Hepatic SPARC Expression Is Associated with Inflammasome Activation during the Progression of Non-Alcoholic Fatty Liver Disease in Both Mice and Morbidly Obese Patients
Abstract
:1. Introduction
2. Results
2.1. SPARC and Lipotoxic Free Fatty Acids Promote IL-1β Synthesis in Macrophages and Hepatocytes
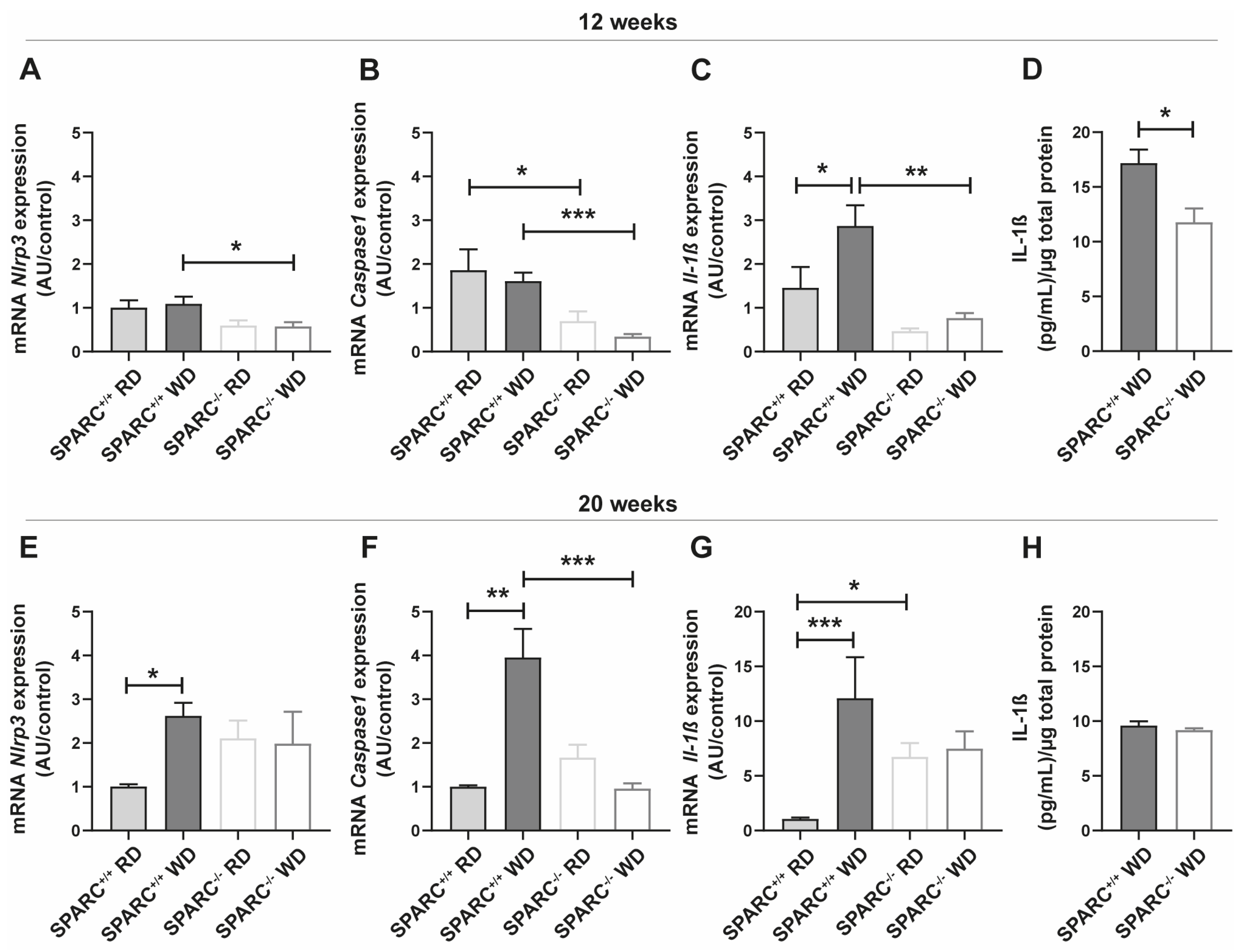
2.2. The Absence of SPARC Decreases the Expression of Inflammasome Pathway-Related Genes in the Liver
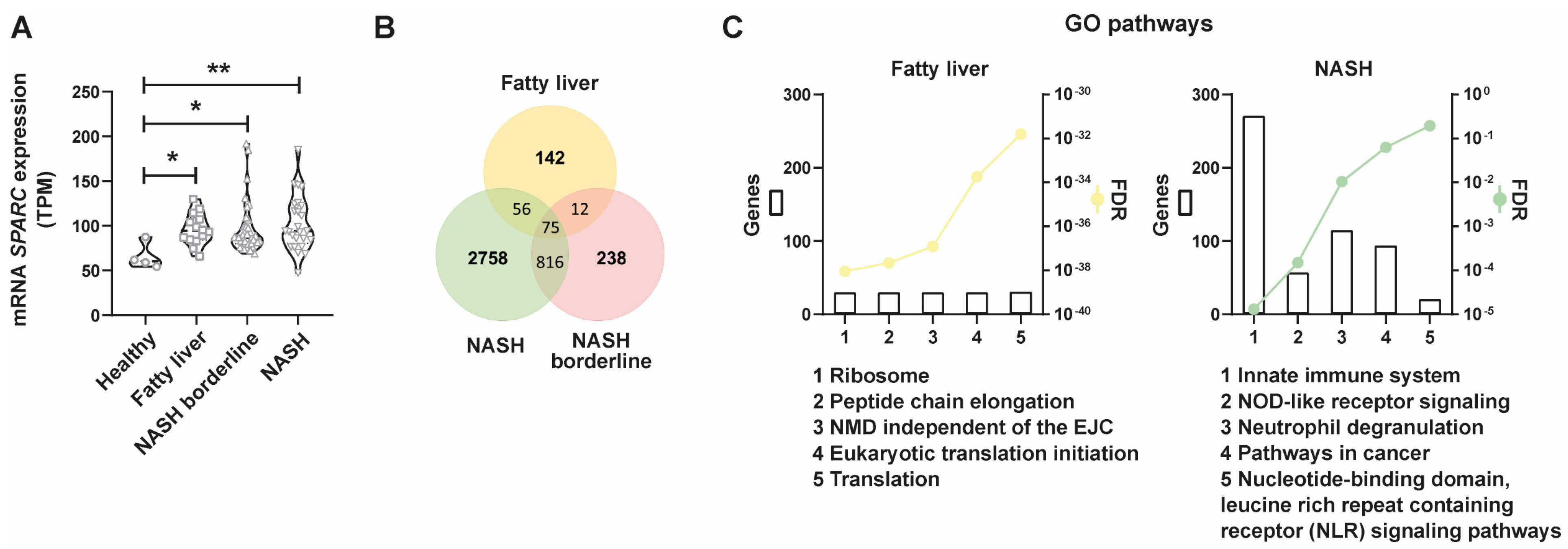
2.3. SPARC Is Differentially Expressed in NAFLD Patients and SPARC-Positively Correlated Genes Are Associated with Inflammasome-Related Pathways throughout the Progression of NAFLD
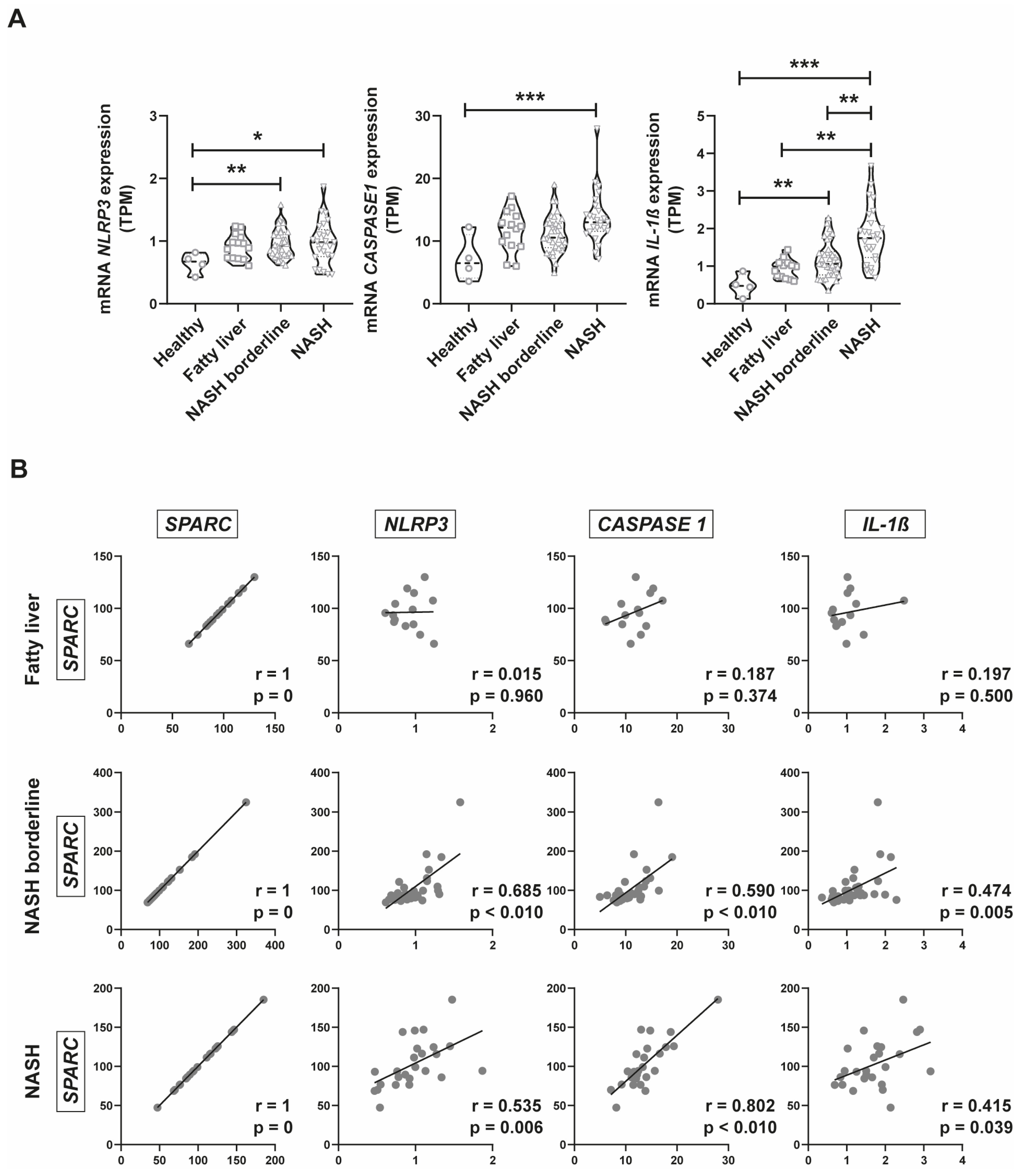
2.4. Inflammasome Gene Expression Analysis in Public Databases of NAFLD Patients
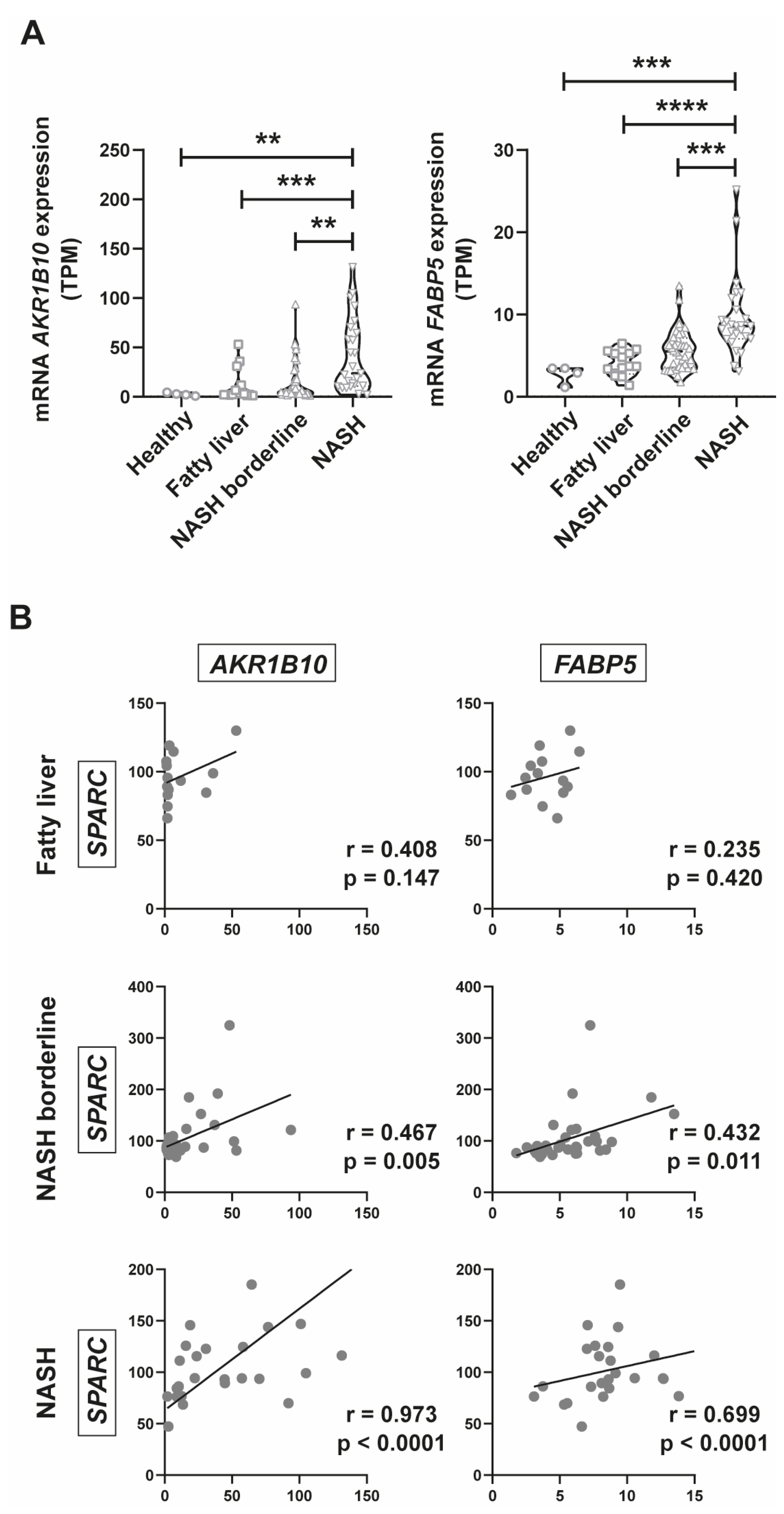
2.5. AKR1B10 and FABP5 Positively Correlate with SPARC Expression
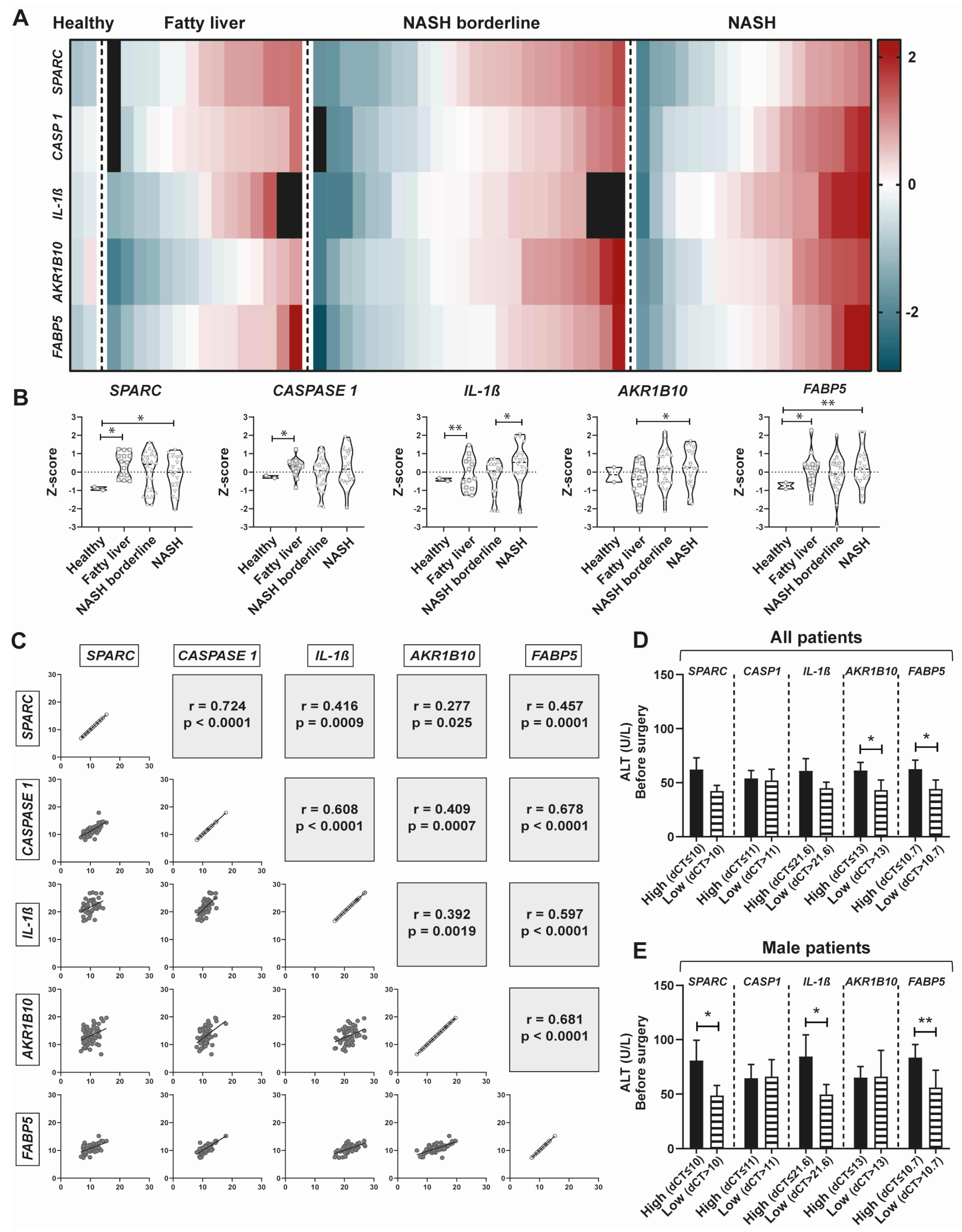
2.6. SPARC expression positively correlates with CASP1, IL-1β, AKR1B10, and FABP5 Expression in a New Prospective Patient Cohort
3. Discussion
4. Materials and Methods
4.1. J774 Culture, Primary Hepatocyte Isolation, and In Vitro Steatosis Assay
4.2. Animal Experimental Design
4.3. Quantitative Real-Time PCR (qPCR)
4.4. ELISA
4.5. Bioinformatic Analysis
4.6. Patients’ Cohort and Study Protocol
4.7. Clinical–Biochemical Assessment and Histopathology
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.; Tacke, F.; Arrese, M.; Sharma, B.C.; Mostafa, I.; Bugianesi, E.; Wong, V.W.-S.; Yilmaz, Y.; George, J.; Fan, J.; et al. Global Perspectives on Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Hepatology 2019, 69, 2672–2682. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.; Oakley, F.; Anstee, Q.M.; Day, C.P. Nonalcoholic Fatty Liver Disease: Pathogenesis and Disease Spectrum. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 451–496. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y. Histopathology of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J. Gastroenterol. 2014, 20, 15539–15548. [Google Scholar] [CrossRef]
- Schuster, S.; Cabrera, D.; Arrese, M.; Feldstein, A.E. Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 349–364. [Google Scholar] [CrossRef]
- Mazzolini, G.; Sowa, J.-P.; Atorrasagasti, C.; Kücükoglu, Ö.; Syn, W.-K.; Canbay, A. Significance of Simple Steatosis: An Update on the Clinical and Molecular Evidence. Cells 2020, 9, 2458. [Google Scholar] [CrossRef]
