.A.; Simmons, M.; Bedinger, P.; Goff, S.A.; Ringli, C. Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 2003, 131, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Draeger, C.; Ndinyanka Fabrice, T.; Gineau, E.; Mouille, G.; Kuhn, B.M.; Moller, I.; Abdou, M.-T.; Frey, B.; Pauly, M.; Bacic, A. Arabidopsis leucine-rich repeat extensin (LRX) proteins modify cell wall composition and influence plant growth. BMC plant Biol. 2015, 15, 155. [Google Scholar] [CrossRef]
- Cannon, M.C.; Terneus, K.; Hall, Q.; Tan, L.; Wang, Y.; Wegenhart, B.L.; Chen, L.; Lamport, D.T.; Chen, Y.; Kieliszewski, M.J. Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 2008, 105, 2226–2231. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Behrens, B.X.; West, P.R.; Mort, A.J. Solubilization and partial characterization of extensin fragments from cell walls of cotton suspension cultures (evidence for a covalent cross-link between extensin and pectin). Plant Physiol. 1995, 108, 1691–1701. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Ringli, C.; Keller, B. The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 2001, 15, 1128–1139. [Google Scholar] [CrossRef]
- Baumberger, N.; Steiner, M.; Ryser, U.; Keller, B.; Ringli, C. Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 2003, 35, 71–81. [Google Scholar] [CrossRef]
- Dünser, K.; Gupta, S.; Herger, A.; Feraru, M.I.; Ringli, C.; Kleine-Vehn, J. Extracellular matrix sensing by FERONIA and Leucine-Rich Repeat Extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J. 2019, 38, e100353. [Google Scholar] [CrossRef]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.-C.; Zhang, L.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.-K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef]
- Mecchia, M.A.; Santos-Fernandez, G.; Duss, N.N.; Somoza, S.C.; Boisson-Dernier, A.; Gagliardini, V.; Martínez-Bernardini, A.; Fabrice, T.N.; Ringli, C.; Muschietti, J.P. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 2017, 358, 1600–1603. [Google Scholar] [CrossRef]
- Covey, P.A.; Subbaiah, C.C.; Parsons, R.L.; Pearce, G.; Lay, F.T.; Anderson, M.A.; Ryan, C.A.; Bedinger, P.A. A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 2010, 153, 703–715. [Google Scholar] [CrossRef]
- Yun, D.J.; Bressan, R.A.; Hasegawa, P.M. Plant antifungal proteins. Plant Breed. Rev. 1997, 14, 39–88. [Google Scholar]
- van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- van der Wel, H.; Loeve, K. Isolation and characterization of thaumatin I and II, the sweet-tasting proteins from Thaumatococcus daniellii Benth. Eur. J. Biochem. 1972, 31, 221–225. [Google Scholar] [PubMed]
- Jami, S.K.; Anuradha, T.S.; Guruprasad, L.; Kirti, P.B. Molecular, biochemical and structural characterization of osmotin-like protein from black nightshade (Solanum nigrum). J. Plant Physiol. 2007, 164, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Tachi, H.; Fukuda-Yamada, K.; Kojima, T.; Shiraiwa, M.; Takahara, H. Molecular characterization of a novel soybean gene encoding a neutral PR-5 protein induced by high-salt stress. Plant Physiol. Biochem. 2009, 47, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.W.; Tung, W.L.; Griffith, M.; Chow, K.-C. Cloning of a cDNA encoding the thaumatin-like protein of winter rye (Secale cereale L. Musketeer) and its functional characterization. J. Exp. Bot. 1999, 50, 1627–1628. [Google Scholar]
- Ghosh, R.; Chakrabarti, C. Crystal structure analysis of NP24-I: A thaumatin-like protein. Planta 2008, 228, 883–890. [Google Scholar] [CrossRef]
