TOR Complex 1: Orchestrating Nutrient Signaling and Cell Cycle Progression
Abstract
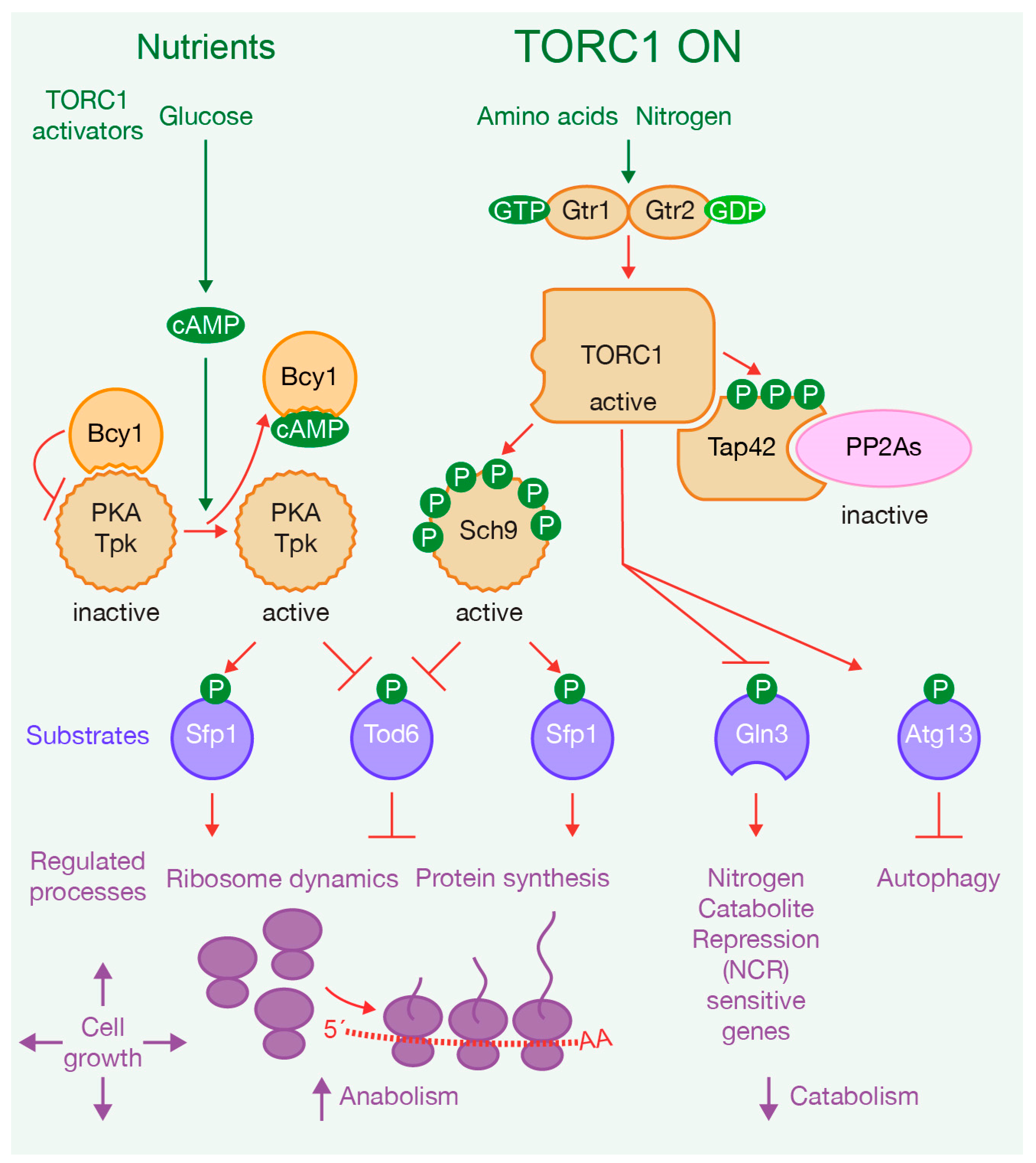
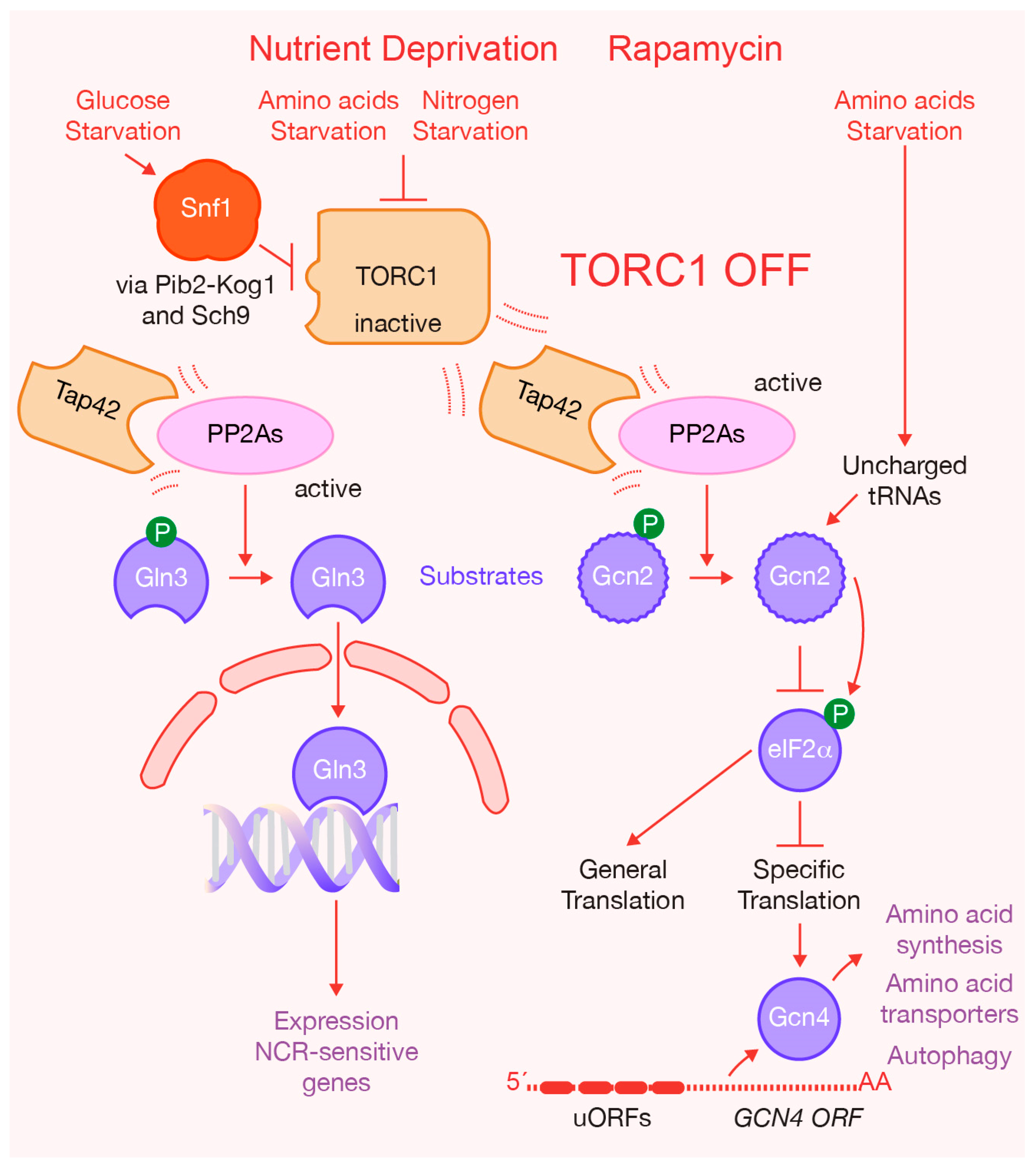
:1. Introduction
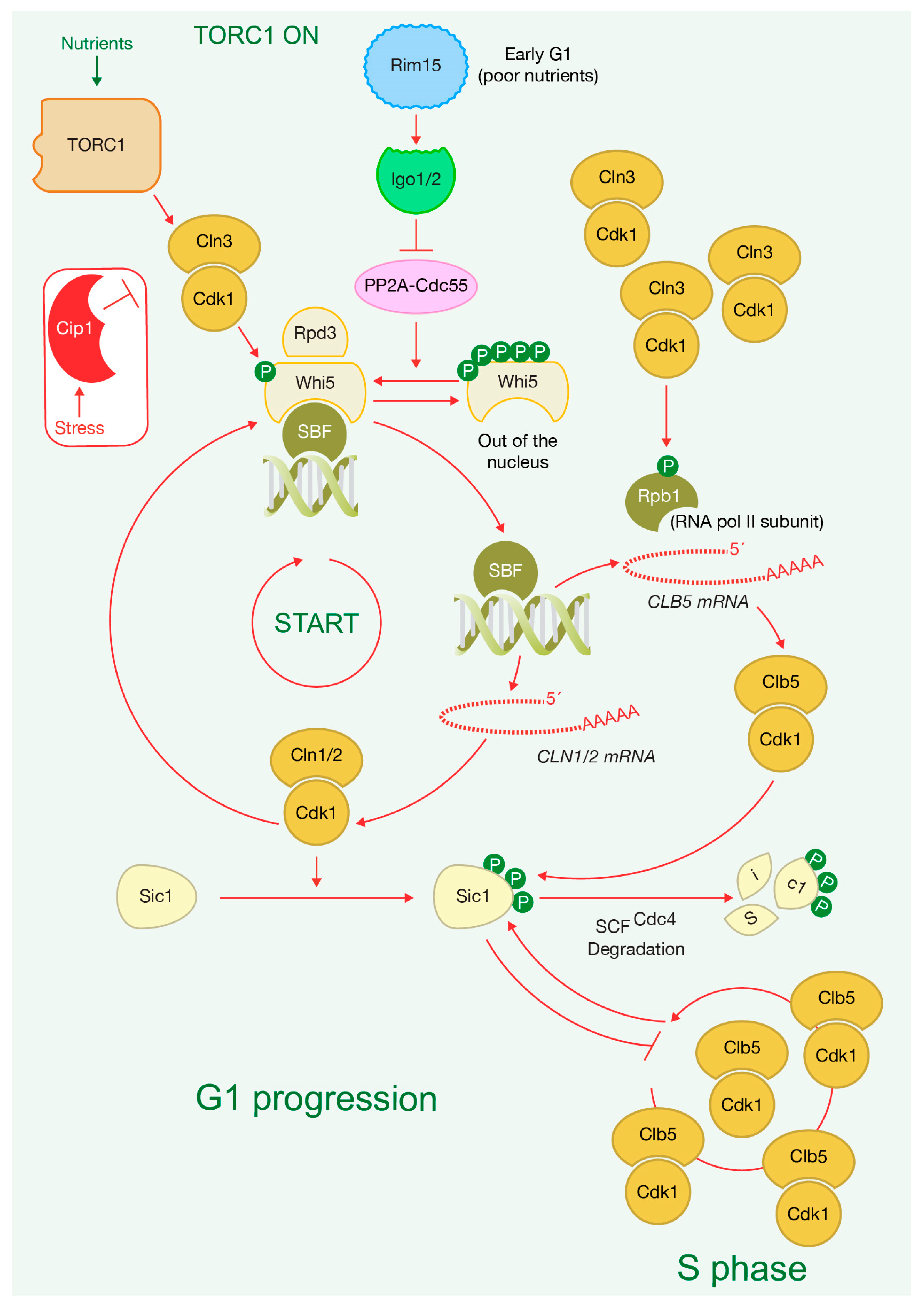
2. TORC1 Promotes G1 Progression by Inducing G1 Cyclin Activity
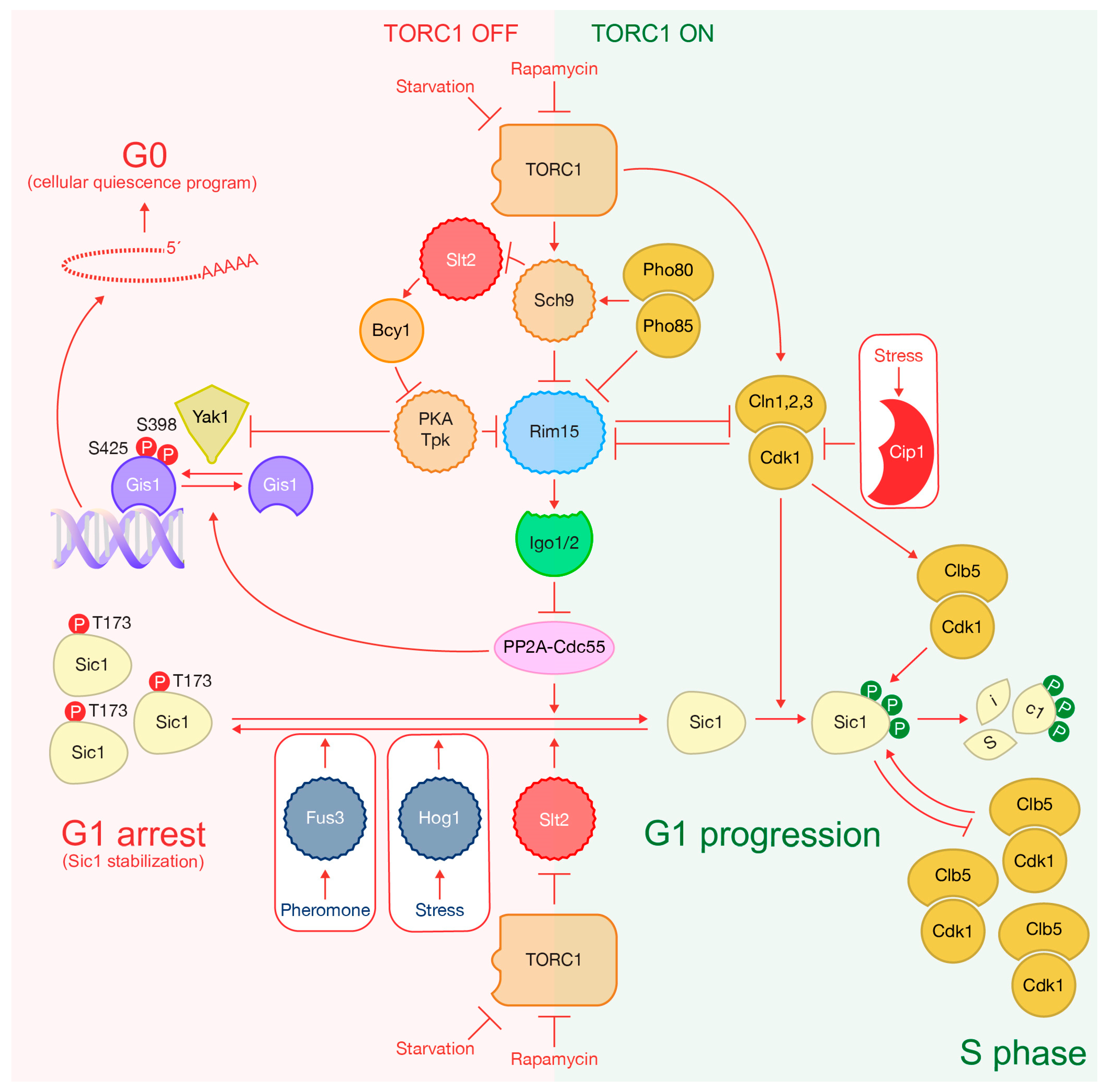
3. TORC1 Activity Ultimately Determines Sic1 Degradation to Drive Cells into S Phase
4. Inactivation of TORC1 Promotes Sic1 Stabilization to Arrest the Cell Cycle
5. TORC1 Regulates Mitosis Progression
6. TORC1 Blocks Separation between Mother and Daughter Cells
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Tapon, N.; Moberg, K.H.; Hariharan, I.K. The coupling of cell growth to the cell cycle. Curr. Opin. Cell Biol. 2001, 13, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Gonzalez, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Battaglioni, S.; Benjamin, D.; Walchli, M.; Maier, T.; Hall, M.N. mTOR substrate phosphorylation in growth control. Cell 2022, 185, 1814–1836. [Google Scholar] [CrossRef] [PubMed]
- Thorner, J. TOR complex 2 is a master regulator of plasma membrane homeostasis. Biochem. J. 2022, 479, 1917–1940. [Google Scholar] [CrossRef]
- Yip, C.K.; Murata, K.; Walz, T.; Sabatini, D.M.; Kang, S.A. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell 2010, 38, 768–774. [Google Scholar] [CrossRef]
- Aylett, C.H.; Sauer, E.; Imseng, S.; Boehringer, D.; Hall, M.N.; Ban, N.; Maier, T. Architecture of human mTOR complex 1. Science 2016, 351, 48–52. [Google Scholar] [CrossRef]
- Baretic, D.; Berndt, A.; Ohashi, Y.; Johnson, C.M.; Williams, R.L. Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat. Commun. 2016, 7, 11016. [Google Scholar] [CrossRef]
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; Latek, R.R.; Guntur, K.V.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 2003, 11, 895–904. [Google Scholar] [CrossRef]
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef]
- Hara, K.; Maruki, Y.; Long, X.; Yoshino, K.; Oshiro, N.; Hidayat, S.; Tokunaga, C.; Avruch, J.; Yonezawa, K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002, 110, 177–189. [Google Scholar] [CrossRef]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Adami, A.; Garcia-Alvarez, B.; Arias-Palomo, E.; Barford, D.; Llorca, O. Structure of TOR and its complex with KOG1. Mol. Cell 2007, 27, 509–516. [Google Scholar] [CrossRef]
- Reinke, A.; Anderson, S.; McCaffery, J.M.; Yates, J., 3rd; Aronova, S.; Chu, S.; Fairclough, S.; Iverson, C.; Wedaman, K.P.; Powers, T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 14752–14762. [Google Scholar] [CrossRef] [PubMed]
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar] [CrossRef]
