Micromechanical Study of Hyperacetylated Nucleosomes Using Single Molecule Transverse Magnetic Tweezers
Abstract
1. Introduction
2. Results
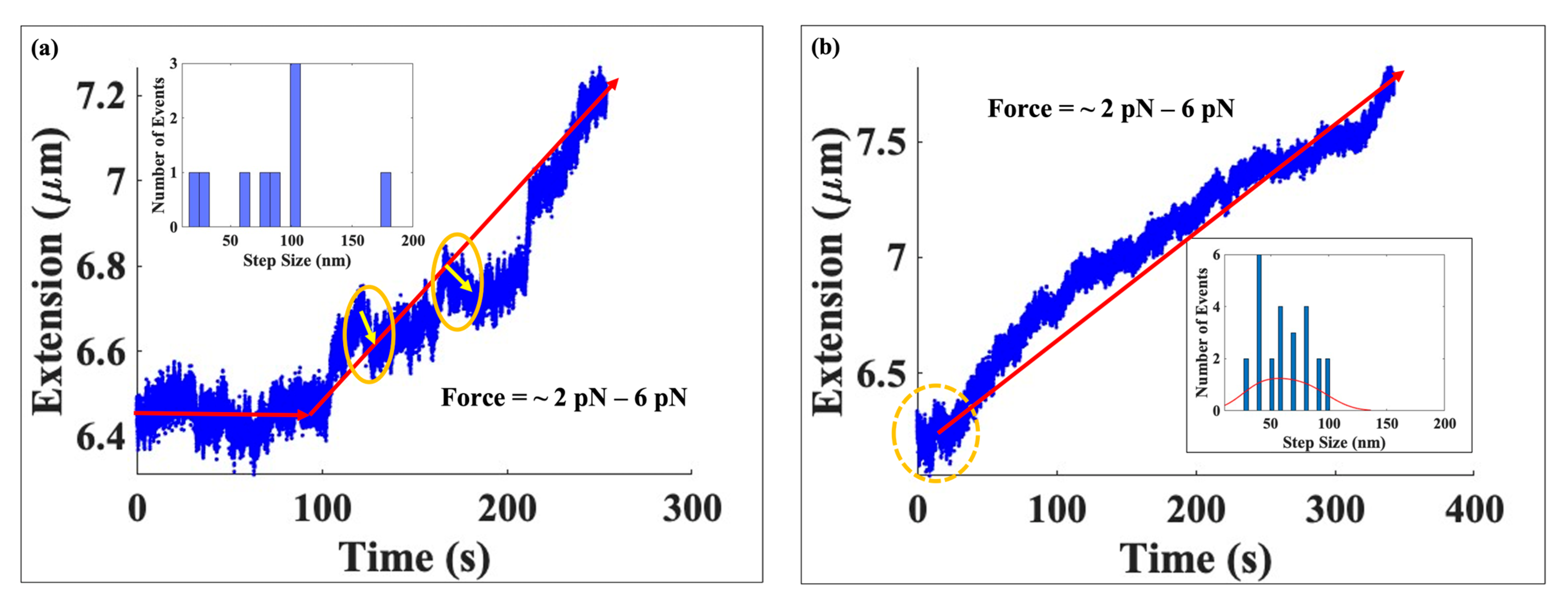
2.1. A Typical Experiment Using Hyperacetylated Histones
2.2. Comparison of the Mechanical Response of Native and Hyperacetylated Nucleosomes
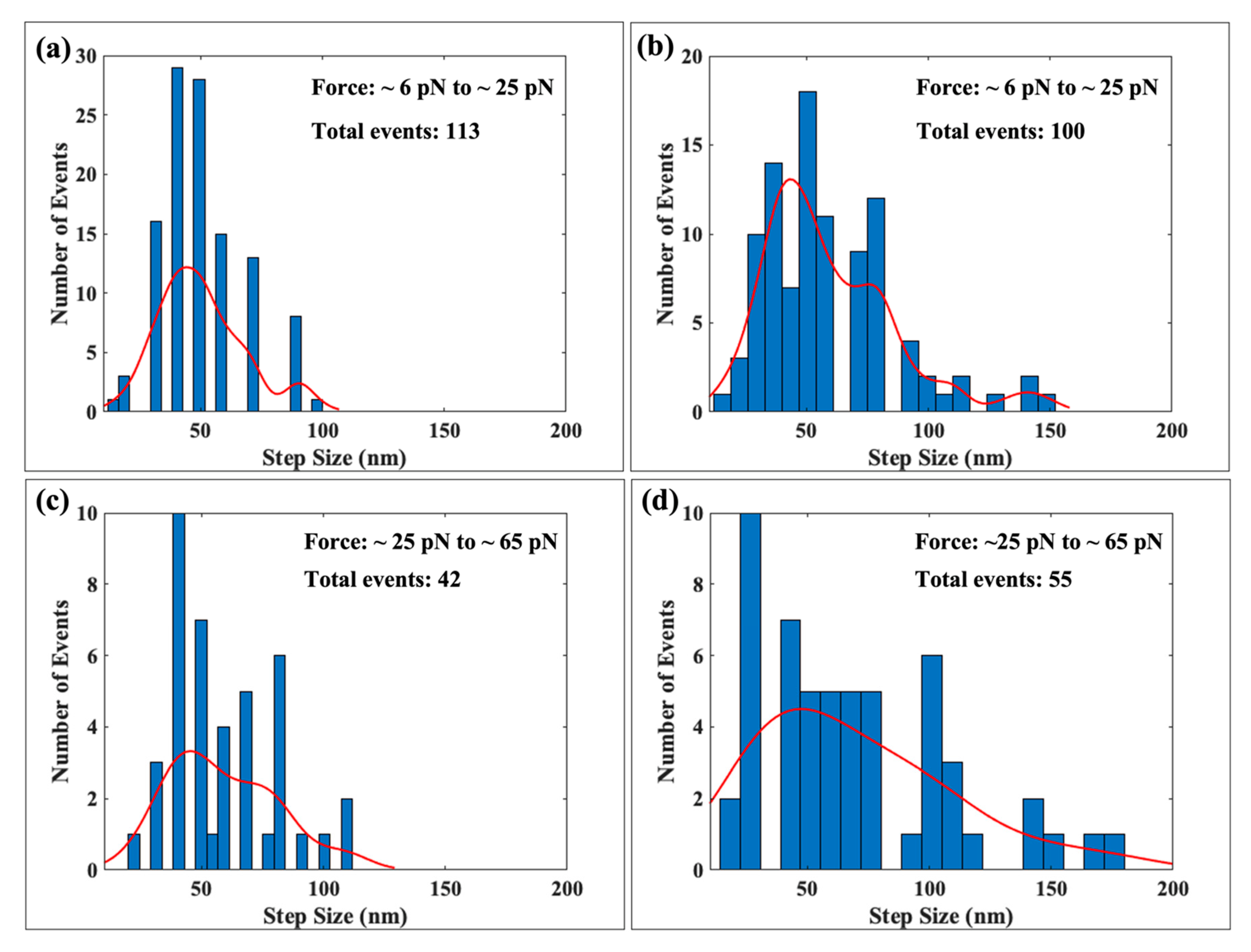
2.2.1. Comparison at Low Force Range
2.2.2. Comparison at Moderate and High Forces
3. Discussion
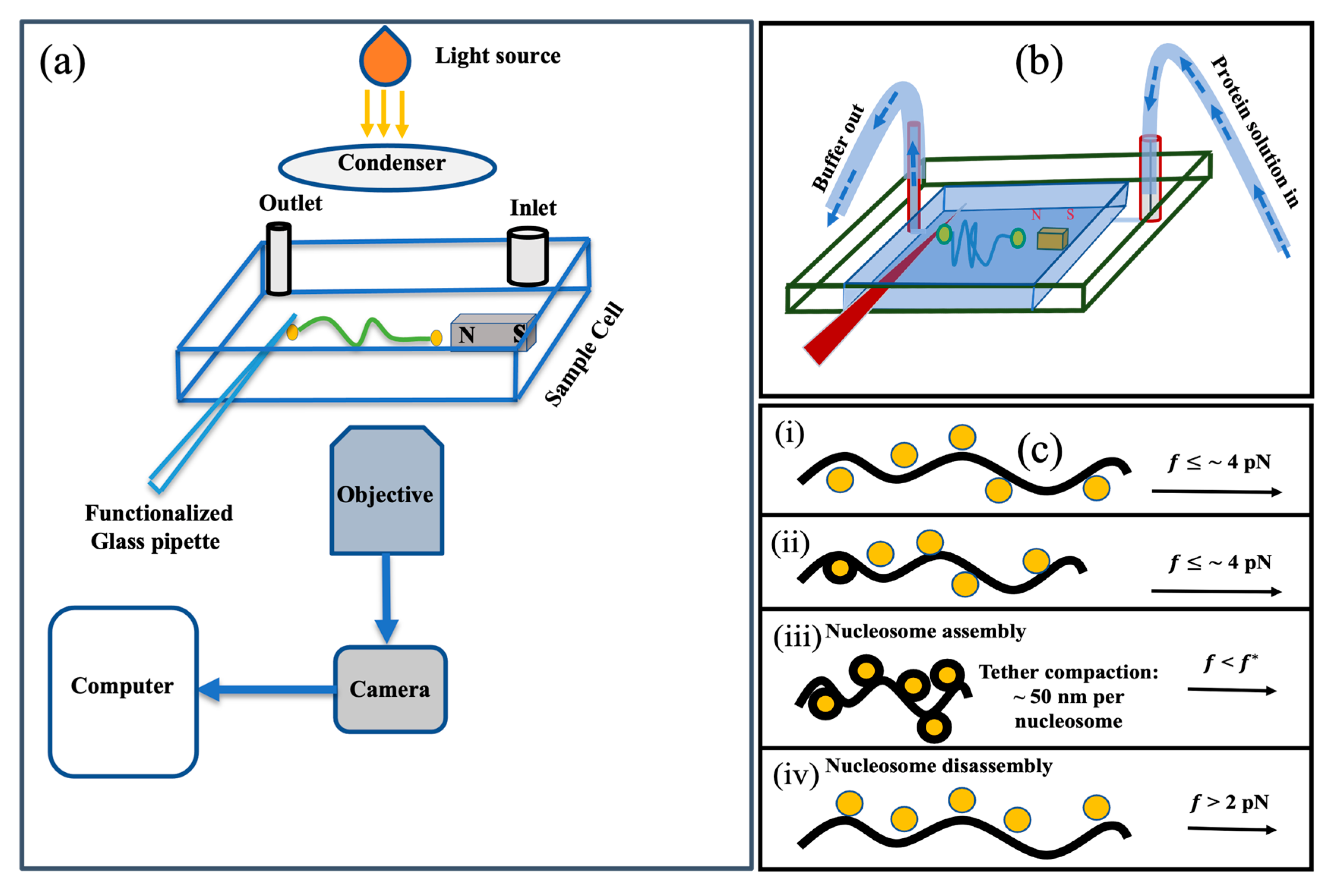
4. Materials and Methods
4.1. Micromanipulation Experiments with Horizontal (Transverse) Magnetic Tweezers
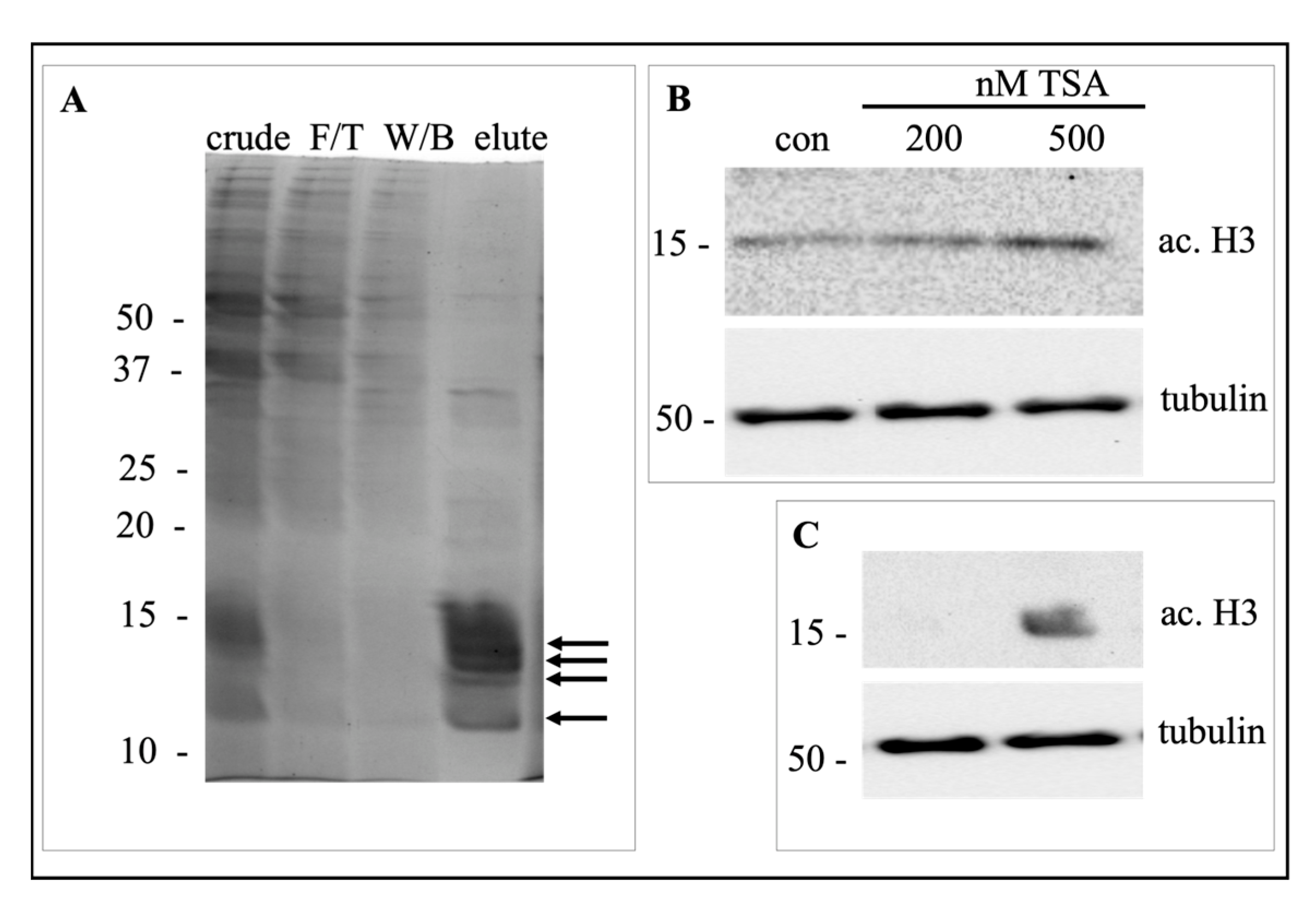
4.2. Histone Purification and Storage
4.3. Steps Detection and Data Analysis
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simon, M.; North, J.A.; Shimko, J.C.; Forties, R.A.; Ferdinand, M.B.; Manohar, M.; Zhang, M.; Fishel, R.; Ottesen, J.J.; Poirier, M.G. Histone fold modifications control nucleosome unwrapping and disassembly. Proc. Natl. Acad. Sci. USA 2011, 108, 12711–12716. [Google Scholar] [CrossRef]
- Kornberg, R. Chromatin Structure: A Repeating Unit of Histones and DNA Chromatin structure is based on a repeating unit of eight. Science 1974, 184, 868–871. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Ranjith, P.; Yan, J.; Marko, J.F. Nucleosome hopping and sliding kinetics determined from dynamics of single chromatin fibers in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA 2007, 104, 13649–13654. [Google Scholar] [CrossRef]
