Myocardial Fibrosis in Hypertrophic Cardiomyopathy: A Perspective from Fibroblasts
Abstract
1. Introduction
2. Cellular Effectors of Myocardial Fibrosis
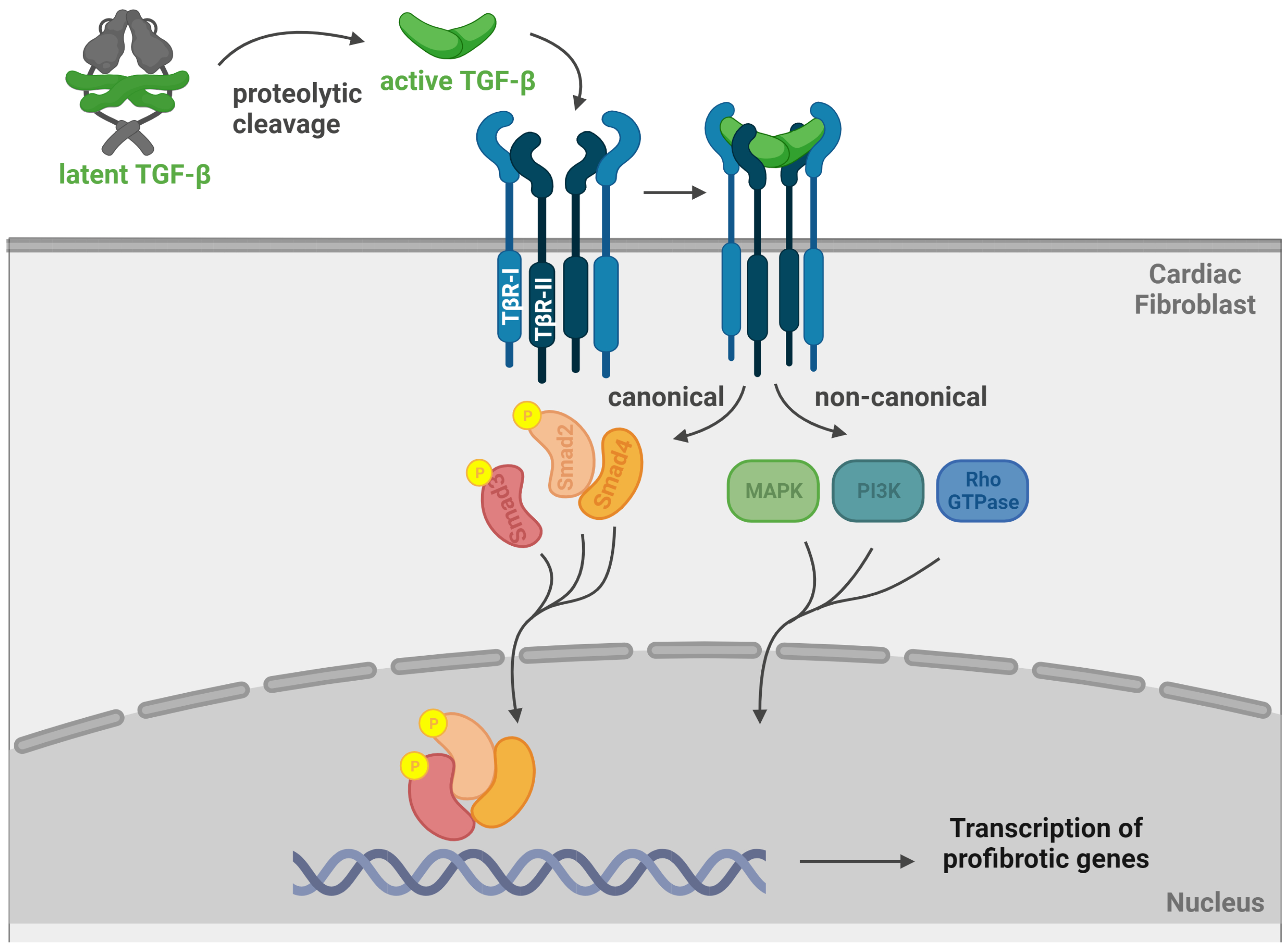
3. TGF-β Signalling
3.1. TGF-β Signalling in HCM Myocardial Fibrosis
3.2. Canonical and Non-Canonical TGF-β Signalling
4. Interactions between Cardiomyocytes and Cardiac Fibroblasts in HCM Myocardial Fibrosis
5. Profibrotic Genes Cascade in HCM
6. Novel HCM Genetic and Nongenetic Actors
7. Conclusions and Future Research Scenarios
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arbelo, E.; Protonotarios, A.; Gimeno, J.R.; Arbustini, E.; Barriales-Villa, R.; Basso, C.; Bezzina, C.R.; Biagini, E.; Blom, N.A.; de Boer, R.A.; et al. ESC Scientific Document Group. 2023 ESC Guidelines for the management of cardiomyopathies. Eur. Heart J. 2023, ehad194. [Google Scholar] [CrossRef]
- Maron, B.J.; Maron, M.S. Hypertrophic cardiomyopathy. Lancet 2013, 381, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Moody, W.E.; Elliott, P.M. Changing concepts in heart muscle disease: The evolving understanding of hypertrophic cardiomyopathy. Heart 2022, 108, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Ingles, J.; Burns, C.; Bagnall, R.D.; Lam, L.; Yeates, L.; Sarina, T.; Puranik, R.; Briffa, T.; Atherton, J.J.; Driscoll, T.; et al. Nonfamilial Hypertrophic Cardiomyopathy: Prevalence, Natural History, and Clinical Implications. Circ. Cardiovasc. Genet. 2017, 10, e001620. [Google Scholar] [CrossRef] [PubMed]
- Aziz, A.; Musiol, S.K.; Moody, W.E.; Pickup, L.; Cooper, R.; Lip, G.Y.H. Clinical prediction of genotypes in hypertrophic cardiomyopathy: A systematic review. Eur. J. Clin. Invest. 2021, 51, e13593. [Google Scholar] [CrossRef]
- Alfares, A.A.; Kelly, M.A.; McDermott, G.; Funke, B.H.; Lebo, M.S.; Baxter, S.B.; Shen, J.; McLaughlin, H.M.; Clark, E.H.; Babb, L.J.; et al. Results of clinical genetic testing of 2912 probands with hypertrophic cardiomyopathy: Expanded panels offer limited additional sensitivity. Genet. Med. 2015, 17, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.; Elliott, P. The genetics of hypertrophic cardiomyopathy. Glob. Cardiol. Sci. Pract. 2018, 36. [Google Scholar] [CrossRef]
- Marian, A.J. Molecular Genetic Basis of Hypertrophic Cardiomyopathy. Circ. Res. 2021, 128, 1533–1553. [Google Scholar] [CrossRef]
- Gerull, B.; Klaassen, S.; Brodehl, A. The genetic landscape of cardiomyopathies. In Genetic Causes of Cardiac Disease, 1st ed.; Erdmann, J., Moretti, A., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 45–91. [Google Scholar] [CrossRef]
- Wolf, C.M. Hypertrophic cardiomyopathy: Genetics and clinical perspectives. Cardiovasc. Diagn. Ther. 2019, 9 (Suppl. S2), S388–S415. [Google Scholar] [CrossRef]
- Stafford, F.; Thomson, K.; Butters, A.; Ingles, J. Hypertrophic Cardiomyopathy: Genetic Testing and Risk Stratification. Curr. Cardiol. Rep. 2021, 23, 9. [Google Scholar] [CrossRef]
- Chou, C.; Chin, M.T. Pathogenic Mechanisms of Hypertrophic Cardiomyopathy beyond Sarcomere Dysfunction. Int. J. Mol. Sci. 2021, 22, 8933. [Google Scholar] [CrossRef] [PubMed]
- Ingles, J.; Goldstein, J.; Thaxton, C.; Caleshu, C.; Corty, E.W.; Crowley, S.B.; Dougherty, K.; Harrison, S.M.; McGlaughon, J.; Milko, L.V.; et al. Evaluating the Clinical Validity of Hypertrophic Cardiomyopathy Genes. Circ. Genom. Precis. Med. 2019, 12, e002460. [Google Scholar] [CrossRef] [PubMed]
