Structural Insights of the DciA Helicase Loader in Its Relationship with DNA
Abstract
1. Introduction
2. Results and Discussion
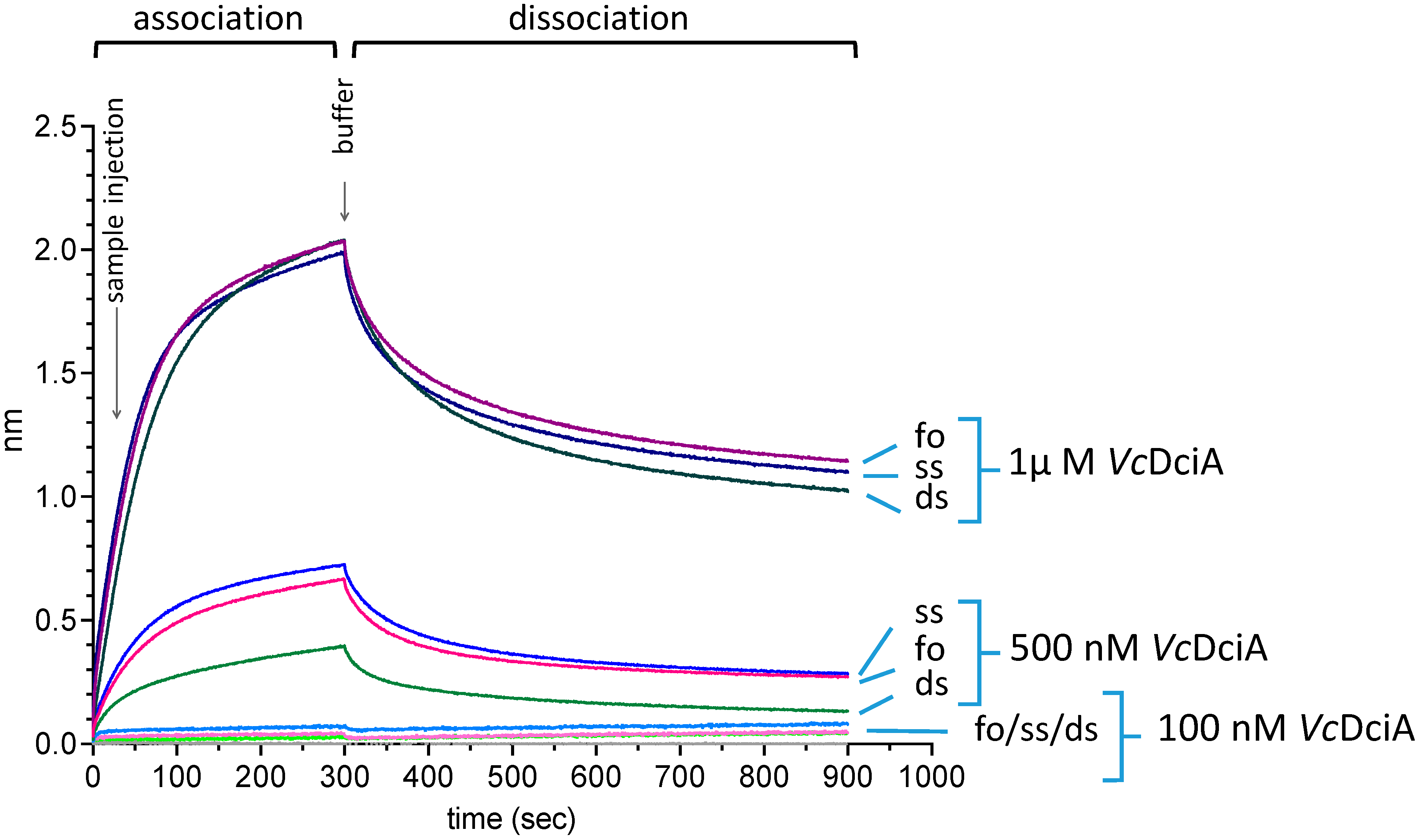
2.1. VcDciA Binds to Single- and Double-Stranded Oligodeoxynucleotide Substrates
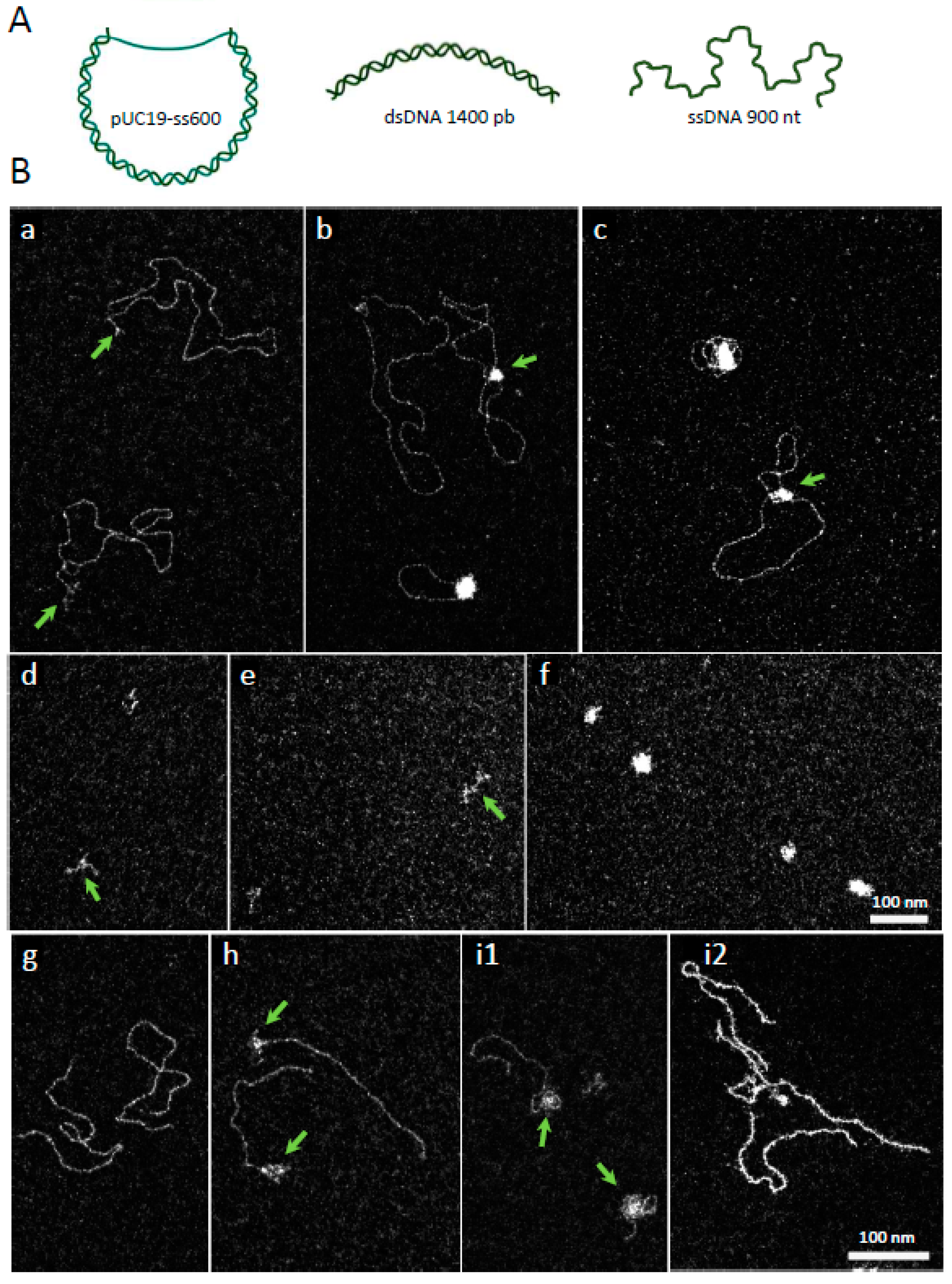
2.2. VcDciA Binds to Single- and Double-Stranded DNA in Intermediate Structures
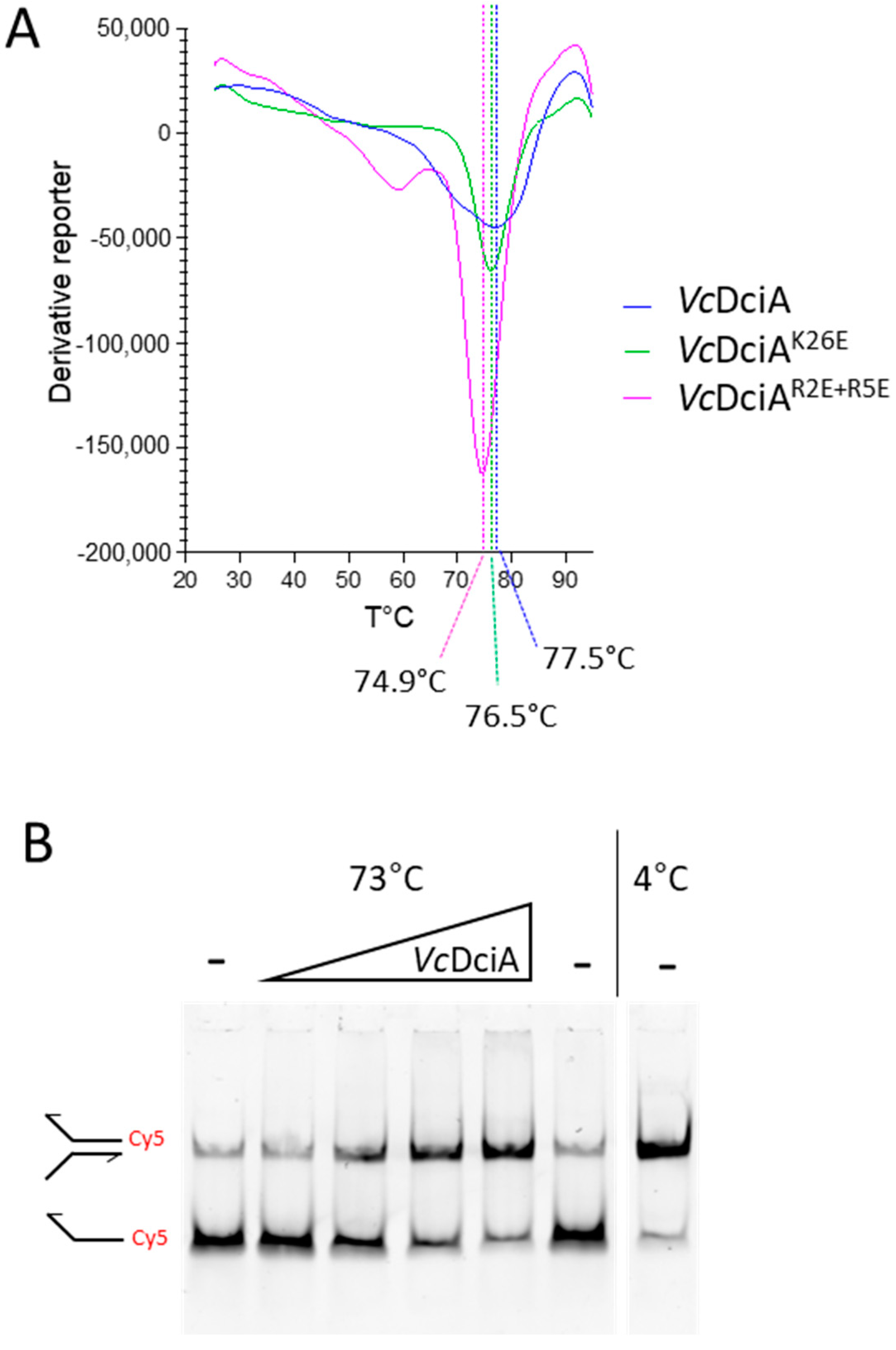
2.3. VcDciA Protects Double-Stranded DNA Oligonucleotide Substrates from Thermal Denaturation
2.4. The Long α-Helix 1 of VcDciA Interacts with Different DNA Substrates
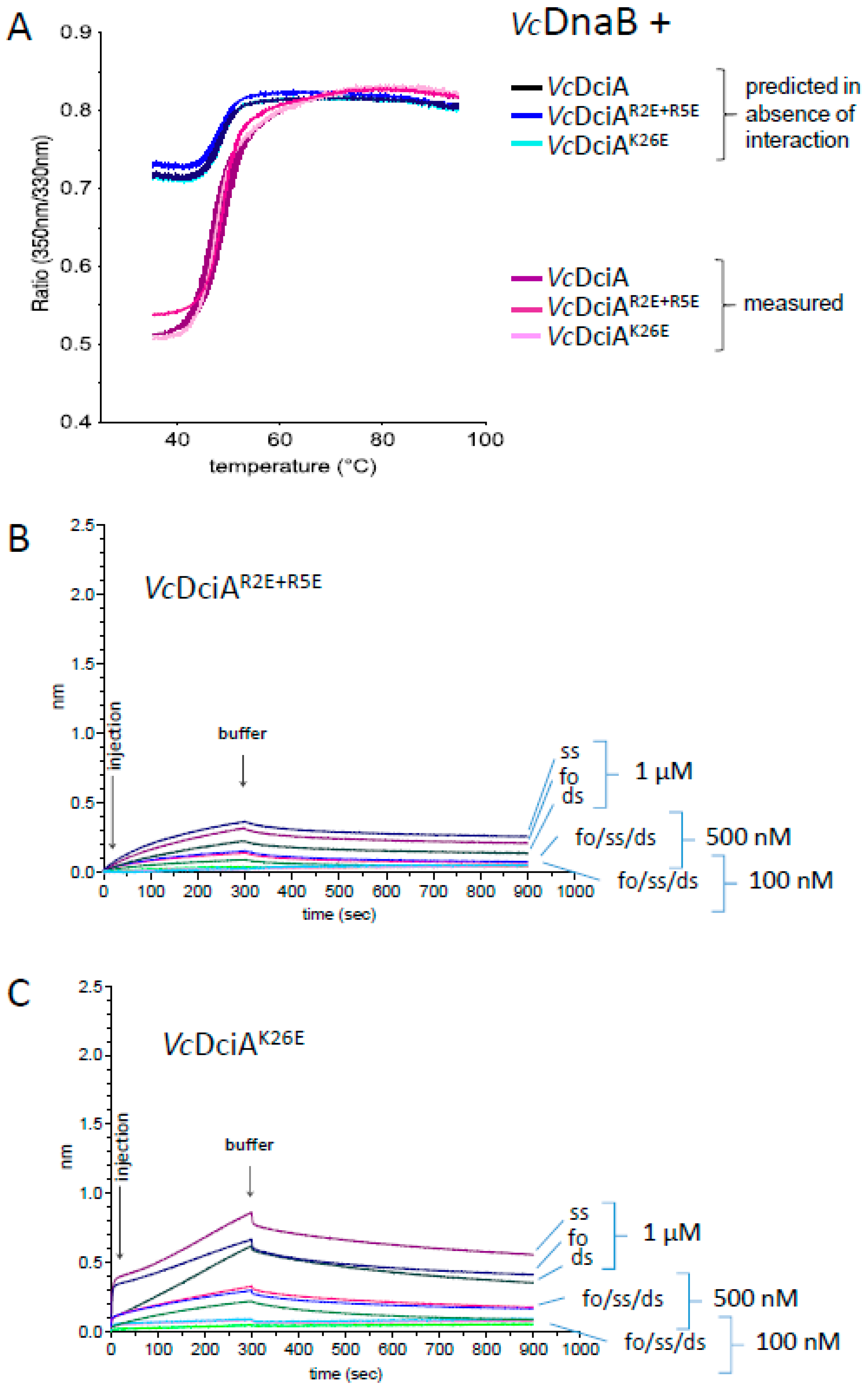
2.5. The R2, R5, and K26 Residues Involved in the Interaction of VcDciA with the DNA Have a Different Impact on the Helicase Loading
3. Materials and Methods
3.1. Protein Samples Preparation and Site-Directed Mutagenesis
3.2. Measurement of VcDciA–DNA Interaction by Bio-Layer Interferometry (BLI)
3.3. Electron Microscopy
3.4. Measurement of the Melting Point of VcDciA and of the VcDciAR2E+R5E and VcDciAK26E Mutants by Thermal Shift Assay (TSA)
3.5. Protection of the dsDNA from Thermal Denaturation by VcDciA
3.6. NMR Experiments
3.7. Protein–Protein Interaction Analysis by Thermal Shift Assay and Intrinsic Fluorescence Variation (DSF Differential Scanning Fluorescence)