- Loomba, R.; Friedman, S.L.; Shulman, G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021, 184, 2537–2564. [Google Scholar] [CrossRef]
- Singh, S.; Allen, A.M.; Wang, Z.; Prokop, L.J.; Murad, M.H.; Loomba, R. Fibrosis Progression in Nonalcoholic Fatty Liver vs Nonalcoholic Steatohepatitis: A Systematic Review and Meta-analysis of Paired-Biopsy Studies. Clin. Gastroenterol. Hepatol. 2014, 13, 643–654.e9. [Google Scholar] [CrossRef]
- Loomba, R.; Adams, L.A. The 20% Rule of NASH Progression: The Natural History of Advanced Fibrosis and Cirrhosis Caused by NASH. Hepatology 2019, 70, 1885–1888. [Google Scholar] [CrossRef]
- Bradshaw, A.D.; Sage, E.H. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J. Clin. Investig. 2001, 107, 1049–1054. [Google Scholar] [CrossRef]
- Atorrasagasti, C.; Onorato, A.M.; Mazzolini, G. The role of SPARC (secreted protein acidic and rich in cysteine) in the pathogenesis of obesity, type 2 diabetes, and non-alcoholic fatty liver disease. J. Physiol. Biochem. 2022, 78, 1–17. [Google Scholar] [CrossRef]
- Ghanemi, A.; Yoshioka, M.; St-Amand, J. Secreted Protein Acidic and Rich in Cysteine: Metabolic and Homeostatic Properties beyond the Extracellular Matrix Structure. Appl. Sci. 2020, 10, 2388. [Google Scholar] [CrossRef]
- Kos, K.; Wilding, J.P.H. SPARC: A key player in the pathologies associated with obesity and diabetes. Nat. Rev. Endocrinol. 2010, 6, 225–235. [Google Scholar] [CrossRef]
- Atorrasagasti, C.; Peixoto, E.; Aquino, J.B.; Kippes, N.; Malvicini, M.; Alaniz, L.; Garcia, M.; Piccioni, F.; Fiore, E.J.; Bayo, J.; et al. Lack of the Matricellular Protein SPARC (Secreted Protein, Acidic and Rich in Cysteine) Attenuates Liver Fibrogenesis in Mice. PLoS ONE 2013, 8, e54962. [Google Scholar] [CrossRef]
- Atorrasagasti, C.; Onorato, A.; Gimeno, M.L.; Andreone, L.; Garcia, M.; Malvicini, M.; Fiore, E.; Bayo, J.; Perone, M.J.; Mazzolini, G.D. SPARC is required for the maintenance of glucose homeostasis and insulin secretion in mice. Clin. Sci. 2019, 133, 351–365. [Google Scholar] [CrossRef]
- Frizell, E.; Liu, S.-L.; Abraham, A.; Ozaki, I.; Eghbali, M.; Sage, E.H.; Zern, M.A. Expression of SPARC in normal and fibrotic livers. Hepatology 1995, 21, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Onorato, A.M.; Fiore, E.; Bayo, J.; Casali, C.; Fernandez-Tomé, M.; Rodríguez, M.; Domínguez, L.; Argemi, J.; Hidalgo, F.; Favre, C.; et al. SPARC inhibition accelerates NAFLD-associated hepatocellular carcinoma development by dysregulating hepatic lipid metabolism. Liver Int. 2021, 41, 1677–1693. [Google Scholar] [CrossRef]