- Olivotto, I.; Cecchi, F.; Poggesi, C.; Yacoub, M.H. Developmental origins of hypertrophic cardiomyopathy phenotypes: A unifying hypothesis. Nat. Rev. Cardiol. 2009, 6, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J. Clinical Course and Management of Hypertrophic Cardiomyopathy. N. Engl. J. Med. 2018, 379, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Chute, M.; Aujla, P.; Jana, S.; Kassiri, Z. The Non-Fibrillar Side of Fibrosis: Contribution of the Basement Membrane, Proteoglycans, and Glycoproteins to Myocardial Fibrosis. J. Cardiovasc. Dev. Dis. 2019, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Transforming growth factor–β in tissue fibrosis. J. Exp. Med. 2020, 217, e20190103. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Afzal, J.; Vakrou, S.; Greenland, G.V.; Talbot, C.C.; Hebl, V.B.; Guan, Y.; Karmali, R.; Tardiff, J.C.; Leinwand, L.A.; et al. Differences in microRNA-29 and Pro-fibrotic Gene Expression in Mouse and Human Hypertrophic Cardiomyopathy. Front. Cardiovasc. Med. 2019, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Teekakirikul, P.; Eminaga, S.; Toka, O.; Alcalai, R.; Wang, L.; Wakimoto, H.; Nayor, M.; Konno, T.; Gorham, J.M.; Wolf, C.M.; et al. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-β. J. Clin. Invest. 2010, 120, 3520–3529. [Google Scholar] [CrossRef]
- Larson, A.; Codden, C.J.; Huggins, G.S.; Rastegar, H.; Chen, F.Y.; Maron, B.J.; Rowin, E.J.; Maron, M.S.; Chin, M.T. Altered intercellular communication and extracellular matrix signaling as a potential disease mechanism in human hypertrophic cardiomyopathy. Sci. Rep. 2022, 12, 5211. [Google Scholar] [CrossRef]
- Axelsson Raja, A.; Wakimoto, H.; DeLaughter, D.M.; Reichart, D.; Gorham, J.; Conner, D.A.; Lun, M.; Probst, C.K.; Sakai, N.; Knipe, R.S.; et al. Ablation of lysophosphatidic acid receptor 1 attenuates hypertrophic cardiomyopathy in a mouse model. Proc. Natl. Acad. Sci. USA 2022, 119, e2204174119. [Google Scholar] [CrossRef]
- Shirani, J.; Pick, R.; Roberts, W.C.; Maron, B.J. Morphology and significance of the left ventricular collagen network in young patients with hypertrophic cardiomyopathy and sudden cardiac death. J. Am. Coll. Cardiol. 2000, 35, 36–44. [Google Scholar] [CrossRef]
- Lombardi, R.; Betocchi, S.; Losi, M.A.; Tocchetti, C.G.; Aversa, M.; Miranda, M.; D’Alessandro, G.; Cacace, A.; Ciampi, Q.; Chiariello, M. Myocardial collagen turnover in hypertrophic cardiomyopathy. Circulation 2003, 108, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.Y.; López, B.; Coelho-Filho, O.R.; Lakdawala, N.K.; Cirino, A.L.; Jarolim, P.; Kwong, R.; González, A.; Colan, S.D.; Seidman, J.G.; et al. Myocardial Fibrosis as an Early Manifestation of Hypertrophic Cardiomyopathy. N. Engl. J. Med. 2010, 363, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Eijgenraam, T.R.; Silljé, H.H.W.; de Boer, R.A. Current understanding of fibrosis in genetic cardiomyopathies. Trends Cardiovasc. Med. 2020, 30, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Corrado, D.; Zorzi, A.; Cipriani, A.; Bauce, B.; Bariani, R.; Brunetti, G.; Graziano, F.; De Lazzari, M.; Mattesi, G.; Migliore, F.; et al. Scarring/arrhythmogenic cardiomyopathy. Eur. Heart J. Suppl. 2023, 25, C144–C154. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Cardiac fibrosis. Cardiovasc. Res. 2021, 117, 1450–1488. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Porreca, G.J.; Song, L.; Greenway, S.C.; Gorham, J.M.; Church, G.M.; Seidman, C.E.; Seidman, J.G. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science 2007, 316, 1481–1484. [Google Scholar] [CrossRef]