- Binda, M.; Peli-Gulli, M.P.; Bonfils, G.; Panchaud, N.; Urban, J.; Sturgill, T.W.; Loewith, R.; De Virgilio, C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 2009, 35, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tsang, C.K.; Watkins, M.; Bertram, P.G.; Zheng, X.F. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 2006, 442, 1058–1061. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef]
- Gaubitz, C.; Prouteau, M.; Kusmider, B.; Loewith, R. TORC2 Structure and Function. Trends Biochem. Sci. 2016, 41, 532–545. [Google Scholar] [CrossRef]
- Roelants, F.M.; Leskoske, K.L.; Martinez Marshall, M.N.; Locke, M.N.; Thorner, J. The TORC2-Dependent Signaling Network in the Yeast Saccharomyces cerevisiae. Biomolecules 2017, 7, 66. [Google Scholar] [CrossRef]
- Martin, D.E.; Soulard, A.; Hall, M.N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 2004, 119, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Lippman, S.I.; Broach, J.R. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl. Acad. Sci. USA 2009, 106, 19928–19933. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R. TORC1 Signaling in Budding Yeast. Enzymes 2010, 27, 147–175. [Google Scholar] [CrossRef]
- Yerlikaya, S.; Meusburger, M.; Kumari, R.; Huber, A.; Anrather, D.; Costanzo, M.; Boone, C.; Ammerer, G.; Baranov, P.V.; Loewith, R. TORC1 and TORC2 work together to regulate ribosomal protein S6 phosphorylation in Saccharomyces cerevisiae. Mol. Biol. Cell 2016, 27, 397–409. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Shetty, S.; Hofstetter, J.; Battaglioni, S.; Ritz, D.; Hall, M.N. TORC1 phosphorylates and inhibits the ribosome preservation factor Stm1 to activate dormant ribosomes. EMBO J. 2023, 42, e112344. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, X.; Li, G.; Wang, X. TORC1 Signaling in Fungi: From Yeasts to Filamentous Fungi. Microorganisms 2023, 11, 218. [Google Scholar] [CrossRef]
- Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. eLife 2015, 4, e09181. [Google Scholar] [CrossRef]
- Sullivan, A.; Wallace, R.L.; Wellington, R.; Luo, X.; Capaldi, A.P. Multilayered regulation of TORC1-body formation in budding yeast. Mol. Biol. Cell 2019, 30, 400–410. [Google Scholar] [CrossRef]
- Caligaris, M.; Nicastro, R.; Hu, Z.; Tripodi, F.; Hummel, J.E.; Pillet, B.; Deprez, M.A.; Winderickx, J.; Rospert, S.; Coccetti, P.; et al. Snf1/AMPK fine-tunes TORC1 signaling in response to glucose starvation. eLife 2023, 12, e84319. [Google Scholar] [CrossRef]
- Zurita-Martinez, S.A.; Cardenas, M.E. Tor and cyclic AMP-protein kinase A: Two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 2005, 4, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.; Lippman, S.I.; Schneper, L.; Slonim, N.; Broach, J.R. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 2009, 5, 245. [Google Scholar] [CrossRef]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef]