- Mazzolini, G.; Atorrasagasti, C.; Onorato, A.; Peixoto, E.; Schlattjan, M.; Sowa, J.-P.; Sydor, S.; Gerken, G.; Canbay, A. SPARC expression is associated with hepatic injury in rodents and humans with non-alcoholic fatty liver disease. Sci. Rep. 2018, 8, 725. [Google Scholar] [CrossRef]
- Nie, J.; Bradshaw, A.D.; Delany, A.M.; Sage, E.H. Inactivation of SPARC enhances high-fat diet-induced obesity in mice. Connect. Tissue Res. 2010, 52, 99–108. [Google Scholar] [CrossRef]
- Ryu, S.; Sidorov, S.; Ravussin, E.; Artyomov, M.; Iwasaki, A.; Wang, A.; Dixit, V.D. The matricellular protein SPARC induces inflammatory interferon-response in macrophages during aging. Immunity 2022, 55, 1609–1626.e7. [Google Scholar] [CrossRef]
- Gong, T.; Liu, L.; Jiang, W.; Zhou, R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2019, 20, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 inflammasome: An overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [PubMed]
- Nolan, C.J.; Larter, C.Z. Lipotoxicity: Why do saturated fatty acids cause and monounsaturates protect against it? J. Gastroenterol. Hepatol. 2009, 24, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Szabo, G.; Csak, T. Inflammasomes in liver diseases. J. Hepatol. 2012, 57, 642–654. [Google Scholar] [CrossRef]
- Wree, A.; McGeough, M.D.; Peña, C.A.; Schlattjan, M.; Li, H.; Inzaugarat, M.E.; Messer, K.; Canbay, A.; Hoffman, H.M.; Feldstein, A.E. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J. Mol. Med. 2014, 92, 1069–1082. [Google Scholar] [CrossRef]
- Hoang, S.A.; Oseini, A.; Feaver, R.E.; Cole, B.K.; Asgharpour, A.; Vincent, R.; Siddiqui, M.; Lawson, M.J.; Day, N.C.; Taylor, J.M.; et al. Gene Expression Predicts Histological Severity and Reveals Distinct Molecular Profiles of Nonalcoholic Fatty Liver Disease. Sci. Rep. 2019, 9, 12541. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef]
- Enooku, K.; Tsutsumi, T.; Kondo, M.; Fujiwara, N.; Sasako, T.; Shibahara, J.; Kado, A.; Okushin, K.; Fujinaga, H.; Nakagomi, R.; et al. Hepatic FATP5 expression is associated with histological progression and loss of hepatic fat in NAFLD patients. J. Gastroenterol. 2019, 55, 227–243. [Google Scholar] [CrossRef]
- Park, A.; Choi, S.J.; Park, S.; Kim, S.M.; Lee, H.E.; Joo, M.; Kim, K.K.; Kim, D.; Chung, D.H.; Im, J.B.; et al. Plasma Aldo-Keto Reductase Family 1 Member B10 as a Biomarker Performs Well in the Diagnosis of Nonalcoholic Steatohepatitis and Fibrosis. Int. J. Mol. Sci. 2022, 23, 5035. [Google Scholar] [CrossRef]