- Morrison, A.J.; Shen, X. Chromatin remodelling beyond transcription: The INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 2009, 10, 373–384. [Google Scholar] [CrossRef]
- Cosgrove, M.S.; Boeke, J.D.; Wolberger, C. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 2004, 11, 1037–1043. [Google Scholar] [CrossRef]
- Mersfelder, E.L.; Parthun, M.R. The tale beyond the tail: Histone core domain modifications and the regulation of chromatin structure. Nucleic. Acids Res. 2006, 34, 2653–2662. [Google Scholar] [CrossRef]
- Zhang, L.; Eugeni, E.E.; Parthun, M.R.; Freitas, M.A. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 2003, 112, 77–86. [Google Scholar] [CrossRef]
- Manohar, M.; Mooney, A.M.; North, J.A.; Nakkula, R.J.; Picking, J.W.; Edon, A.; Fishel, R.; Poirier, M.G.; Ottesen, J.J. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J. Biol. Chem. 2009, 284, 23312–23321. [Google Scholar] [CrossRef]
- English, C.M.; Adkins, M.W.; Carson, J.J.; Churchill, M.E.A.; Tyler, J.K. Structural Basis for the Histone Chaperone Activity of Asf1. Cell 2006, 127, 495–508. [Google Scholar] [CrossRef]
- Rothbart, S.B.; Strahl, B.D. Interpreting the language of histone and DNA modification. Biochim. Biophys. Acta 2014, 1839, 627–643. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Muthurajan, U.M.; Park, Y.-J.; Edayathumangalam, R.S.; Suto, R.K.; Chakravarthy, S.; Dyer, P.N.; Luger, K. Structure and dynamics of nucleosomal DNA. Biopolymers 2003, 68, 547–556. [Google Scholar] [CrossRef]