- Litviňuková, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Worth, C.L.; Lindberg, E.L.; Kanda, M.; Polanski, K.; Heinig, M.; Lee, M.; et al. Cells of the adult human heart. Nature 2020, 588, 466–472. [Google Scholar] [CrossRef]
- Khan, S.; Joyce, J.; Margulies, K.B.; Tsuda, T. Enhanced bioactive myocardial transforming growth factor-β in advanced human heart failure. Circ. J. 2014, 78, 2711–2718. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. Cardiac fibrosis: Cell biological mechanisms, molecular pathways and therapeutic opportunities. Mol. Aspects. Med. 2019, 65, 70–99. [Google Scholar] [CrossRef]
- Fan, G.P.; Wang, W.; Zhao, H.; Cai, L.; Zhang, P.D.; Yang, Z.H.; Zhang, J.; Wang, X. Pharmacological Inhibition of Focal Adhesion Kinase Attenuates Cardiac Fibrosis in Mice Cardiac Fibroblast and Post-Myocardial-Infarction Models. Cell. Physiol. Biochem. 2015, 37, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Transforming growth factor-β in myocardial disease. Nat. Rev. Cardiol. 2022, 19, 435–455. [Google Scholar] [CrossRef] [PubMed]
- Dobaczewski, M.; Chen, W.; Frangogiannis, N.G. Transforming growth factor (TGF)-β signaling in cardiac remodeling. J. Mol. Cell. Cardiol. 2011, 51, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Murphy-Ullrich, J.E.; Suto, M.J. Thrombospondin-1 regulation of latent TGF-β activation: A therapeutic target for fibrotic disease. Matrix Biol. 2018, 68–69, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Bertaud, A.; Joshkon, A.; Heim, X.; Bachelier, R.; Bardin, N.; Leroyer, A.S.; Blot-Chabaud, M. Signaling Pathways and Potential Therapeutic Strategies in Cardiac Fibrosis. Int. J. Mol. Sci. 2023, 24, 1756. [Google Scholar] [CrossRef] [PubMed]
- Stawowy, P.; Margeta, C.; Kallisch, H.; Seidah, N.G.; Chrétien, M.; Fleck, E.; Graf, K. Regulation of matrix metalloproteinase MT1-MMP/MMP-2 in cardiac fibroblasts by TGF-beta1 involves furin-convertase. Cardiovasc. Res. 2004, 63, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Hu, C.; Song, Q.; Li, Y.; Da, X.; Yu, Y.; Li, H.; Clark, I.M.; Chen, Q.; Wang, Q.K. ADAMTS16 activates latent TGF-β, accentuating fibrosis and dysfunction of the pressure-overloaded heart. Cardiovasc. Res. 2020, 116, 956–969. [Google Scholar] [CrossRef]
- Vistnes, M.; Erusappan, P.M.; Sasi, A.; Nordén, E.S.; Bergo, K.K.; Romaine, A.; Lunde, I.G.; Zhang, L.; Olsen, M.B.; Øgaard, J.; et al. Inhibition of the extracellular enzyme A disintegrin and metalloprotease with thrombospondin motif 4 prevents cardiac fibrosis and dysfunction. Cardiovasc. Res. 2023, 119, 1915–1927. [Google Scholar] [CrossRef]
- Rifkin, D.; Sachan, N.; Singh, K.; Sauber, E.; Tellides, G.; Ramirez, F. The role of LTBPs in TGF beta signaling. Dev. Dyn. 2022, 251, 95–104. [Google Scholar] [CrossRef]
- Aashaq, S.; Batool, A.; Mir, S.A.; Beigh, M.A.; Andrabi, K.I.; Shah, Z.A. TGF-β signaling: A recap of SMAD-independent and SMAD-dependent pathways. J. Cell. Physiol. 2022, 237, 59–85. [Google Scholar] [CrossRef]