- Shi, L.; Guo, S.; Zhang, S.; Gao, X.; Liu, A.; Wang, Q.; Zhang, T.; Zhang, Y.; Wen, A. Glycyrrhetinic acid attenuates disturbed vitamin a metabolism in non-alcoholic fatty liver disease through AKR1B10. Eur. J. Pharmacol. 2020, 883, 173167. [Google Scholar] [CrossRef]
- Yang, F.; Ni, B.; Lian, Q.; Qiu, X.; He, Y.; Zhang, Q.; Zou, X.; He, F.; Chen, W. Key genes associated with non-alcoholic fatty liver disease and hepatocellular carcinoma with metabolic risk factors. Front. Genet. 2023, 14, 1066410. [Google Scholar] [CrossRef] [PubMed]
- Brekken, R.A.; Sage, E. SPARC, a matricellular protein: At the crossroads of cell-matrix communication. Matrix Biol. 2001, 19, 815–827. [Google Scholar] [CrossRef]
- Kato, Y.; Lewalle, J.-M.; Baba, Y.; Tsukuda, M.; Sakai, N.; Baba, M.; Kobayashi, K.; Koshika, S.; Nagashima, Y.; Frankenne, F.; et al. Induction of SPARC by VEGF in Human Vascular Endothelial Cells. Biochem. Biophys. Res. Commun. 2001, 287, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, E.; Atorrasagasti, C.; Aquino, J.B.; Militello, R.; Bayo, J.; Fiore, E.; Piccioni, F.; Salvatierra, E.; Alaniz, L.; García, M.G.; et al. SPARC (secreted protein acidic and rich in cysteine) knockdown protects mice from acute liver injury by reducing vascular endothelial cell damage. Gene Ther. 2014, 22, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Atorrasagasti, C.; Aquino, J.B.; Hofman, L.; Alaniz, L.; Malvicini, M.; Garcia, M.; Benedetti, L.; Friedman, S.L.; Podhajcer, O.; Mazzolini, G. SPARC downregulation attenuates the profibrogenic response of hepatic stellate cells induced by TGF-β1and PDGF. Am. J. Physiol. Liver Physiol. 2011, 300, G739–G748. [Google Scholar] [CrossRef]
- Nakatani, K.; Seki, S.; Kawada, N.; Kitada, T.; Yamada, T.; Sakaguchi, H.; Kadoya, H.; Ikeda, K.; Kaneda, K. Expression of SPARC by activated hepatic stellate cells and its correlation with the stages of fibrogenesis in human chronic hepatitis. Virchows Arch. 2002, 441, 466–474. [Google Scholar] [CrossRef]
- Chaly, Y.; Fu, Y.; Marinov, A.; Hostager, B.; Yan, W.; Campfield, B.; Kellum, J.A.; Bushnell, D.; Wang, Y.; Vockley, J.; et al. Follistatin-like protein 1 enhances NLRP3 inflammasome-mediated IL-1β secretion from monocytes and macrophages. Eur. J. Immunol. 2014, 44, 1467–1479. [Google Scholar] [CrossRef]
