The Effect of Gene Editing by CRISPR-Cas9 of miR-21 and the Indirect Target MMP9 in Metastatic Prostate Cancer
Abstract
:1. Introduction
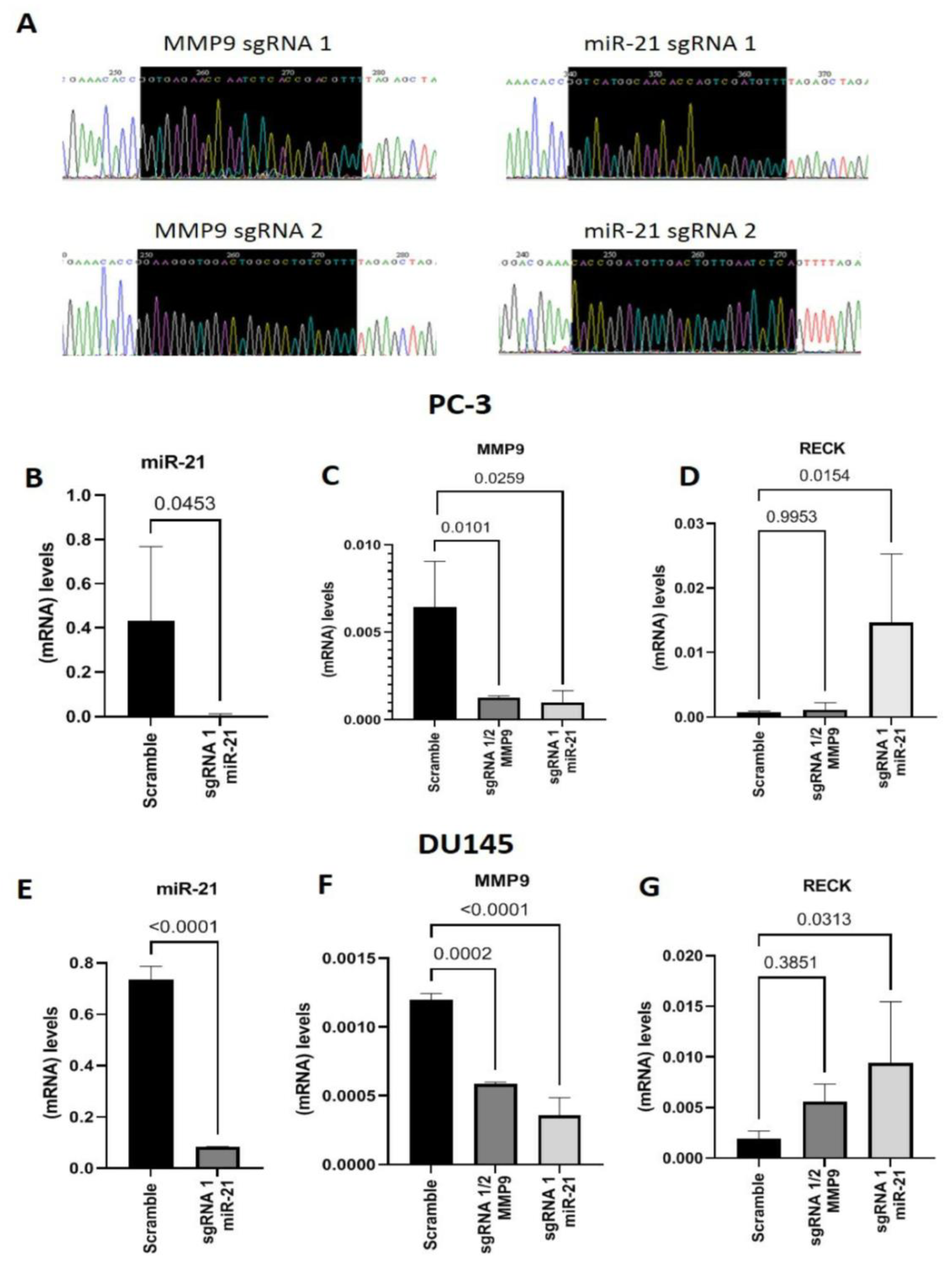
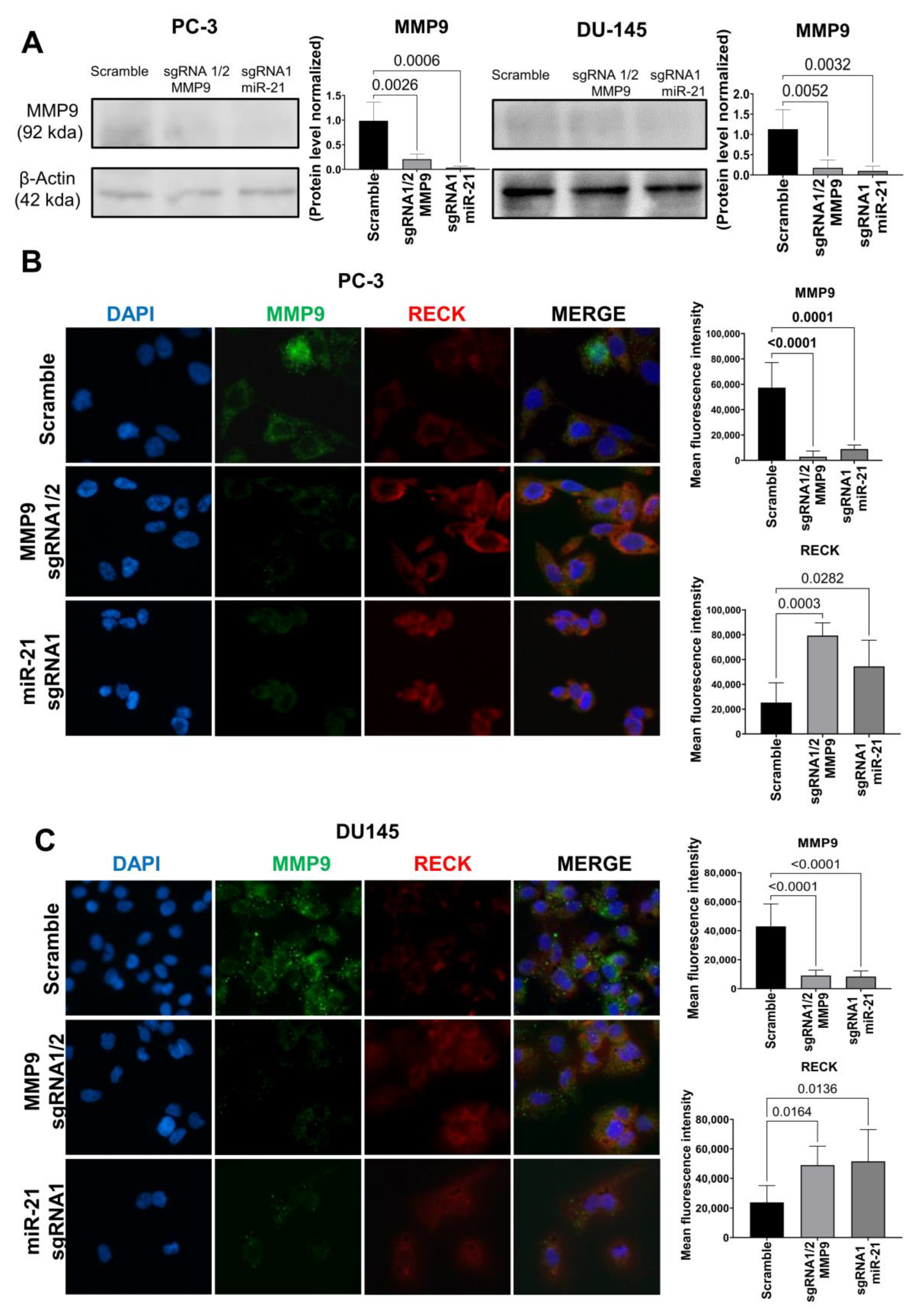
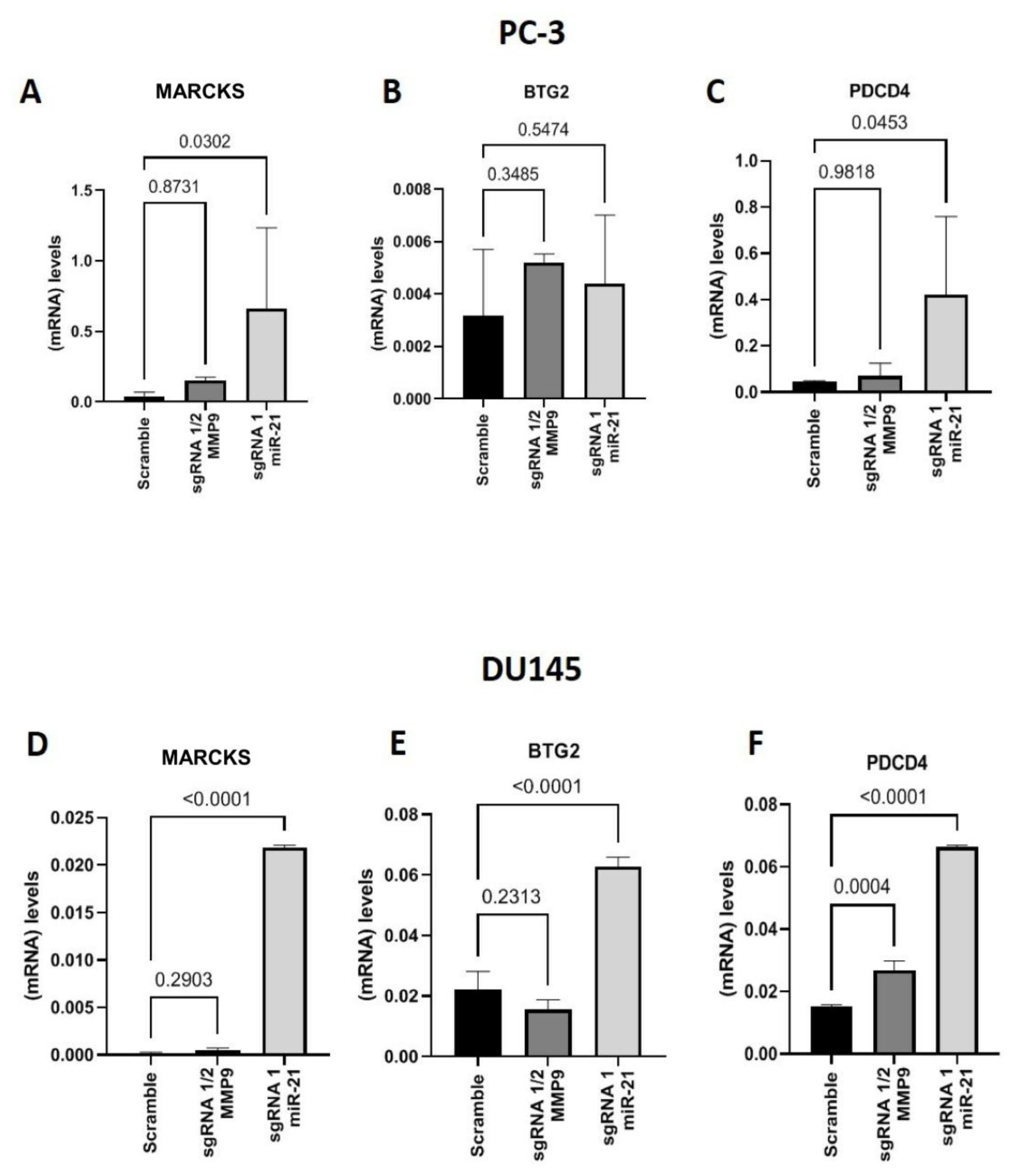
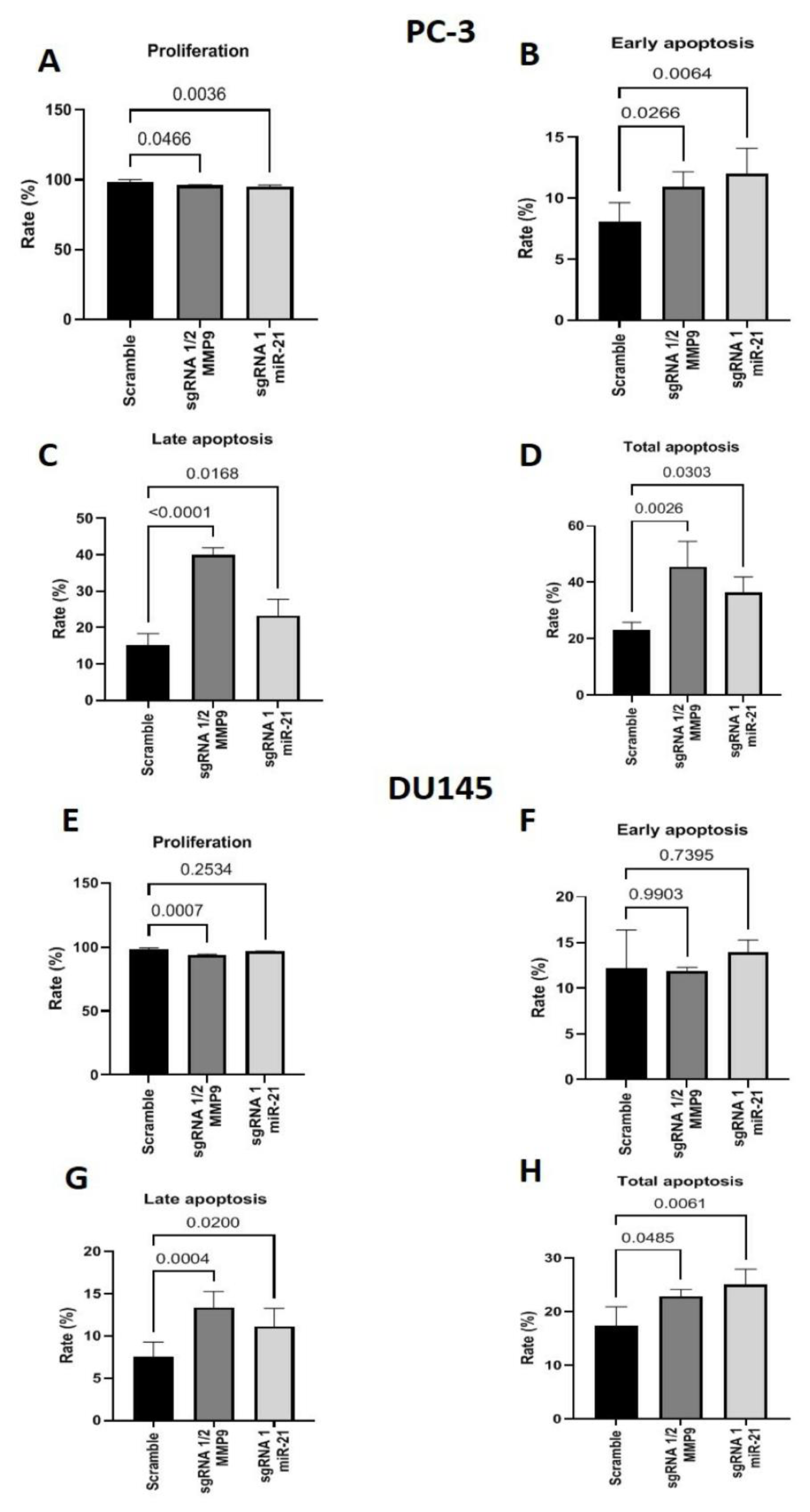
2. Results
Gene Editing with CRISPR/Cas9
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. CRISPR-Cas9
4.3. RNA and MicroRNA Isolation and Real-Time PCR
4.4. Flow Cytometry for Cell Proliferation and Apoptosis
4.5. Western Blotting
4.6. Immunofluorescence
4.7. Invasion Assays
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Lavaud, P.; Dumont, C.; Thibault, C.; Albiges, L.; Baciarello, G.; Colomba, E.; Flippot, R.; Fuerea, A.; Loriot, Y.; Fizazi, K. Next-generation androgen receptor inhibitors in non-metastatic castration-resistant prostate cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835920978134. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Chippada-Venkata, U.D.; Oh, W.K. Roles of matrix metalloproteinases and their natural inhibitors in prostate cancer progression. Cancers 2014, 6, 1298–1327. [Google Scholar] [CrossRef]