- Blossey, R.; Schiessel, H. The dynamics of the nucleosome: Thermal effects, external forces and ATP. FEBS J. 2011, 278, 3619–3632. [Google Scholar] [CrossRef]
- Hyland, E.M.; Cosgrove, M.S.; Molina, H.; Wang, D.; Pandey, A.; Cotter, R.J.; Boeke, J.D. Insights into the Role of Histone H3 and Histone H4 Core Modifiable Residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005, 25, 10060–10070. [Google Scholar] [CrossRef]
- Muthurajan, U.M.; Bao, Y.; Forsberg, L.J.; Edayathumangalam, R.S.; Dyer, P.N.; White, C.L.; Luger, K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004, 23, 260–271. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Makowski, M.M.; Gräwe, C.; Foster, B.M.; Nguyen N, V.; Bartke, T.; Vermeulen, M. Global profiling of protein-DNA and protein-nucleosome binding affinities using quantitative mass spectrometry. Nat. Commun. 2018, 9, 1653. [Google Scholar] [CrossRef]
- Tse, C.; Sera, T.; Wolffe, A.P.; Hansen, J.C. Disruption of Higher-Order Folding by Core Histone Acetylation Dramatically Enhances Transcription of Nucleosomal Arrays by RNA Polymerase III. Mol. Cell. Biol. 1998, 18, 4629–4638. [Google Scholar] [CrossRef]
- Bowman, G.D.; Poirier, M.G. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2014, 115, 2274–2295. [Google Scholar] [CrossRef]
- Rippe, K.; Mazurkiewicz, J.; Kepper, N. Interactions of Histones with DNA: Nucleosome Assembly, Stability, Dynamics, and Higher Order Structure. In DNA Interactions with Polymers and Surfactants; John and Wiley and Sons: Hoboken, NJ, USA, 2008; pp. 135–172. [Google Scholar]