- Cai, C.; Zhu, X.; Li, P.; Li, J.; Gong, J.; Shen, W.; He, K. NLRP3 Deletion Inhibits the Non-alcoholic Steatohepatitis Development and Inflammation in Kupffer Cells Induced by Palmitic Acid. Inflammation 2017, 40, 1875–1883. [Google Scholar] [CrossRef]
- Csak, T.; Ganz, M.; Pespisa, J.; Kodys, K.; Dolganiuc, A.; Szabo, G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology 2011, 54, 133–144. [Google Scholar] [CrossRef]
- de Haan, J.J.; Smeets, M.B.; Pasterkamp, G.; Arslan, F. Danger signals in the initiation of the inflammatory response after myocardial infarction. Mediat. Inflamm. 2013, 2013, 1–13. [Google Scholar] [CrossRef]
- Frevert, C.W.; Felgenhauer, J.; Wygrecka, M.; Nastase, M.V.; Schaefer, L. Danger-Associated Molecular Patterns Derived From the Extracellular Matrix Provide Temporal Control of Innate Immunity. J. Histochem. Cytochem. 2018, 66, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.d.C.; Szabo, G. Role of the Inflammasome in Liver Disease. Annu. Rev. Pathol. Mech. Dis. 2022, 17, 345–365. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Sage, E.H. SPARC functions as an inhibitor of adipogenesis. J. Cell Commun. Signal. 2009, 3, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Blazejewski, S.; Le Bail, B.; Boussarie, L.; Blanc, J.F.; Malaval, L.; Okubo, K.; Saric, J.; Bioulac-Sage, P.; Rosenbaum, J. Osteonectin (SPARC) expression in human liver and in cultured human liver myofibroblasts. . Am. J. Pathol. 1997, 151, 651–657. [Google Scholar] [PubMed]
- Larsen, F.T.; Hansen, D.; Terkelsen, M.K.; Bendixen, S.M.; Avolio, F.; Wernberg, C.W.; Lauridsen, M.E.; Grønkjaer, L.L.; Jacobsen, B.G.; Klinggaard, E.G.; et al. Stellate cell expression of SPARC-related modular calcium-binding protein 2 is associated with human non-alcoholic fatty liver disease severity. JHEP Rep. 2022, 5, 100615. [Google Scholar] [CrossRef]
- Inzaugarat, M.E.; Johnson, C.D.; Holtmann, T.M.; McGeough, M.D.; Trautwein, C.; Papouchado, B.G.; Schwabe, R.; Hoffman, H.M.; Wree, A.; Feldstein, A.E. NLR Family Pyrin Domain-Containing 3 Inflammasome Activation in Hepatic Stellate Cells Induces Liver Fibrosis in Mice. Hepatology 2019, 69, 845–859. [Google Scholar] [CrossRef]
- Mridha, A.R.; Wree, A.; Robertson, A.A.; Yeh, M.M.; Johnson, C.D.; Van Rooyen, D.M.; Haczeyni, F.; Teoh, N.C.-H.; Savard, C.; Ioannou, G.N.; et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J. Hepatol. 2017, 66, 1037–1046. [Google Scholar] [CrossRef]
- Park, I.; Kim, N.; Lee, S.; Park, K.; Son, M.-Y.; Cho, H.-S.; Kim, D.-S. Characterization of signature trends across the spectrum of non-alcoholic fatty liver disease using deep learning method. Life Sci. 2023, 314, 121195. [Google Scholar] [CrossRef]