- Barillari, G. The Impact of Matrix Metalloproteinase-9 on the Sequential Steps of the Metastatic Process. Int. J. Mol. Sci. 2020, 21, 4526. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Li, W.; Zhu, J.; Deng, S.; Tao, X. Low expression of RECK in oral squamous cell carcinoma patients induces a shorter survival rate through an imbalance of RECK/MMPs. Int. J. Clin. Exp. Pathol. 2020, 13, 501–508. [Google Scholar]
- Reis, S.T.; Pontes-Junior, J.; Antunes, A.A.; de Sousa-Canavez, J.M.; Dall’Oglio, M.F.; Passerotti, C.C.; Abe, D.K.; Crippa, A.; Da Cruz, J.A.S.; Timoszczuk, L.M.; et al. MMP-9 overexpression due to TIMP-1 and RECK underexpression is associated with prognosis in prostate cancer. Int. J. Biol. Markers 2011, 26, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wang, R.; Shen, D.; Cheng, S.; Wang, H.; Lu, Z.; Zheng, Q.; Wang, L.; Xia, L.; Li, G. Role of noncoding RNA in drug resistance of prostate cancer. Cell Death Dis. 2021, 12, 590. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef]
- McDonald, A.C.; Vira, M.; Walter, V.; Shen, J.; Raman, J.D.; Sanda, M.G.; Patil, D.; Taioli, E. Circulating microRNAs in plasma among men with low-grade and high-grade prostate cancer at prostate biopsy. Prostate 2019, 79, 961–968. [Google Scholar] [CrossRef]
- Zhao, W.; Ning, L.; Wang, L.; Ouyang, T.; Qi, L.; Yang, R.; Wu, Y. miR-21 inhibition reverses doxorubicin-resistance and inhibits PC3 human prostate cancer cells proliferation. Andrologia 2021, 53, e14016. [Google Scholar] [CrossRef]