- Kunkel, J.; Luo, X.; Capaldi, A.P. Integrated TORC1 and PKA signaling control the temporal activation of glucose-induced gene expression in yeast. Nat. Commun. 2019, 10, 3558. [Google Scholar] [CrossRef]
- Stracka, D.; Jozefczuk, S.; Rudroff, F.; Sauer, U.; Hall, M.N. Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 2014, 289, 25010–25020. [Google Scholar] [CrossRef]
- De Virgilio, C. The essence of yeast quiescence. FEMS Microbiol. Rev. 2012, 36, 306–339. [Google Scholar] [CrossRef]
- Jorgensen, P.; Rupes, I.; Sharom, J.R.; Schneper, L.; Broach, J.R.; Tyers, M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004, 18, 2491–2505. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Bodenmiller, B.; Uotila, A.; Stahl, M.; Wanka, S.; Gerrits, B.; Aebersold, R.; Loewith, R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009, 23, 1929–1943. [Google Scholar] [CrossRef]
- Lee, J.; Moir, R.D.; Willis, I.M. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J. Biol. Chem. 2009, 284, 12604–12608. [Google Scholar] [CrossRef]
- Huber, A.; French, S.L.; Tekotte, H.; Yerlikaya, S.; Stahl, M.; Perepelkina, M.P.; Tyers, M.; Rougemont, J.; Beyer, A.L.; Loewith, R. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011, 30, 3052–3064. [Google Scholar] [CrossRef]
- Di Como, C.J.; Arndt, K.T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996, 10, 1904–1916. [Google Scholar] [CrossRef]
- Jiang, Y.; Broach, J.R. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999, 18, 2782–2792. [Google Scholar] [CrossRef]
- Yan, G.; Shen, X.; Jiang, Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 2006, 25, 3546–3555. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Jiang, Y. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol. Biol. Cell 2003, 14, 4342–4351. [Google Scholar] [CrossRef]
- Bertram, P.G.; Choi, J.H.; Carvalho, J.; Ai, W.; Zeng, C.; Chan, T.F.; Zheng, X.F. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 2000, 275, 35727–35733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Du, G.; Zhou, J.; Chen, J. Regulation of Sensing, Transportation, and Catabolism of Nitrogen Sources in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2018, 82, e00040-17. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.J.; Tolley, E.A.; Cooper, T.G. Sit4 and PP2A Dephosphorylate Nitrogen Catabolite Repression-Sensitive Gln3 When TorC1 Is Up- as Well as Downregulated. Genetics 2019, 212, 1205–1225. [Google Scholar] [CrossRef]