- Rosso, N.; Stephenson, A.M.; Giraudi, P.J.; Tiribelli, C. Diagnostic management of nonalcoholic fatty liver disease: A transformational period in the development of diagnostic and predictive tools—A narrative review. Ann. Transl. Med. 2021, 9, 727. [Google Scholar] [CrossRef]
- Sumida, Y.; Nakajima, A.; Itoh, Y. Limitations of liver biopsy and non-invasive diagnostic tests for the diagnosis of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J. Gastroenterol. 2014, 20, 475–485. [Google Scholar] [CrossRef]
- Brunt, E.M. Nonalcoholic fatty liver disease and the ongoing role of liver biopsy evaluation. Hepatol. Commun. 2017, 1, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Lemmer, P.; Selbach, N.; Baars, T.; Porsch-Özcürümez, M.; Heider, D.; Canbay, A.; Sowa, J.-P. Transaminase Concentrations Cannot Separate Non-Alcoholic Fatty Liver and Non-Alcoholic Steatohepatitis in Morbidly Obese Patients Irrespective of Histological Algorithm. Dig. Dis. 2021, 40, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Schindhelm, R.K.; Diamant, M.; Dekker, J.M.; Tushuizen, M.E.; Teerlink, T.; Heine, R.J. Alanine aminotransferase as a marker of non-alcoholic fatty liver disease in relation to type 2 diabetes mellitus and cardiovascular disease. Diabetes/Metab. Res. Rev. 2006, 22, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Fiore, E.J.; Bayo, J.M.; Garcia, M.G.; Malvicini, M.; Lloyd, R.; Piccioni, F.; Rizzo, M.; Peixoto, E.; Sola, M.B.; Atorrasagasti, C.; et al. Mesenchymal stromal cells engineered to produce IGF-I by recombinant adenovirus ameliorate liver fibrosis in mice. Stem Cells Dev. 2015, 24, 791–801. [Google Scholar] [CrossRef]
- Win, S.; Than, T.A.; Le, B.H.A.; García-Ruiz, C.; Fernandez-Checa, J.C.; Kaplowitz, N. Sab (Sh3bp5) dependence of JNK mediated inhibition of mitochondrial respiration in palmitic acid induced hepatocyte lipotoxicity. J. Hepatol. 2015, 62, 1367–1374. [Google Scholar] [CrossRef]
- Ibrahim, S.H.; Hirsova, P.; Malhi, H.; Gores, G.J. Animal Models of Nonalcoholic Steatohepatitis: Eat, Delete, and Inflame. Dig. Dis. Sci. 2015, 61, 1325–1336. [Google Scholar] [CrossRef]