3.8. Measurement of the Activation of VcDnaB Loading by VcDciA or Mutants by Interaction by Bio-Layer Interferometry (BLI)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mott, M.L.; Berger, J.M. DNA replication initiation: Mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007, 5, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Arias-Palomo, E.; Puri, N.; O’Shea Murray, V.L.; Yan, Q.; Berger, J.M. Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA. Mol. Cell 2019, 74, 173–184.e4. [Google Scholar] [CrossRef] [PubMed]
- Arias-Palomo, E.; O’Shea, V.L.; Hood, I.V.; Berger, J.M. The bacterial DnaC helicase loader is a DnaB ring breaker. Cell 2013, 153, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Funnell, B.E.; Baker, T.A.; Kornberg, A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem. 1987, 262, 10327–10334. [Google Scholar] [CrossRef] [PubMed]
- Chodavarapu, S.; Kaguni, J.M. Replication Initiation in Bacteria. Enzymes 2016, 39, 1–30. [Google Scholar] [PubMed]
- Brézellec, P.; Vallet-Gely, I.; Possoz, C.; Quevillon-Cheruel, S.; Ferat, J.L. DciA is an ancestral replicative helicase operator essential for bacterial replication initiation. Nat. Commun. 2016, 7, 13271. [Google Scholar] [CrossRef]