- Tate, J.J.; Rai, R.; De Virgilio, C.; Cooper, T.G. N- and C-terminal Gln3-Tor1 interaction sites: One acting negatively and the other positively to regulate nuclear Gln3 localization. Genetics 2021, 217, iyab017. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005, 59, 407–450. [Google Scholar] [CrossRef]
- Cherkasova, V.A.; Hinnebusch, A.G. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003, 17, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Lempiainen, H.; Uotila, A.; Urban, J.; Dohnal, I.; Ammerer, G.; Loewith, R.; Shore, D. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell 2009, 33, 704–716. [Google Scholar] [CrossRef]
- Kamada, Y.; Yoshino, K.; Kondo, C.; Kawamata, T.; Oshiro, N.; Yonezawa, K.; Ohsumi, Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell Biol. 2010, 30, 1049–1058. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Guerra, P.; Vuillemenot, L.P.E.; van Oppen, Y.B.; Been, M.; Milias-Argeitis, A. TORC1 and PKA activity towards ribosome biogenesis oscillates in synchrony with the budding yeast cell cycle. J. Cell Sci. 2022, 135, jcs260378. [Google Scholar] [CrossRef]
- Barbet, N.C.; Schneider, U.; Helliwell, S.B.; Stansfield, I.; Tuite, M.F.; Hall, M.N. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 1996, 7, 25–42. [Google Scholar] [CrossRef]
- Pedruzzi, I.; Dubouloz, F.; Cameroni, E.; Wanke, V.; Roosen, J.; Winderickx, J.; De Virgilio, C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 2003, 12, 1607–1613. [Google Scholar] [CrossRef] [PubMed]
- Takhaveev, V.; Ozsezen, S.; Smith, E.N.; Zylstra, A.; Chaillet, M.L.; Chen, H.; Papagiannakis, A.; Milias-Argeitis, A.; Heinemann, M. Temporal segregation of biosynthetic processes is responsible for metabolic oscillations during the budding yeast cell cycle. Nat. Metab. 2023, 5, 294–313. [Google Scholar] [CrossRef] [PubMed]
- Blank, H.M.; Reuse, C.; Schmidt-Hohagen, K.; Hammer, S.E.; Hiller, K.; Polymenis, M. Branched-chain amino acid synthesis is coupled to TOR activation early in the cell cycle in yeast. EMBO Rep. 2023, 24, e57372. [Google Scholar] [CrossRef] [PubMed]
- Schmelzle, T.; Hall, M.N. TOR, a central controller of cell growth. Cell 2000, 103, 253–262. [Google Scholar] [CrossRef]
- Jorgensen, P.; Tyers, M. How cells coordinate growth and division. Curr. Biol. 2004, 14, R1014–R1027. [Google Scholar] [CrossRef]
- Cuyas, E.; Corominas-Faja, B.; Joven, J.; Menendez, J.A. Cell cycle regulation by the nutrient-sensing mammalian target of rapamycin (mTOR) pathway. Methods Mol. Biol. 2014, 1170, 113–144. [Google Scholar] [CrossRef]
- Perez-Hidalgo, L.; Moreno, S. Coupling TOR to the Cell Cycle by the Greatwall-Endosulfine-PP2A-B55 Pathway. Biomolecules 2017, 7, 59. [Google Scholar] [CrossRef]