- Zhang, Y.E. Non-Smad Signaling Pathways of the TGF-β Family. Cold Spring Harb. Perspect. Biol. 2017, 9, a022129. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.J.; Raita, Y.; Liang, L.W.; Maurer, M.S.; Hasegawa, K.; Fifer, M.A.; Reilly, M.P. Comprehensive Proteomics Profiling Reveals Circulating Biomarkers of Hypertrophic Cardiomyopathy. Circ. Heart Fail. 2021, 14, e007849. [Google Scholar] [CrossRef] [PubMed]
- Ayça, B.; Sahin, I.; Kucuk, S.H.; Akin, F.; Kafadar, D.; Avşar, M.; Avci, I.I.; Gungor, B.; Okuyan, E.; Dinckal, M.H. Increased Transforming Growth Factor-β Levels Associated with Cardiac Adverse Events in Hypertrophic Cardiomyopathy. Clin. Cardiol. 2015, 38, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Borger, M.A.; Williams, W.G.; Weisel, R.D.; Mickle, D.A.; Wigle, E.D.; Li, R.K. Regional overexpression of insulin-like growth factor-I and transforming growth factor-beta1 in the myocardium of patients with hypertrophic obstructive cardiomyopathy. J. Thorac. Cardiovasc. Surg. 2002, 123, 89–95. [Google Scholar] [CrossRef][Green Version]
- Ibrahim, A.M.; Roshdy, M.; Elshorbagy, S.; Hosny, M.; Halawa, S.; Yehia, D.; Elfawy, H.A.; Eldessouki, A.; Mohamed, F.; Ellithy, A.; et al. An Investigation of Fibulin-2 in Hypertrophic Cardiomyopathy. Int. J. Mol. Sci. 2020, 21, 7176. [Google Scholar] [CrossRef]
- Kuwahara, F.; Kai, H.; Tokuda, K.; Kai, M.; Takeshita, A.; Egashira, K.; Imaizumi, T. Transforming growth factor-beta function blocking prevents myocardial fibrosis and diastolic dysfunction in pressure-overloaded rats. Circulation 2002, 106, 130–135. [Google Scholar] [CrossRef]
- Rosenkranz, S.; Flesch, M.; Amann, K.; Haeuseler, C.; Kilter, H.; Seeland, U.; Schlüter, K.D.; Böhm, M. Alterations of beta-adrenergic signaling and cardiac hypertrophy in transgenic mice overexpressing TGF-beta(1). Am. J. Physiol. Heart Circ. Physiol. 2002, 283, H1253–H1262. [Google Scholar] [CrossRef]
- Brooks, W.W.; Conrad, C.H. Myocardial fibrosis in transforming growth factor beta(1) heterozygous mice. J. Mol. Cell. Cardiol. 2000, 32, 187–195. [Google Scholar] [CrossRef]
- Lucas, J.A.; Zhang, Y.; Li, P.; Gong, K.; Miller, A.P.; Hassan, E.; Hage, F.; Xing, D.; Wells, B.; Oparil, S.; et al. Inhibition of transforming growth factor-β signaling induces left ventricular dilation and dysfunction in the pressure-overloaded heart. Am. J. Physiol. Heart Circ. Physiol. 2010, 298, H424–H432. [Google Scholar] [CrossRef]
- Khalil, H.; Kanisicak, O.; Prasad, V.; Correll, R.N.; Fu, X.; Schips, T.; Vagnozzi, R.J.; Liu, R.; Huynh, T.; Lee, S.J.; et al. Fibroblast-specific TGF-β–Smad2/3 signaling underlies cardiac fibrosis. J. Clin. Invest. 2017, 127, 3770–3783. [Google Scholar] [CrossRef]
- Engebretsen, K.V.T.; Skårdal, K.; Bjørnstad, S.; Marstein, H.S.; Skrbic, B.; Sjaastad, I.; Christensen, G.; Bjørnstad, J.L.; Tønnessen, T. Attenuated development of cardiac fibrosis in left ventricular pressure overload by SM16, an orally active inhibitor of ALK5. J. Mol. Cell. Cardiol. 2014, 76, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Russo, I.; Cavalera, M.; Huang, S.; Su, Y.; Hanna, A.; Chen, B.; Shinde, A.V.; Conway, S.J.; Graff, J.; Frangogiannis, N.G. Protective Effects of Activated Myofibroblasts in the Pressure-Overloaded Myocardium Are Mediated Through Smad-Dependent Activation of a Matrix-Preserving Program. Circ. Res. 2019, 124, 1214–1227. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Bhandary, B.; Bhuiyan, M.S.; James, J.; Osinska, H.; Valiente-Alandi, I.; Shay-Winkler, K.; Gulick, J.; Molkentin, J.D.; Blaxall, B.C.; et al. Myofibroblast-Specific TGFβ Receptor II Signaling in the Fibrotic Response to Cardiac Myosin Binding Protein C-Induced Cardiomyopathy. Circ. Res. 2018, 123, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Razzaque, M.A.; Gupta, M.; Osinska, H.; Gulick, J.; Blaxall, B.C.; Robbins, J. An Endogenously Produced Fragment of Cardiac Myosin-Binding Protein C Is Pathogenic and Can Lead to Heart Failure. Circ. Res. 2013, 113, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Bhandary, B.; Osinska, H.; James, J.; Xu, N.; Shay-Winkler, K.; Gulick, J.; Willis, M.S.; Lander, C.; Robbins, J. MMI-0100 Inhibits Cardiac Fibrosis in a Mouse Model Overexpressing Cardiac Myosin Binding Protein C. J. Am. Heart Assoc. 2017, 6, e006590. [Google Scholar] [CrossRef] [PubMed]
- Vullaganti, S.; Levine, J.; Raiker, N.; Syed, A.A.; Collins, J.D.; Carr, J.C.; Bonow, R.O.; Choudhury, L. Fibrosis in Hypertrophic Cardiomyopathy Patients with and Without Sarcomere Gene Mutations. Heart Lung Circ. 2021, 30, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Ellims, A.H.; Iles, L.M.; Ling, L.H.; Chong, B.; Macciocca, I.; Slavin, G.S.; Hare, J.L.; Kaye, D.M.; Marasco, S.F.; McLean, C.A.; et al. A comprehensive evaluation of myocardial fibrosis in hypertrophic cardiomyopathy with cardiac magnetic resonance imaging: Linking genotype with fibrotic phenotype. Eur. Heart J. Cardiovasc. Imaging 2014, 15, 1108–1116. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, R.; Fujino, N.; Konno, T.; Nomura, A.; Nagata, Y.; Tsuda, T.; Tada, H.; Sakata, K.; Yamagishi, M.; Hayashi, K.; et al. Late Gadolinium Enhancement for Prediction of Mutation-Positive Hypertrophic Cardiomyopathy on the Basis of Panel-Wide Sequencing. Circ. J. 2018, 82, 1139–1148. [Google Scholar] [CrossRef]
- Ho, C.Y.; Abbasi, S.A.; Neilan, T.G.; Shah, R.V.; Chen, Y.; Heydari, B.; Cirino, A.L.; Lakdawala, N.K.; Orav, E.J.; González, A.; et al. T1 measurements identify extracellular volume expansion in hypertrophic cardiomyopathy sarcomere mutation carriers with and without left ventricular hypertrophy. Circ. Cardiovasc. Imaging 2013, 6, 415–422. [Google Scholar] [CrossRef]
- Neubauer, S.; Kolm, P.; Ho, C.Y.; Kwong, R.Y.; Desai, M.Y.; Dolman, S.F.; Appelbaum, E.; Desvigne-Nickens, P.; DiMarco, J.P.; Friedrich, M.G.; et al. Distinct Subgroups in Hypertrophic Cardiomyopathy in the NHLBI HCM Registry. J. Am. Coll. Cardiol. 2019, 74, 2333–2345. [Google Scholar] [CrossRef]
- Rowin, E.J.; Maron, M.S.; Lesser, J.R.; Maron, B.J. CMR with late gadolinium enhancement in genotype positive-phenotype negative hypertrophic cardiomyopathy. JACC Cardiovasc. Imaging 2012, 5, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Ouyang, H.; Lin, F.; Zhang, H.; Yang, Y.; Pang, D.; Han, R.; Tang, X. MYBPC3 deficiency in cardiac fibroblasts drives their activation and contributes to fibrosis. Cell Death Dis. 2022, 13, 948. [Google Scholar] [CrossRef] [PubMed]