- Marsin, S.; Adam, Y.; Cargemel, C.; Andreani, J.; Baconnais, S.; Legrand, P.; Li de la Sierra-Gallay, I.; Humbert, A.; Aumont-Nicaise, M.; Velours, C.; et al. Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase. Nucleic Acids Res. 2021, 49, 6569–6586. [Google Scholar] [CrossRef]
- Chan-Yao-Chong, M.; Marsin, S.; Quevillon-Cheruel, S.; Durand, D.; Ha-Duong, T. Structural ensemble and biological activity of DciA intrinsically disordered region. J. Struct. Biol. 2020, 212, 107573. [Google Scholar] [CrossRef] [PubMed]
- Cargemel, C.; Marsin, S.; Noiray, M.; Legrand, P.; Bounoua, H.; Li de la Sierra-Gallay, I.; Walbott, H.; Quevillon-Cheruel, S. The LH-DH module of the bacterial replicative helicases is the common binding site for DciA and other helicase loaders. bioRxiv 2022. [Google Scholar] [CrossRef]
- Blaine, H.C.; Burke, J.T.; Ravi, J.; Stallings, C.L. DciA Helicase Operators Exhibit Diversity across Bacterial Phyla. J. Bacteriol. 2022, 204, e0016322. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Okada, A.; Ohtsuka, J.; Ohkuri, T.; Akama, Y.; Sakiyama, Y.; Miyazaki, E.; Horita, S.; Katayama, T.; Ueda, T.; et al. Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: Structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding. J. Biochem. 