- Gonzalez, S.; Rallis, C. The TOR Signaling Pathway in Spatial and Temporal Control of Cell Size and Growth. Front. Cell Dev. Biol. 2017, 5, 61. [Google Scholar] [CrossRef]
- Ewald, J.C. How yeast coordinates metabolism, growth and division. Curr. Opin. Microbiol. 2018, 45, 1–7. [Google Scholar] [CrossRef]
- Lamm, N.; Rogers, S.; Cesare, A.J. The mTOR pathway: Implications for DNA replication. Prog. Biophys. Mol. Biol. 2019, 147, 17–25. [Google Scholar] [CrossRef]
- Morgan, D.O. Principles of CDK regulation. Nature 1995, 374, 131–134. [Google Scholar] [CrossRef]
- Morgan, D.O. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 1997, 13, 261–291. [Google Scholar] [CrossRef]
- Nurse, P. A long twentieth century of the cell cycle and beyond. Cell 2000, 100, 71–78. [Google Scholar] [CrossRef]
- Swaffer, M.P.; Jones, A.W.; Flynn, H.R.; Snijders, A.P.; Nurse, P. CDK Substrate Phosphorylation and Ordering the Cell Cycle. Cell 2016, 167, 1750–1761.e1716. [Google Scholar] [CrossRef]
- Loog, M.; Morgan, D.O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 2005, 434, 104–108. [Google Scholar] [CrossRef]
- Ord, M.; Loog, M. How the cell cycle clock ticks. Mol. Biol. Cell 2019, 30, 169–172. [Google Scholar] [CrossRef]
- Stern, B.; Nurse, P. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 1996, 12, 345–350. [Google Scholar] [CrossRef]
- Uhlmann, F.; Bouchoux, C.; Lopez-Aviles, S. A quantitative model for cyclin-dependent kinase control of the cell cycle: Revisited. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3572–3583. [Google Scholar] [CrossRef]
- Pirincci Ercan, D.; Chretien, F.; Chakravarty, P.; Flynn, H.R.; Snijders, A.P.; Uhlmann, F. Budding yeast relies on G(1) cyclin specificity to couple cell cycle progression with morphogenetic development. Sci. Adv. 2021, 7, eabg0007. [Google Scholar] [CrossRef]
- Basu, S.; Greenwood, J.; Jones, A.W.; Nurse, P. Core control principles of the eukaryotic cell cycle. Nature 2022, 607, 381–386. [Google Scholar] [CrossRef]
- Ghiara, J.B.; Richardson, H.E.; Sugimoto, K.; Henze, M.; Lew, D.J.; Wittenberg, C.; Reed, S.I. A cyclin B homolog in S. cerevisiae: Chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell 1991, 65, 163–174. [Google Scholar] [CrossRef]
- Lew, D.J.; Reed, S.I. Morphogenesis in the yeast cell cycle: Regulation by Cdc28 and cyclins. J. Cell Biol. 1993, 120, 1305–1320. [Google Scholar] [CrossRef]
- Stegmeier, F.; Amon, A. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004, 38, 203–232. [Google Scholar] [CrossRef]
- Weiss, E.L. Mitotic Exit and Separation of Mother and Daughter Cells. Genetics 2012, 192, 1165–1202. [Google Scholar] [CrossRef]