- Lieber, C.S.; Leo, M.A.; Mak, K.M.; Xu, Y.; Cao, Q.; Ren, C.; Ponomarenko, A.; DeCarli, L.M. Model of nonalcoholic steatohepatitis. Am. J. Clin. Nutr. 2004, 79, 502–509. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.-C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Sumida, Y.; Yoneda, M.; Hyogo, H.; Itoh, Y.; Ono, M.; Fujii, H.; Eguchi, Y.; Suzuki, Y.; Aoki, N.; Kanemasa, K.; et al. Validation of the FIB4 index in a Japanese nonalcoholic fatty liver disease population. BMC Gastroenterol. 2012, 12, 2. [Google Scholar] [CrossRef]

| Hospital Universitario Austral cohort | Variable | Healthy | Fatty liver | NASH borderline | NASH | |
| n (Total = 59) | 2 (3.4%) | 15 (25.4%) | 24 (40.7%) | 18 (30.5%) | ||
| Sex | M | 1 (50%) | 6 (40%) | 13 (54.2%) | 10 (55.6%) | |
| F | 1 (50%) | 9 (60%) | 11 (45.8%) | 8 (44.4%) | ||
| Age (mean) (SD) | M | 60 (0.0) | 47.7 (12.5) | 47.2 (9.6) | 42.1 (4.5) | |
| F | 43 (0.0) | 46.7 (10.4) | 40.3 (12.3) | 47.8 (7.8) | ||
| Height (mean in metres) (SD) | M | 1.65 (0.0) | 1.75 (0.05) | 1.74 (0.06) | 1.75 (0.07) | |
| F | 1.54 (0.0) | 1.59 (0.03) | 1.60 (0.07) | 1.61 (0.03) | ||
| Weight (mean in Kg) (SD) | M | 117 (0.0) | 121.5 (25.5) | 127.2 (22.0) | 123.2 (20.1) | |
| F | 94 (0.0) | 100.2 (16.1) | 108.1 (17.5) | 105.4 (9.1) | ||
| Body mass index (BMI) | M | 42.8 (0.0) | 40.0 (8.7) | 42.4 (9.8) | 40.4 (8.1) | |
| F | 39.6 (0.0) | 39.3 (5.7) | 42.5 (6.7) | 40.8 (2.7) | ||
| NAFLD activity score | 0 | 1 & 2 | 3 & 4 | 5, 6 & 7 | ||
| Steatosis grade | 0 | 2 (100%) | 3 (20%) | 0 | 0 | |
| 1 | 0 | 12 (80%) | 16 (66.7%) | 2 (11.1%) | ||
| 2 | 0 | 0 | 8 (33.3%) | 8 (44.4%) | ||
| 3 | 0 | 0 | 0 | 8 (44.4%) | ||
| Lobular inflammation grade | 0 | 2 (100%) | 7 (46.7%) | 1 (4.2%) | 0 | |
| 1 | 0 | 7 (46.7%) | 22 (91.6%) | 5 (27.8%) | ||
| 2 | 0 | 1 (6.6%) | 1 (4.2%) | 12 (66.7%) | ||
| 3 | 0 | 0 | 0 | 1 (5.5%) | ||
| Cytological ballooning grade | 0 | 2 (100%) | 11 (73.3%) | 0 | 0 | |
| 1 | 0 | 4 (26.7%) | 21 (87.5%) | 6 (33.3%) | ||
| 2 | 0 | 0 | 3 (12.5%) | 12 (66.7%) | ||
| Fibrosis stage | ||||||
| 0 | 2 (100%) | 8 (53.3%) | 5 (20.8%) | 5 (27.8%) | ||
| 1 | 0 | 3 (20%) | 11 (45.8%) | 9 (50%) | ||
| 2 | 0 | 0 | 2 (8.3%) | 1 (5.5%) | ||
| 3 | 0 | 0 | 3 (12.5%) | 3 (16.7%) | ||
| 4 | 0 | 0 | 1 (4.3%) | 0 | ||
| No fibrosis data available | 0 | 4 (26.7%) | 2 (8.3%) | 0 | ||
| Biochemical analysis (Serum sample taken at the time of bariatric surgery) | ||||||
| Albumin (mean in g/dL) (SD) | M | 3.74 (0) | 4.14 (0.3) | 3.89 (0.7) | 4.27 (0.4) | |
| F | 4.69 (0) | 3.97 (0.5) | 4.08 (0.4) | 3.40 (0.8) | ||
| Alkaline phosphatase (mean in U/L) (SD) | M | 42.3 (0) | 63.86 (8.7) | 57.00 (23.6) | 41.92 (22.9) | |
| F | 99.1 (0) | 53.26 (21.3) | 59.42 (19.4) | 56.48 (31.2) | ||