2020, 167, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Grishin, N.V. KH domain: One motif, two folds. Nucleic Acids Res. 2020, 29, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Gopal, B.; Haire, L.F.; Gamblin, S.J.; Dodson, E.J.; Lane, A.N.; Papavinasasundaram, K.G.; Colston, M.J.; Dodson, G. Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 A resolution. J. Mol. Biol. 2021, 314, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Valverde, R.; Edwards, L.; Regan, L. Structure and function of KH domains. FEBS J. 2008, 275, 2712–2726. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.R.; Laine, P.S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990, 54, 342–380. [Google Scholar] [CrossRef] [PubMed]
- Dupaigne, P.; Le Breton, C.; Fabre, F.; Gangloff, S.; Le Cam, E.; Veaute, X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol. Cell 2008, 29, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Beuth, B.; Pennell, S.; Arnvig, K.B.; Martin, S.R.; Taylor, I.A. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005, 24, 3576–3587. [Google Scholar] [CrossRef] [PubMed]
- Benureau, Y.; Moreira Tavares, E.; Muhammad, A.A.; Baconnais, S.; Le Cam, E.; Dupaigne, P. Method combining BAC film and positive staining for the characterization of DNA intermediates by dark-field electron microscopy. Biol. Methods Protoc. 2020, 5, bpaa012. [Google Scholar] [CrossRef] [PubMed]
- Farmer, B.T., II; Constantine, K.L.; Goldfarb, V.; Friedrichs, M.S.; Wittekind, M.; Yanchunas, J., Jr.; Robertson, J.G.; Mueller, L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat. Struct. Biol. 1996, 3, 995–997. [Google Scholar] [CrossRef] [PubMed]


| Name | Size (nt) | Sequence (5′→3′) |
|---|---|---|
| oso3 | 50 | Cy5-GCAGGCTCGTTACGTAGCTGTACCG-dT(25) |
| oso4 | 50 | dT(25)-CGGTACAGCTACGTAACGAGCCTGC |
| oso18 | 88 | CCAGGAATACGGCAAGTTGGAGGCCGGGCTGGATGGAGACTAAGCTTTGGAAGTGAAGGTTTCGAATCAGAGGTAGGTTTCACCACGC |
| oso23 | 80 | Biotin-AAGCGTGGTGAAACCTACCTCTGATTCGAAACCTTCACTTTACGTGGTCTGGCGTGGTGAATGTTCGTCGGCGTGCTCGA |
| oso24 | 78 | TCGAGCACGCCGACGAACATTCACCACGCCAGACCACGTAAAGTGAAGGTTTCGAATCAGAGGTAGGTTTCACCACGC |
| foDNANMR | 18/18 | GCTGTACCGTTTTTTTTT and TTTTTTTTTCGGTACAGC |
| dsDNANMR | 18 | GCTGTACCGACTAGCAAT and ATTGCTAGTCGGTACAGC |
| ssDNANMR | 18 | GCTGTACCGACTAGCAAT |
| 5′extDNANMR | 18/9 | TTTTTTTTTCGGTACAGC and GCTGTACCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cargemel, C.; Baconnais, S.; Aumont-Nicaise, M.; Noiray, M.; Maurin, L.; Andreani, J.; Walbott, H.; Le