- Sanchez-Diaz, A.; Nkosi, P.J.; Murray, S.; Labib, K. The Mitotic Exit Network and Cdc14 phosphatase initiate cytokinesis by counteracting CDK phosphorylations and blocking polarised growth. EMBO J. 2012, 31, 3620–3634. [Google Scholar] [CrossRef]
- van Werven, F.J.; Amon, A. Regulation of entry into gametogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3521–3531. [Google Scholar] [CrossRef]
- Bell, S.P.; Labib, K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics 2016, 203, 1027–1067. [Google Scholar] [CrossRef] [PubMed]
- Solaki, M.; Ewald, J.C. Fueling the Cycle: CDKs in Carbon and Energy Metabolism. Front. Cell Dev. Biol. 2018, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Bahler, J. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 2005, 39, 69–94. [Google Scholar] [CrossRef]
- McInerny, C.J. Cell cycle regulated gene expression in yeasts. Adv. Genet. 2011, 73, 51–85. [Google Scholar] [CrossRef]
- Haase, S.B.; Wittenberg, C. Topology and control of the cell-cycle-regulated transcriptional circuitry. Genetics 2014, 196, 65–90. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, C.; Skotheim, J.M.; de Bruin, R.A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef]
- Yahya, G.; Parisi, E.; Flores, A.; Gallego, C.; Aldea, M. A Whi7-anchored loop controls the G1 Cdk-cyclin complex at start. Mol. Cell 2014, 53, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Zapata, J.; Dephoure, N.; Macdonough, T.; Yu, Y.; Parnell, E.J.; Mooring, M.; Gygi, S.P.; Stillman, D.J.; Kellogg, D.R. PP2ARts1 is a master regulator of pathways that control cell size. J. Cell Biol. 2014, 204, 359–376. [Google Scholar] [CrossRef]
- Sommer, R.A.; DeWitt, J.T.; Tan, R.; Kellogg, D.R. Growth-dependent signals drive an increase in early G1 cyclin concentration to link cell cycle entry with cell growth. eLife 2021, 10, e64364. [Google Scholar] [CrossRef]
- Parviz, F.; Heideman, W. Growth-independent regulation of CLN3 mRNA levels by nutrients in Saccharomyces cerevisiae. J. Bacteriol. 1998, 180, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Zinzalla, V.; Graziola, M.; Mastriani, A.; Vanoni, M.; Alberghina, L. Rapamycin-mediated G1 arrest involves regulation of the Cdk inhibitor Sic1 in Saccharomyces cerevisiae. Mol. Microbiol. 2007, 63, 1482–1494. [Google Scholar] [CrossRef]
- Polymenis, M.; Schmidt, E.V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997, 11, 2522–2531. [Google Scholar] [CrossRef] [PubMed]
- Anthony, C.; Zong, Q.; De Benedetti, A. Overexpression of eIF4E in Saccharomyces cerevisiae causes slow growth and decreased alpha-factor response through alterations in CLN3 expression. J. Biol. Chem. 2001, 276, 39645–39652. [Google Scholar] [CrossRef]