- Velazhahan, R.; Datta, S.K.; Muthukrishnan, S. The PR-5 family: Thaumatin-like proteins. In Pathogenesis-Related Proteins in Plants; CRC Press: Boca Raton, FL, USA, 1999; pp. 107–129. [Google Scholar]
- Wang, X.; Zafian, P.; Choudhary, M.; Lawton, M. The PR5K receptor protein kinase from Arabidopsis thaliana is structurally related to a family of plant defense proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 2598–2602. [Google Scholar] [CrossRef]
- Liu, J.-J.; Sturrock, R.; Ekramoddoullah, A.K. The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Rep. 2010, 29, 419–436. [Google Scholar] [CrossRef]
- Clarke, J.D.; Volko, S.M.; Ledford, H.; Ausubel, F.M.; Dong, X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 2000, 12, 2175–2190. [Google Scholar] [CrossRef]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Kim, M.C.; Kumar, D.; Park, B.; Cheong, M.S.; Choi, W.; Park, H.C.; Chun, H.J.; Park, H.J.; Lee, S.Y. AtPR5K2, a PR5-like receptor kinase, modulates plant responses to drought stress by phosphorylating protein phosphatase 2Cs. Front. Plant Sci. 2019, 10, 1146. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Lee, J.H.; Yoon, G.M.; Cho, H.S.; Park, S.-W.; Suh, M.C.; Choi, D.; Ha, H.J.; Liu, J.R.; Pai, H.-S. CHRK1, a chitinase-related receptor-like kinase in tobacco. Plant Physiol. 2000, 123, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Takei, K.; Sakakibara, H.; Sun Cho, H.; Kim, D.M.; Kim, Y.S.; Min, S.R.; Kim, W.T.; Sohn, D.Y.; Lim, Y.P. CHRK1, a chitinase-related receptor-like kinase, plays a role in plant development and cytokinin homeostasis in tobacco. Plant Mol. Biol. 2003, 53, 877–890. [Google Scholar] [CrossRef]
- Feuillet, C.; Schachermayr, G.; Keller, B. Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J. 1997, 11, 45–52. [Google Scholar] [CrossRef]
- Veley, K.M.; Michaels, S.D. Functional redundancy and new roles for genes of the autonomous floral-promotion pathway. Plant Physiol. 2008, 147, 682–695. [Google Scholar] [CrossRef][Green Version]
- Lim, C.W.; Yang, S.H.; Shin, K.H.; Lee, S.C.; Kim, S.H. The AtLRK10L1. 2, Arabidopsis ortholog of wheat LRK10, is involved in ABA-mediated signaling and drought resistance. Plant Cell Rep. 2015, 34, 447–455. [Google Scholar] [CrossRef]
- Jinjun, Z.; Peina, J.; Fang, Z.; Chongke, Z.; Bo, B.; Yaping, L.; Haifeng, W.; Fan, C.; Xianzhi, X. OsSRK1, an atypical S-receptor-like kinase positively regulates leaf width and salt tolerance in rice. Rice Sci. 2020, 27, 133–142. [Google Scholar] [CrossRef]
- Vaid, N.; Pandey, P.; Srivastava, V.K.; Tuteja, N. Pea lectin receptor-like kinase functions in salinity adaptation without yield penalty, by alleviating osmotic and ionic stresses and upregulating stress-responsive genes. Plant Mol. Biol. 2015, 88, 193–206. [Google Scholar] [CrossRef]
- Li, C.-H.; Wang, G.; Zhao, J.-L.; Zhang, L.-Q.; Ai, L.-F.; Han, Y.-F.; Sun, D.-Y.; Zhang, S.-W.; Sun, Y. The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef]
- Van der Does, D.; Boutrot, F.; Engelsdorf, T.; Rhodes, J.; McKenna, J.F.; Vernhettes, S.; Koevoets, I.; Tintor, N.; Veerabagu, M.; Miedes, E. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 2017, 13, e1006832. [Google Scholar] [CrossRef]