Cam, E.; Ochsenbein, F.; Marsin, S.; et al. Structural Insights of the DciA Helicase Loader in Its Relationship with DNA. Int. J. Mol. Sci. 2023, 24, 1427. https://doi.org/10.3390/ijms24021427
Cargemel C, Baconnais S, Aumont-Nicaise M, Noiray M, Maurin L, Andreani J, Walbott H, Le Cam E, Ochsenbein F, Marsin S, et al. Structural Insights of the DciA Helicase Loader in Its Relationship with DNA. International Journal of Molecular Sciences. 2023; 24(2):1427. https://doi.org/10.3390/ijms24021427
Chicago/Turabian StyleCargemel, Claire, Sonia Baconnais, Magali Aumont-Nicaise, Magali Noiray, Lia Maurin, Jessica Andreani, Hélène Walbott, Eric Le Cam, Françoise Ochsenbein, Stéphanie Marsin, and et al. 2023. "Structural Insights of the DciA Helicase Loader in Its Relationship with DNA" International Journal of Molecular Sciences 24, no. 2: 1427. https://doi.org/10.3390/ijms24021427
APA StyleCargemel, C., Baconnais, S., Aumont-Nicaise, M., Noiray, M., Maurin, L., Andreani, J., Walbott, H., Le Cam, E., Ochsenbein, F., Marsin, S., & Quevillon-Cheruel, S. (2023). Structural Insights of the DciA Helicase Loader in Its Relationship with DNA. International Journal of Molecular Sciences, 24(2), 1427. https://doi.org/10.3390/ijms24021427