- Coppola, V.; Musumeci, M.; Patrizii, M.; Cannistraci, A.; Addario, A.; Maugeri-Saccà, M.; Biffoni, M.; Francescangeli, F.; Cordenonsi, M.; Piccolo, S.; et al. BTG2 loss and miR-21 upregulation contribute to prostate cell transformation by inducing luminal markers expression and epithelial-mesenchymal transition. Oncogene 2013, 32, 1843–1853. [Google Scholar] [CrossRef]
- Li, T.; Li, D.; Sha, J.; Sun, P.; Huang, Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem. Biophys. Res. Commun. 2009, 383, 280–285. [Google Scholar] [CrossRef]
- Leite, K.R.; Reis, S.T.; Viana, N.; Morais, D.R.; Moura, C.M.; Silva, I.A.; Katz, B.; Srougi, M. Controlling RECK miR21 Promotes Tumor Cell Invasion and Is Related to Biochemical Recurrence in Prostate Cancer. J. Cancer 2015, 6, 292–301. [Google Scholar] [CrossRef]
- Lentsch, E.; Li, L.; Pfeffer, S.; Ekici, A.B.; Taher, L.; Pilarsky, C.; Grützmann, R. CRISPR/Cas9-Mediated Knock-Out of Kras. Int. J. Mol. Sci. 2019, 20, 5706. [Google Scholar] [CrossRef]
- Bodey, B.; Siegel, S.E.; Kaiser, H.E. Immunocytochemical detection of matrix metalloproteinase expression in prostate cancer. In Vivo 2001, 15, 65–70. [Google Scholar]