- Hansen, J.C. Conformational dynamics of the chromatin fiber in solution: Determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 361–392. [Google Scholar] [CrossRef]
- Flaus, A.; Rencurel, C.; Ferreira, H.; Wiechens, N.; Owen-Hughes, T. Sin mutations alter inherent nucleosome mobility. EMBO J. 2004, 23, 343–353. [Google Scholar] [CrossRef]
- Kurumizaka, H.; Wolffe, A.P. Sin mutations of histone H3: Influence on nucleosome core structure and function. Mol. Cell. Biol. 1997, 17, 6953–6969. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, K.; Grunstein, M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 2005, 121, 375–385. [Google Scholar] [CrossRef]
- Chen, C.-C.; Carson, J.J.; Feser, J.; Tamburini, B.; Zabaronick, S.; Linger, J.; Tyler, J.K. Acetylated Lysine 56 on Histone H3 Drives Chromatin Assembly after Repair and Signals for the Completion of Repair. Cell 2008, 134, 231–243. [Google Scholar] [CrossRef]
- Krajewski, W.A. On the role of inter-nucleosomal interactions and intrinsic nucleosome dynamics in chromatin function. Biochem. Biophys. Reports 2016, 5, 492–501. [Google Scholar] [CrossRef][Green Version]
- Marcuello, C.; Frempong, G.A.; Balsera, M.; Medina, M.; Lostao, A. Atomic force microscopy to elicit conformational transitions of ferredoxin-dependent flavin thioredoxin reductases. Antioxidants 2021, 10, 1437. [Google Scholar] [CrossRef]