| ALT (mean in U/L) (SD) | M | 3 (0) | 13.88 (7.6) | 18.12 (14.9) | 20.16 (12.7) | |
| F | 9.6 (0) | 21.10 (32.2) | 11.01 (2.7) | 13.95 (9.7) | ||
| AST (mean in U/L) (SD) | M | 12.9 (0) | 30.40 (10.3) | 32.68 (14.6) | 54.36 (30.2) | |
| F | 16 (0) | 44.93 (47.2) | 49.64 (46.9) | 47.83 (45.3) | ||
| Direct bilirubin (mean in mg/dL) (SD) | M | 0.24 (0) | 0.28 (0.09) | 0.29 (0.14) | 0.24 (0.07) | |
| F | 0.38 (0) | 0.25 (0.13) | 0.21 (0.1) | 0.22 (0.1) | ||
| Total bilirubin (mean in mg/dL) (SD) | M | 0.57 (0) | 0.75 (0.2) | 0.74 (0.43) | 0.71 (0.28) | |
| F | 1.03 (0) | 0.59 (0.25) | 0.46 (0.24) | 0.47 (0.26) | ||
| Cholesterol (mean in mg/dL) (SD) | M | 137.4 (0) | 143.6 (41.3) | 134.4 (37.4) | 144.5 (27.0) | |
| F | 206 (0) | 161.0 (35.5) | 147.7 (20.1) | 144.2 (49.2) | ||
| Blood glucose (mean in mg/dL) (SD) | M | 95.1 (0) | 119 (36.4) | 107.0 (31.1) | 112.3 (21.7) | |
| F | 104.2 (0) | 102.3 (23.2) | 126.3 (33.5) | 128.1 (41.7) | ||
| Prothrombin time (mean in g/dL) (SD) | M | 6.36 (0) | 7.47 (0.4) | 6.85 (1.4) | 8.20 (1.7) | |
| F | 8.8 (0) | 7.47 (0.9) | 7.68 (1.7) | 6.69 (1.7) | ||
| No biochemical data available (n) | 0 | 3 | 2 | 0 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onorato, A.M.; Lameroli Mauriz, L.; Bayo, J.; Fiore, E.; Cantero, M.J.; Bueloni, B.; García, M.; Lagües, C.; Martínez-Duartez, P.; Menaldi, G.; et al. Hepatic SPARC Expression Is Associated with Inflammasome Activation during the Progression of Non-Alcoholic Fatty Liver Disease in Both Mice and Morbidly Obese Patients. Int. J. Mol. Sci. 2023, 24, 14843. https://doi.org/10.3390/ijms241914843
Onorato AM, Lameroli Mauriz L, Bayo J, Fiore E, Cantero MJ, Bueloni B, García M, Lagües C, Martínez-Duartez P, Menaldi G, et al. Hepatic SPARC Expression Is Associated with Inflammasome Activation during the Progression of Non-Alcoholic Fatty Liver Disease in Both Mice and Morbidly Obese Patients. International Journal of Molecular Sciences. 2023; 24(19):14843. https://doi.org/10.3390/ijms241914843
Chicago/Turabian StyleOnorato, Agostina M., Lucía Lameroli Mauriz, Juan Bayo, Esteban Fiore, María José Cantero, Barbara Bueloni, Mariana García, Cecilia Lagües, Pedro Martínez-Duartez, Gabriel Menaldi, and et al. 2023. "Hepatic SPARC Expression Is Associated with Inflammasome Activation during the Progression of Non-Alcoholic Fatty Liver Disease in Both Mice and Morbidly Obese Patients" International Journal of Molecular Sciences 24, no. 19: 14843. https://doi.org/10.3390/ijms241914843