- Giri, J.; Vij, S.; Dansana, P.K.; Tyagi, A.K. Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 2011, 191, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Rai, K.M.; Balasubramanian, V.K.; Upadhyay, S.K.; Luo, H.; Mendu, V. Genome-wide identification and characterization of LRR-RLKs reveal functional conservation of the SIF subfamily in cotton (Gossypium hirsutum). BMC Plant Biol. 2018, 18, 185. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.L.; Cui, W.N.; Zhao, Q.; Zhao, J.; Hou, X.N.; Li, D.Y.; Chen, Z.L.; Shen, Y.Z.; Huang, Z.J. Functional study of a salt-inducible TaSR gene in Triticum aestivum. Physiol. Plant. 2016, 156, 40–53. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, Z.; Wang, J.; Cong, B.; Chen, K.; Liu, S. A novel receptor-like kinase (PnRLK-1) from the Antarctic moss Pohlia nutans enhances salt and oxidative stress tolerance. Plant Mol. Biol. Rep. 2015, 33, 1156–1170. [Google Scholar] [CrossRef]
- Sun, X.-L.; Yu, Q.-Y.; Tang, L.-L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.-F.; Ding, X.-D.; Zhu, Y.-M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xiao, F.; Zheng, Y.; Liu, G.; Zhuang, Y.; Wang, Z.; Zhang, Y.; He, J.; Fu, C.; Lin, H. Pamp-induced secreted peptide 3 modulates salt tolerance through receptor-like kinase 7 in plants. Plant Cell 2022, 34, 927–944. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-B.; Liu, C.; Tang, D.-Y.; Yan, L.; Wang, D.; Yang, Y.-Z.; Gui, J.-S.; Zhao, X.-Y.; Li, L.-G.; Tang, X.-D. The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, X.; Gao, Y.; Jiao, J.; Sun, Y.; Zhu, D.; Yang, J.; Wu, F.; Su, H. Genome-wide analysis of lectin receptor-like kinases (LecRLKs) in sweet cherry (Prunus avium) and reveals PaLectinL16 enhances sweet cherry resistance with salt stress. Environ. Exp. Bot. 2022, 194, 104751. [Google Scholar] [CrossRef]
- Shi, C.-C.; Feng, C.-C.; Yang, M.-M.; Li, J.-L.; Li, X.-X.; Zhao, B.-C.; Huang, Z.-J.; Ge, R.-C. Overexpression of the receptor-like protein kinase genes AtRPK1 and OsRPK1 reduces the salt tolerance of Arabidopsis thaliana. Plant Science 2014, 217, 63–70. [Google Scholar] [CrossRef]
- Sade, N.; Weng, F.; Tajima, H.; Zeron, Y.; Zhang, L.; Rubio Wilhelmi, M.d.M.; Day, G.; Peleg, Z.; Blumwald, E. A cytoplasmic receptor-like kinase contributes to salinity tolerance. Plants 2020, 9, 1383. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-s.; Liang, C.-c.; Hou, S.-g.; Wang, X.; Chen, D.-h.; Shen, J.-l.; Zhang, W.; Wang, M. The LRR-RLK protein HSL3 regulates stomatal closure and the drought stress response by modulating hydrogen peroxide homeostasis. Front. Plant Sci. 2020, 11, 548034. [Google Scholar] [CrossRef] [PubMed]
- Ramegowda, V.; Basu, S.; Krishnan, A.; Pereira, A. Rice GROWTH UNDER DROUGHT KINASE is required for drought tolerance and grain yield under normal and drought stress conditions. Plant Physiol. 2014, 166, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Gao, Y.; Zhang, Z.; Chen, T.; Guo, W.; Zhang, T. A receptor-like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought-stress tolerance in Arabidopsis. BMC Plant Biol. 2013, 13, 110. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Q.; Yu, Q.; Peng, L.; Wang, J.; Dai, Q.; Yang, Y.; Li, X. CARK6 is involved in abscisic acid to regulate stress responses in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2019, 513, 460–464. [Google Scholar] [CrossRef]
- Feng, L.; Gao, Z.; Xiao, G.; Huang, R.; Zhang, H. Leucine-rich repeat receptor-like kinase FON1 regulates drought stress and seed germination by activating the expression of ABA-responsive genes in rice. Plant Mol. Biol. Rep. 2014, 32, 1158–1168. [Google Scholar] [CrossRef]
- Wu, F.; Sheng, P.; Tan, J.; Chen, X.; Lu, G.; Ma, W.; Heng, Y.; Lin, Q.; Zhu, S.; Wang, J. Plasma membrane receptor-like kinase leaf panicle 2 acts downstream of the DROUGHT AND SALT TOLERANCE transcription factor to regulate drought sensitivity in rice. J. Exp. Bot. 2015, 66, 271–281. [Google Scholar] [CrossRef]