- Dever, T.E. Gene-specific regulation by general translation factors. Cell 2002, 108, 545–556. [Google Scholar] [CrossRef]
- Brenner, C.; Nakayama, N.; Goebl, M.; Tanaka, K.; Toh-e, A.; Matsumoto, K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol. Cell. Biol. 1988, 8, 3556–3559. [Google Scholar] [CrossRef]
- Danaie, P.; Altmann, M.; Hall, M.N.; Trachsel, H.; Helliwell, S.B. CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E. Biochem. J. 1999, 340 Pt 1, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Nash, R.; Tokiwa, G.; Anand, S.; Erickson, K.; Futcher, A.B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988, 7, 4335–4346. [Google Scholar] [CrossRef]
- Costanzo, M.; Nishikawa, J.L.; Tang, X.; Millman, J.S.; Schub, O.; Breitkreuz, K.; Dewar, D.; Rupes, I.; Andrews, B.; Tyers, M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 2004, 117, 899–913. [Google Scholar] [CrossRef]
- de Bruin, R.A.; McDonald, W.H.; Kalashnikova, T.I.; Yates, J., 3rd; Wittenberg, C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 2004, 117, 887–898. [Google Scholar] [CrossRef]
- Wagner, M.V.; Smolka, M.B.; de Bruin, R.A.; Zhou, H.; Wittenberg, C.; Dowdy, S.F. Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS ONE 2009, 4, e4300. [Google Scholar] [CrossRef]
- Doncic, A.; Falleur-Fettig, M.; Skotheim, J.M. Distinct interactions select and maintain a specific cell fate. Mol. Cell 2011, 43, 528–539. [Google Scholar] [CrossRef]
- Chang, Y.L.; Tseng, S.F.; Huang, Y.C.; Shen, Z.J.; Hsu, P.H.; Hsieh, M.H.; Yang, C.W.; Tognetti, S.; Canal, B.; Subirana, L.; et al. Yeast Cip1 is activated by environmental stress to inhibit Cdk1-G1 cyclins via Mcm1 and Msn2/4. Nat. Commun. 2017, 8, 56. [Google Scholar] [CrossRef]
- Tyers, M.; Tokiwa, G.; Futcher, B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993, 12, 1955–1968. [Google Scholar] [CrossRef] [PubMed]
- Eser, U.; Falleur-Fettig, M.; Johnson, A.; Skotheim, J.M. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol. Cell 2011, 43, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Skotheim, J.M.; Di Talia, S.; Siggia, E.D.; Cross, F.R. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 2008, 454, 291–296. [Google Scholar] [CrossRef]
- Koivomagi, M.; Swaffer, M.P.; Turner, J.J.; Marinov, G.; Skotheim, J.M. G(1) cyclin-Cdk promotes cell cycle entry through localized phosphorylation of RNA polymerase II. Science 2021, 374, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Carey, L.B.; Cai, Y.; Wijnen, H.; Futcher, B. Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoS Biol. 2009, 7, e1000189. [Google Scholar] [CrossRef] [PubMed]