- Chaffin, M.; Papangeli, I.; Simonson, B.; Akkad, A.D.; Hill, M.C.; Arduini, A.; Fleming, S.J.; Melanson, M.; Hayat, S.; Kost-Alimova, M.; et al. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy. Nature 2022, 608, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Bagnall, R.D.; Tsoutsman, T.; Shephard, R.E.; Ritchie, W.; Semsarian, C. Global microRNA profiling of the mouse ventricles during development of severe hypertrophic cardiomyopathy and heart failure. PLoS ONE 2012, 7, e44744. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhou, Y.; Wang, C.X. LncRNA-MIAT regulates fibrosis in hypertrophic cardiomyopathy (HCM) by mediating the expression of miR-29a-3p. J. Cell. Biochem. 2019, 120, 7265–7275. [Google Scholar] [CrossRef] [PubMed]
- Scolari, F.L.; Faganello, L.S.; Garbin, H.I.; Piva, E.; Mattos, B.; Biolo, A. A systematic review of microRNAs in patients with hypertrophic cardiomyopathy. Int. J. Cardiol. 2021, 327, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.A.; Wang, R.S.; Shevtsov, S.; Drakos, S.G.; Arons, E.; Wever-Pinzon, O.; Huggins, G.S.; Samokhin, A.O.; Oldham, W.M.; Aguib, Y.; et al. Individualized interactomes for network-based precision medicine in hypertrophic cardiomyopathy with implications for other clinical pathophenotypes. Nat. Commun. 2021, 12, 873. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.Y.; Day, S.M.; Ashley, E.A.; Michels, M.; Pereira, A.C.; Jacoby, D.; Cirino, A.L.; Fox, J.C.; Lakdawala, N.K.; Ware, J.S.; et al. Genotype and Lifetime Burden of Disease in Hypertrophic Cardiomyopathy: Insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation 2018, 138, 1387–1398. [Google Scholar] [CrossRef]
- Watkins, H. Time to Think Differently About Sarcomere-Negative Hypertrophic Cardiomyopathy. Circulation 2021, 143, 2415–2417. [Google Scholar] [CrossRef]
- Mazzarotto, F.; Olivotto, I.; Boschi, B.; Girolami, F.; Poggesi, C.; Barton, P.J.R.; Walsh, R. Contemporary Insights into the Genetics of Hypertrophic Cardiomyopathy: Toward a New Era in Clinical Testing? J. Am. Heart Assoc. 2020, 9, e015473. [Google Scholar] [CrossRef]
- Harper, A.R.; Goel, A.; Grace, C.; Thomson, K.L.; Petersen, S.E.; Xu, X.; Waring, A.; Ormondroyd, E.; Kramer, C.M.; Ho, C.Y.; et al. Common genetic variants and modifiable risk factors underpin hypertrophic cardiomyopathy susceptibility and expressivity. Nat. Genet. 2021, 53, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Tadros, R.; Francis, C.; Xu, X.; Vermeer, A.M.C.; Harper, A.R.; Huurman, R.; Kelu Bisabu, K.; Walsh, R.; Hoorntje, E.T.; Rijdt, W.P.T.; et al. Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nat. Genet. 2021, 53, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.; Offerhaus, J.A.; Tadros, R.; Bezzina, C.R. Minor hypertrophic cardiomyopathy genes, major insights into the genetics of cardiomyopathies. Nat. Rev. Cardiol. 2022, 19, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.A.; Wang, R.S.; Carnethon, M.R.; Rowin, E.J.; Loscalzo, J.; Maron, B.J.; Maron, M.S. What Causes Hypertrophic Cardiomyopathy? Am. J. Cardiol. 2022, 179, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, I.; Kozin, M.; Baulina, N.; Pisklova, M.; Danilova, L.; Zotov, A.; Chumakova, O.; Zateyshchikov, D.; Favorova, O. Novel Genes Involved in Hypertrophic Cardiomyopathy: Data of Transcriptome and Methylome Profiling. Int. J. Mol. Sci. 2022, 23, 15280. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Schuldt, M.; Nagyova, E.; Gu, Z.; El Bouhaddani, S.; Yiangou, L.; Jansen, M.; Calis, J.J.A.; Dorsch, L.M.; Blok, C.S.; et al. Multi-omics integration identifies key upstream regulators of pathomechanisms in hypertrophic cardiomyopathy due to truncating MYBPC3 mutations. Clin. Epigenet. 2021, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Sartorio, C.L.; Lazzeroni, D.; Bertoli, G.; Camici, P.G. Theranostic biomarkers in hypertrophic cardiomyopathy: Insights in a long road ahead. Front. Biosci.(Landmark Ed). 2017, 22, 1724–1749. [Google Scholar]
- Luo, F.; Liu, W.; Bu, H. MicroRNAs in hypertrophic cardiomyopathy: Pathogenesis, diagnosis, treatment potential and roles as clinical biomarkers. Heart Fail. Rev. 2022, 27, 2211–2221. [Google Scholar] [CrossRef]
- Angelopoulos, A.; Oikonomou, E.; Vogiatzi, G.; Antonopoulos, A.; Tsalamandris, S.; Georgakopoulos, C.; Papanikolaou, P.; Lazaros, G.; Charalambous, G.; Siasos, G.; et al. MicroRNAs as Biomarkers in Hypertrophic Cardiomyopathy: Current State of the Art. Curr. Med. Chem. 2021, 28, 7400–7412. [Google Scholar] [CrossRef]
- Bittencourt, M.I.; Cader, S.A.; Araújo, D.V.; Salles, A.L.F.; Albuquerque, F.N.; Spineti, P.P.M.; Albuquerque, D.C.; Mourilhe-Rocha, R. Role of Myocardial Fibrosis in Hypertrophic Cardiomyopathy: A Systematic Review and Updated Meta-Analysis of Risk Markers for Sudden Death. Arq. Bras. Cardiol. 2019, 112, 281–289. [Google Scholar] [CrossRef]
- Pagourelias, E.D.; Alexandridis, G.M.; Vassilikos, V.P. Fibrosis in hypertrophic cardiomyopathy: Role of novel echo techniques and multi-modality imaging assessment. Heart Fail Rev. 2021, 26, 1297–1310. [Google Scholar] [CrossRef]
- Aimo, A.; Spitaleri, G.; Panichella, G.; Lupón, J.; Emdin, M.; Bayes-Genis, A. Pirfenidone as a novel cardiac protective treatment. Heart Fail Rev. 2022, 27, 525–532. [Google Scholar] [CrossRef]
- Lewis, G.A.; Rosala-Hallas, A.; Dodd, S.; Schelbert, E.B.; Williams, S.G.; Cunnington, C.; McDonagh, T.; Miller, C.A. Impact of Myocardial Fibrosis on Cardiovascular Structure, Function and Functional Status in Heart Failure with Preserved Ejection Fraction. J. Cardiovasc. Transl. Res. 2022, 15, 1436–1443. [Google Scholar] [CrossRef]
- Neuber, S.; Ermer, M.R.; Emmert, M.Y.; Nazari-Shafti, T.Z. Treatment of Cardiac Fibrosis with Extracellular Vesicles: What Is Missing for Clinical Translation? Int. J. Mol. Sci. 2023, 24, 10480. [Google Scholar] [CrossRef]
- Pesce, M.; Duda, G.N.; Forte, G.; Girao, H.; Raya, A.; Roca-Cusachs, P.; Sluijter, J.P.G.; Tschöpe, C.; Van Linthout, S. Cardiac fibroblasts and mechanosensation in heart development, health and disease. Nat. Rev. Cardiol. 2023, 20, 309–324. [Google Scholar] [CrossRef]
| Gene | Protein | Function |
|---|---|---|
| ACTC1 | Actin Alpha Cardiac Muscle 1 | Thin filament protein |
| ACTN2 | Actinin Alpha 2 | Z lines |
| ALPK3 | Alpha Kinase 3 | Sarcomere-associated protein |
| ANKRD1 | Cardiac Ankyrin Repeat Protein 1 | Sarcomere-associated protein |
| CALR3 | Calreticulin 3 | Calcium homeostasis |
| CAV3 | Caveolin 3 | Sarcomere-associated protein |
| CSRP3 | Cysteine And Glycine Rich Protein 3 | Z lines |
| FLNC | Filamin C | Sarcomere-associated protein |
| JPH2 | Junctophilin 2 | Calcium homeostasis |