- Novo, N.; Romero-Tamayo, S.; Marcuello, C.; Boneta, S.; Blasco-Machin, I.; Velázquez-Campoy, A.; Villanueva, R.; Moreno-Loshuertos, R.; Lostao, A.; Medina, M.; et al. Beyond a platform protein for the degradosome assembly: The Apoptosis Inducing Factor as efficient nuclease involved in chromatinolysis. PNAS Nexus. 2022, 2, pgac312. [Google Scholar] [CrossRef]
- Simpson, R.T. Structure of chromatin containing extensively acetylated H3 and H4. Cell 1978, 13, 691–699. [Google Scholar] [CrossRef]
- Verweij, E.W.E.; Bosma, R.; Gao, M.; Bor, J.v.d.; Araaj, B.A.; Munnik, S.M.d.; Ma, X.; Leurs, R.; Vischer, H.F. BRET-Based Biosensors to Measure Agonist Efficacies in Histamine H1 Receptor-Mediated G Protein Activation, Signaling and Interactions with GRKs and β-Arrestins. Int. J. Mol. Sci. 2022, 23, 3184. [Google Scholar] [CrossRef]
- Pérez-Domínguez, S.; Caballero-Mancebo, S.; Marcuello, C.; Martínez-Júlvez, M.; Medina, M.; Lostao, A. Nanomechanical Study of Enzyme: Coenzyme Complexes: Bipartite Sites in Plastidic Ferredoxin-NADP+ Reductase for the Interaction with NADP+. Antioxidants 2022, 11, 537. [Google Scholar] [CrossRef]
- Jakobowska, I.; Becker, F.; Minguzzi, S.; Hansen, K.; Henke, B.; Epalle, N.H.; Beitz, E.; Hannus, S. Fluorescence cross-correlation spectroscopy yields true affinity and binding kinetics of plasmodium lactate transport inhibitors. Pharmaceuticals 2021, 14, 757. [Google Scholar] [CrossRef]