- Babichenko, I.I.; Andriukhin, M.I.; Pulbere, S.; Loktev, A. Immunohistochemical expression of matrix metalloproteinase-9 and inhibitor of matrix metalloproteinase-1 in prostate adenocarcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 9090–9098. [Google Scholar] [PubMed]
- Folini, M.; Gandellini, P.; Longoni, N.; Profumo, V.; Callari, M.; Pennati, M.; Colecchia, M.; Supino, R.; Veneroni, S.; Salvioni, R.; et al. miR-21: An oncomir on strike in prostate cancer. Mol. Cancer 2010, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Shen, J.K.; Li, Z.; Choy, E.; Hornicek, F.J.; Duan, Z. Development and potential applications of CRISPR-Cas9 genome editing technology in sarcoma. Cancer Lett. 2016, 373, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Fei, T.; Chen, Y.; Xiao, T.; Li, W.; Cato, L.; Zhang, P.; Cotter, M.B.; Bowden, M.; Lis, R.T.; Zhao, S.G.; et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. USA 2017, 114, E5207–E5215. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wang, F.; Liu, W.; Zhao, W.; Yang, Y.; Li, K.; Xiao, L.; Shen, J. CRISPR/Cas9 targeting of the androgen receptor suppresses the growth of LNCaP human prostate cancer cells. Mol. Med. Rep. 2018, 17, 2901–2906. [Google Scholar] [PubMed]
- Ye, R.; Pi, M.; Cox, J.V.; Nishimoto, S.K.; Quarles, L.D. CRISPR/Cas9 targeting of GPRC6A suppresses prostate cancer tumorigenesis in a human xenograft model. J. Exp. Clin. Cancer Res. 2017, 36, 90. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Arisan, E.D.; Rencuzogullari, O.; Freitas, I.L.; Radzali, S.; Keskin, B.; Kothari, A.; Warford, A.; Uysal-Onganer, P. Upregulated Wnt-11 and miR-21 Expression Trigger Epithelial Mesenchymal Transition in Aggressive Prostate Cancer Cells. Biology 2020, 9, 52. [Google Scholar] [CrossRef]
- Kim, K.; Kim, H.H.; Lee, C.H.; Kim, S.; Cheon, G.J.; Kang, K.W.; Chung, J.-K.; Youn, H. Therapeutic efficacy of modified anti-miR21 in metastatic prostate cancer. Biochem. Biophys. Res. Commun. 2020, 529, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Olson, A.; Le, V.; Aldahl, J.; Yu, E.J.; Hooker, E.; He, Y.; Lee, D.-H.; Kim, W.K.; Cardiff, R.D.; Geradts, J.; et al. The comprehensive role of E-cadherin in maintaining prostatic epithelial integrity during oncogenic transformation and tumor progression. PLoS Genet. 2019, 15, e1008451. [Google Scholar] [CrossRef]
- Page-McCaw, A.; Ewald, A.J.; Werb, Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 2007, 8, 221–233. [Google Scholar] [CrossRef]
- Kurozumi, A.; Goto, Y.; Matsushita, R.; Fukumoto, I.; Kato, M.; Nishikawa, R.; Sakamoto, S.; Enokida, H.; Nakagawa, M.; Ichikawa, T.; et al. Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci. 2016, 107, 84–94. [Google Scholar] [CrossRef]