| KLF10 | Kruppel-like Factor 10 | Transcription factor |
| MYBPC3 | Myosin-binding protein C3 | Thick filament protein |
| MYH6 | Myosin Heavy Chain 6 | Thick filament protein |
| MYH7 | Myosin Heavy Chain 7 | Thick filament protein |
| MYL2 | Myosin Light Chain 2 | Thick filament protein |
| MYL3 | Myosin Light Chain 3 | Thick filament protein |
| MYLK2 | Myosin Light Chain Kinase 2 | Sarcomere-associated protein |
| MYOM1 | Myomesin 1 | M line |
| MYOZ2 | Myozenin 2 | Z lines |
| MYPN | Myopalladin | Sarcomere-associated protein |
| NEXN | Nexilin F-Actin Binding Protein | Sarcomere-associated protein |
| OBSCN | Obscurin, Cytoskeletal Calmodulin and Titin-Interacting RhoGEF | M line |
| PDLIM3 | Alpha-Actinin-2-Associated LIM Protein | Sarcomere-associated protein |
| PLN | Phospholamban | Calcium homeostasis |
| RYR2 | Cardiac Ryanodine Receptor 2 | Calcium homeostasis |
| TCAP | Titin-Cap | Z lines |
| TNNC1 | Troponin C1, Slow Skeletal And Cardiac Type | Thin filament protein |
| TNNI3 | Troponin I3, Cardiac Type | Thin filament protein |
| TNNT2 | Troponin T2, Cardiac Type | Thin filament protein |
| TPM1 | Tropomyosin 1 | Thin filament protein |
| TRIM63 | Tripartite Motif Containing 63 | M line |
| TTN | Titin | Thick filament protein |
| VCL | Vinculin | Sarcomere-associated protein |
| Reference | Model | Genes [Method] |
|---|---|---|
| [19] | Non-myocyte cells isolated from ventricles of an HCM mouse model |
|
| [65] | Ventricles from HCM mouse model |
|
| [54] | Total heart and activated myofibroblasts in Mybpc340kDa transgenic mice |
|
| [66] | Serum samples from HCM patients with or without fibrosis |
|
| [18] | Left ventricle from two mouse models of HCM and septal myectomy of HCM patients |
|
| [46] | HCM myectomy tissues, myocardial cells and interstitial fibroblasts |
|
| [64] | Human cardiac-activated fibroblasts, from patients with advanced HCM |
|
| [68] | myectomy tissue from HCM patients |
|
| [20] | Human HCM myectomy tissues (interventricular septum): cardiomyocytes©, fibroblasts (F) and lymphocytes (L) |
|
| [21] | Left ventricles from HCM transgenic mice and HCM patients: lymphatic endothelial cells (LEC) and fibroblasts (F) |
|
| [58] | MYBPC3 mutant pig and MYBPC3-KO mouse NIH3T3 fibroblasts |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schlittler, M.; Pramstaller, P.P.; Rossini, A.; De Bortoli, M. Myocardial Fibrosis in Hypertrophic Cardiomyopathy: A Perspective from Fibroblasts. Int. J. Mol. Sci. 2023, 24, 14845. https://doi.org/10.3390/ijms241914845
Schlittler M, Pramstaller PP, Rossini A, De Bortoli M. Myocardial Fibrosis in Hypertrophic Cardiomyopathy: A Perspective from Fibroblasts. International Journal of Molecular Sciences. 2023; 24(19):14845. https://doi.org/10.3390/ijms241914845
Chicago/Turabian StyleSchlittler, Maja, Peter P. Pramstaller, Alessandra Rossini, and Marzia De Bortoli. 2023. "Myocardial Fibrosis in Hypertrophic Cardiomyopathy: A Perspective from Fibroblasts" International Journal of Molecular Sciences 24, no. 19: 14845. https://doi.org/10.3390/ijms241914845
APA StyleSchlittler, M., Pramstaller, P. P., Rossini, A., & De Bortoli, M. (2023). Myocardial Fibrosis in Hypertrophic Cardiomyopathy: A Perspective from Fibroblasts. International Journal of Molecular Sciences, 24(19), 14845. https://doi.org/10.3390/ijms241914845