- Fabian, R.; Gaire, S.; Tyson, C.; Adhikari, R.; Pegg, I.; Sarkar, A. A horizontal magnetic tweezers for studying single dna molecules and dna-binding proteins. Molecules 2021, 26, 4781. [Google Scholar] [CrossRef]
- Brower-Toland, B.D.; Smith, C.L.; Yeh, R.C.; Lis, J.T.; Peterson, C.L.; Wang, M.D. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA 2002, 99, 1960–1965. [Google Scholar] [CrossRef]
- Tyson, C.; McAndrew, C.; Tuma, P.L.; Pegg, I.; Sarkar, A. Automated nonparametric method for detection of step-like features in biological data sets. Cytom. Part A 2015, 87, 393–404. [Google Scholar] [CrossRef]
- Smith, S.B.; Finzi, L.; Bustamante, C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science 1992, 258, 1122–1126. [Google Scholar] [CrossRef]
- Yan, J.; Skoko, D.; Marko, J.F. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E—Stat. Physics, Plasmas, Fluids, Relat. Interdiscip. Top. 2004, 70, 011905. [Google Scholar] [CrossRef]
- Fabian, R.; Tyson, C.; Tuma, P.L.; Pegg, I.; Sarkar, A. A horizontal magnetic tweezers and its use for studying single DNA molecules. Micromachines 2018, 9, 188. [Google Scholar] [CrossRef]
- McAndrew, C.P.; Tyson, C.; Zischkau, J.; Mehl, P.; Tuma, P.L.; Pegg, I.L.; Sarkar, A.; Gelsthorpe, A.R.; Gelsthorpe, K.; Sokol, R.J.; et al. Simple horizontal magnetic tweezers for micromanipulation of single DNA molecules and DNA–protein complexes. Biotechniques 2016, 60, 21–27. [Google Scholar] [CrossRef]
- Gaire, S.; Fabian, R.; Pegg, I.; Sarkar, A. Magnetic tweezers: Development and use in single-molecule research. Biotechniques 2022, 72, 65–72. [Google Scholar] [CrossRef]