- Chen, L.-J.; Wuriyanghan, H.; Zhang, Y.-Q.; Duan, K.-X.; Chen, H.-W.; Li, Q.-T.; Lu, X.; He, S.-J.; Ma, B.; Zhang, W.-K. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 2013, 163, 1752–1765. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, G.; Shi, R.; Han, X.; Qi, L.; Wang, R.; Xiong, L.; Li, G. Arabidopsis cysteine-rich receptor-like kinase 45 functions in the responses to abscisic acid and abiotic stresses. Plant Physiol. Biochem. 2013, 67, 189–198. [Google Scholar] [CrossRef]
- Kang, J.; Li, J.; Gao, S.; Tian, C.; Zha, X. Overexpression of the leucine-rich receptor-like kinase gene LRK 2 increases drought tolerance and tiller number in rice. Plant Biotechnol. J. 2017, 15, 1175–1185. [Google Scholar] [CrossRef]
- Pan, J.; Li, Z.; Wang, Q.; Yang, L.; Yao, F.; Liu, W. An S-domain receptor-like kinase, OsESG1, regulates early crown root development and drought resistance in rice. Plant Sci. 2020, 290, 110318. [Google Scholar] [CrossRef]
- Fang, J.; Chai, Z.; Huang, R.; Huang, C.; Ming, Z.; Chen, B.; Yao, W.; Zhang, M. Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis. Front. Plant Sci. 2023, 14, 1191449. [Google Scholar] [CrossRef]
- Wang, K.; Li, S.; Tian, H.; Chen, C.; Hu, Z.; Zhao, Q.; Du, C. Receptor-like cytoplasmic kinase OsRLCK241 functions as an important regulator of abscisic acid synthesis and response in rice. Environ. Exp. Bot. 2022, 194, 104744. [Google Scholar] [CrossRef]
- Yang, L.; Gao, C.; Jiang, L. Leucine-rich repeat receptor-like protein kinase AtORPK1 promotes oxidative stress resistance in an AtORPK1-AtKAPP mediated module in Arabidopsis. Plant Sci. 2022, 315, 111147. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, X.; Cui, Y.; Guo, C.; Chen, L. Overexpression of the receptor-like cytoplasmic kinase gene XCRK enhances Xoc and oxidative stress tolerance in rice. J. Plant Biol. 2017, 60, 523–532. [Google Scholar] [CrossRef]
- Hou, X.; Tong, H.; Selby, J.; DeWitt, J.; Peng, X.; He, Z.-H. Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol. 2005, 139, 1704–1716. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Hou, X. Role of OsWAK124, a rice wall-associated kinase, in response to environmental heavy metal stresses. Pak. J. Bot. 2017, 49, 1255–1261. [Google Scholar]
- Hu, W.; Lv, Y.; Lei, W.; Li, X.; Chen, Y.; Zheng, L.; Xia, Y.; Shen, Z. Cloning and characterization of the Oryza sativa wall-associated kinase gene OsWAK11 and its transcriptional response to abiotic stresses. Plant Soil 2014, 384, 335–346. [Google Scholar] [CrossRef]
- Geng, B.; Wang, Q.; Huang, R.; Liu, Y.; Guo, Z.; Lu, S. A novel LRR-RLK (CTLK) confers cold tolerance through regulation on the C-repeat-binding factor pathway, antioxidants, and proline accumulation. Plant J. 2021, 108, 1679–1689. [Google Scholar] [CrossRef]
- Xu, W.; Gao, S.; Song, J.; Yang, Q.; Wang, T.; Zhang, Y.; Zhang, J.; Li, H.; Yang, C.; Ye, Z. NDW, encoding a receptor-like protein kinase, regulates plant growth, cold tolerance and susceptibility to Botrytis cinerea in tomato. Plant Sci. 2020, 301, 110684. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Pan, Y.; Li, J.; Zhou, L.; Shi, H.; Zeng, Y.; Guo, H.; Yang, S.; Zheng, W. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 2017, 8, 14788. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Han, J.; Kim, Y.-J.; Song, M.; Yang, Z.; He, Y.; Fu, R.; Luo, Z.; Hu, J.; Liang, W. Two rice receptor-like kinases maintain male fertility under changing temperatures. Proc. Natl. Acad. Sci. USA 2017, 114, 12327–12332. [Google Scholar] [CrossRef] [PubMed]