- Bonci, D.; Coppola, V.; Patrizii, M.; Addario, A.; Cannistraci, A.; Francescangeli, F.; Pecci, R.; Muto, G.; Collura, D.; Bedini, R.; et al. A microRNA code for prostate cancer metastasis. Oncogene 2016, 35, 1180–1192. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camargo, J.A.; Viana, N.I.; Pimenta, R.; Guimarães, V.R.; dos Santos, G.A.; Candido, P.; Ghazarian, V.; Romão, P.; Silva, I.A.; Birbrair, A.; et al. The Effect of Gene Editing by CRISPR-Cas9 of miR-21 and the Indirect Target MMP9 in Metastatic Prostate Cancer. Int. J. Mol. Sci. 2023, 24, 14847. https://doi.org/10.3390/ijms241914847
Camargo JA, Viana NI, Pimenta R, Guimarães VR, dos Santos GA, Candido P, Ghazarian V, Romão P, Silva IA, Birbrair A, et al. The Effect of Gene Editing by CRISPR-Cas9 of miR-21 and the Indirect Target MMP9 in Metastatic Prostate Cancer. International Journal of Molecular Sciences. 2023; 24(19):14847. https://doi.org/10.3390/ijms241914847
Chicago/Turabian StyleCamargo, Juliana A., Nayara I. Viana, Ruan Pimenta, Vanessa R. Guimarães, Gabriel A. dos Santos, Patrícia Candido, Vitória Ghazarian, Poliana Romão, Iran A. Silva, Alexander Birbrair, and et al. 2023. "The Effect of Gene Editing by CRISPR-Cas9 of miR-21 and the Indirect Target MMP9 in Metastatic Prostate Cancer" International Journal of Molecular Sciences 24, no. 19: 14847. https://doi.org/10.3390/ijms241914847