- Joseph, R.A.; Shepard, B.D.; Kannarkat, G.T.; Rutledge, T.M.; Tuma, D.J.; Tuma, P.L. Microtubule acetylation and stability may explain alcohol-induced alterations in hepatic protein trafficking. Hepatology 2008, 47, 1745–1753. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaire, S.; Fabian, R.L., Jr.; Adhikari, R.; Tuma, P.L.; Pegg, I.L.; Sarkar, A. Micromechanical Study of Hyperacetylated Nucleosomes Using Single Molecule Transverse Magnetic Tweezers. Int. J. Mol. Sci. 2023, 24, 6188. https://doi.org/10.3390/ijms24076188
Gaire S, Fabian RL Jr., Adhikari R, Tuma PL, Pegg IL, Sarkar A. Micromechanical Study of Hyperacetylated Nucleosomes Using Single Molecule Transverse Magnetic Tweezers. International Journal of Molecular Sciences. 2023; 24(7):6188. https://doi.org/10.3390/ijms24076188
Chicago/Turabian StyleGaire, Santosh, Roberto L. Fabian, Jr., Raghabendra Adhikari, Pamela L. Tuma, Ian L. Pegg, and Abhijit Sarkar. 2023. "Micromechanical Study of Hyperacetylated Nucleosomes Using Single Molecule Transverse Magnetic Tweezers" International Journal of Molecular Sciences 24, no. 7: 6188. https://doi.org/10.3390/ijms24076188
APA StyleGaire, S., Fabian, R. L., Jr., Adhikari, R., Tuma, P. L., Pegg, I. L., & Sarkar, A. (2023). Micromechanical Study of Hyperacetylated Nucleosomes Using Single Molecule Transverse Magnetic Tweezers. International Journal of Molecular Sciences, 24(7), 6188. https://doi.org/10.3390/ijms24076188