- Juneidi, S.; Gao, Z.; Yin, H.; Makunga, N.P.; Chen, W.; Hu, S.; Li, X.; Hu, X. Breaking the summer dormancy of Pinellia ternata by introducing a heat tolerance receptor-like kinase ERECTA gene. Front. Plant Sci. 2020, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Zhong, X.; Zhao, F.; Wang, Y.; Yan, B.; Li, Q.; Chen, G.; Mao, B.; Wang, J.; Li, Y. Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato. Nat. Biotechnol. 2015, 33, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.G.; Hwang, S.-G.; Park, Y.C.; Park, H.M.; Kim, D.S.; Park, D.H.; Jang, C.S. Molecular characterization of the cold-and heat-induced Arabidopsis PXL1 gene and its potential role in transduction pathways under temperature fluctuations. J. plant Physiol. 2015, 176, 138–146. [Google Scholar] [CrossRef]
- Guan, D.; Yang, F.; Xia, X.; Shi, Y.; Yang, S.; Cheng, W.; He, S. CaHSL1 acts as a positive regulator of pepper thermotolerance under high humidity and is transcriptionally modulated by CaWRKY40. Front. Plant Sci. 2018, 9, 1802. [Google Scholar] [CrossRef]
- Wang, H.; Niu, H.; Liang, M.; Zhai, Y.; Huang, W.; Ding, Q.; Du, Y.; Lu, M. A wall-associated kinase gene CaWAKL20 from pepper negatively modulates plant thermotolerance by reducing the expression of ABA-responsive genes. Front. Plant Sci. 2019, 10, 591. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Shang, H.; Chen, X.; Xu, X.; Hu, X. TaXa21, a leucine-rich repeat receptor–like kinase gene associated with TaWRKY76 and TaWRKY62, plays positive roles in wheat high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant-Microbe Interact. 2019, 32, 1526–1535. [Google Scholar] [CrossRef]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Bradford, K.J. A water relations analysis of seed germination rates. Plant Physiol. 1990, 94, 840–849. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Maas, E.; Nieman, R. Physiology of plant tolerance to salinity. Crop Toler. Subopt. Land Cond. 1978, 32, 277–299. [Google Scholar]
- Voxeur, A.; Höfte, H. Cell wall integrity signaling in plants: “To grow or not to grow that’s the question”. Glycobiology 2016, 26, 950–960. [Google Scholar] [CrossRef]
- Kathuria, H.; Giri, J.; Tyagi, H.; Tyagi, A.K. Advances in transgenic rice biotechnology. Crit. Rev. Plant Sci. 2007, 26, 65–103. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, W.-H. Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 2013, 64, 451–476. [Google Scholar] [CrossRef]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Agarwal, R. Salinity stress induced alterations in antioxidant metabolism and nitrogen assimilation in wheat (Triticum aestivum L.) as influenced by potassium supplementation. Plant Physiol. Biochem. 2017, 115, 449–460. [Google Scholar] [CrossRef]
- Sun, Y.; Kong, X.; Li, C.; Liu, Y.; Ding, Z. Potassium retention under salt stress is associated with natural variation in salinity tolerance among Arabidopsis accessions. PLoS ONE 2015, 10, e0124032. [Google Scholar] [CrossRef]
- Tao, R.; Ding, J.; Li, C.; Zhu, X.; Guo, W.; Zhu, M. Evaluating and screening of agro-physiological indices for salinity stress tolerance in wheat at the seedling stage. Front. Plant Sci. 2021, 12, 646175. [Google Scholar] [CrossRef]
- Botella, M.; Martinez, V.; Pardines, J.; Cerda, A. Salinity induced potassium deficiency in maize plants. J. Plant Physiol. 1997, 150, 200–205. [Google Scholar] [CrossRef]
- Shabala, S.; Cuin, T.A. Potassium transport and plant salt tolerance. Physiol. Plant. 2008, 133, 651–669. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Halfter, U.; Ishitani, M.; Zhu, J.-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef]
- Lin, H.; Yang, Y.; Quan, R.; Mendoza, I.; Wu, Y.; Du, W.; Zhao, S.; Schumaker, K.S.; Pardo, J.M.; Guo, Y. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 2009, 21, 1607–1619. [Google Scholar] [CrossRef]
- Zhou, X.; Hao, H.; Zhang, Y.; Bai, Y.; Zhu, W.; Qin, Y.; Yuan, F.; Zhao, F.; Wang, M.; Hu, J. SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol. 2015, 168, 659–676. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Shankar, A.; Nalini Chandran, A.K.; Sharma, M.; Jung, K.-H.; Suprasanna, P.; Pandey, G.K. Emerging concepts of potassium homeostasis in plants. J. Exp. Bot. 2020, 71, 608–619. [Google Scholar] [CrossRef]
- Rabara, R.C.; Tripathi, P.; Reese, R.N.; Rushton, D.L.; Alexander, D.; Timko, M.P.; Shen, Q.J.; Rushton, P.J. Tobacco drought stress responses reveal new targets for Solanaceae crop improvement. BMC Genom. 2015, 16, 484. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, H.; Hong, J.-W.; Lee, Y.-N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.-D.; Lee, S.; Lee, S.C.; Kim, B.-G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef]
- Li, H.; Han, X.; Liu, X.; Zhou, M.; Ren, W.; Zhao, B.; Ju, C.; Liu, Y.; Zhao, J. A leucine-rich repeat-receptor-like kinase gene SbER2–1 from sorghum (Sorghum bicolor L.) confers drought tolerance in maize. BMC Genom. 2019, 20, 737. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, B.C.; Oelmüller, R. Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Dat, J.; Vandenabeele, S.; Vranova, E.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. CMLS 2000, 57, 779–795. [Google Scholar] [CrossRef]
- Kimura, S.; Waszczak, C.; Hunter, K.; Wrzaczek, M. Bound by fate: The role of reactive oxygen species in receptor-like kinase signaling. Plant Cell 2017, 29, 638–654. [Google Scholar] [CrossRef]
- Levitt, J. Responses of Plants to Environmental Stress, Chilling, Freezing, and High Temperature Stresses; Academic Press: New York, NY, USA, 1980. [Google Scholar]
- Theocharis, A.; Clément, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef]
- Janská, A.; Maršík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation–what is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef]
- Zhen, Y.; Ungerer, M.C. Clinal variation in freezing tolerance among natural accessions of Arabidopsis thaliana. New Phytol. 2008, 177, 419–427. [Google Scholar] [CrossRef]
- Shpak, E.D. Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol. 2013, 55, 1238–1250. [Google Scholar] [CrossRef]
- Nazar, R.; Iqbal, N.; Masood, A.; Khan, M.I.R.; Syeed, S.; Khan, N.A. Cadmium toxicity in plants and role of mineral nutrients in its alleviation. Am. J. Plant Sci. 2012, 3, 1476–1489. [Google Scholar] [CrossRef]
- Rascio, N.; Navari-Izzo, F. Heavy metal hyperaccumulating plants: How and why do they do it? And what makes them so interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Zeng, Z.H.; Yan, J.Y.; Fan, W.; Bian, H.W.; Zhu, M.Y.; Yang, J.L.; Zheng, S.J. Association of specific pectin methylesterases with Al-induced root elongation inhibition in rice. Physiol. Plant. 2013, 148, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Yin, S.; Zhang, K.; Shi, X.; Lian, C.; Zhang, H.; Hu, Z.; Shen, Z. OsWAK11, a rice wall-associated kinase, regulates Cu detoxification by alteration the immobilization of Cu in cell walls. Environ. Exp. Bot. 2018, 150, 99–105. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).