- Schmoller, K.M.; Turner, J.J.; Koivomagi, M.; Skotheim, J.M. Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size. Nature 2015, 526, 268–272. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, G.; Zahumensky, J.; Honey, S.; Futcher, B. Differential Scaling of Gene Expression with Cell Size May Explain Size Control in Budding Yeast. Mol. Cell 2020, 78, 359–370.e356. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, X.; Yang, X.; Liu, S.; Jiang, L.; Qu, Y.; Hu, L.; Ouyang, Q.; Tang, C. Reliable cell cycle commitment in budding yeast is ensured by signal integration. eLife 2015, 4, e03977. [Google Scholar] [CrossRef] [PubMed]
- Litsios, A.; Huberts, D.; Terpstra, H.M.; Guerra, P.; Schmidt, A.; Buczak, K.; Papagiannakis, A.; Rovetta, M.; Hekelaar, J.; Hubmann, G.; et al. Differential scaling between G1 protein production and cell size dynamics promotes commitment to the cell division cycle in budding yeast. Nat. Cell Biol. 2019, 21, 1382–1392. [Google Scholar] [CrossRef]
- Litsios, A.; Goswami, P.; Terpstra, H.M.; Coffin, C.; Vuillemenot, L.A.; Rovetta, M.; Ghazal, G.; Guerra, P.; Buczak, K.; Schmidt, A.; et al. The timing of Start is determined primarily by increased synthesis of the Cln3 activator rather than dilution of the Whi5 inhibitor. Mol. Biol. Cell 2022, 33, rp2. [Google Scholar] [CrossRef]
- Schmoller, K.M.; Lanz, M.C.; Kim, J.; Koivomagi, M.; Qu, Y.; Tang, C.; Kukhtevich, I.V.; Schneider, R.; Rudolf, F.; Moreno, D.F.; et al. Whi5 is diluted and protein synthesis does not dramatically increase in pre-Start G1. Mol. Biol. Cell 2022, 33, lt1. [Google Scholar] [CrossRef]
- Talarek, N.; Gueydon, E.; Schwob, E. Homeostatic control of START through negative feedback between Cln3-Cdk1 and Rim15/Greatwall kinase in budding yeast. eLife 2017, 6, e26233. [Google Scholar] [CrossRef] [PubMed]
- Mendenhall, M.D. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science 1993, 259, 216–219. [Google Scholar] [CrossRef]
- Schwob, E.; Bohm, T.; Mendenhall, M.D.; Nasmyth, K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 1994, 79, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Donovan, J.D.; Toyn, J.H.; Johnson, A.L.; Johnston, L.H. p40(sdb25), a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 1994, 8, 1640–1653. [Google Scholar] [CrossRef]
- Jaspersen, S.L.; Charles, J.F.; Tinker-Kulberg, R.L.; Morgan, D.O. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 1998, 9, 2803–2817. [Google Scholar] [CrossRef]
- Lengronne, A.; Schwob, E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 2002, 9, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Tyers, M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc. Natl. Acad. Sci. USA 1996, 93, 7772–7776. [Google Scholar] [CrossRef] [PubMed]
- Koivomagi, M.; Valk, E.; Venta, R.; Iofik, A.; Lepiku, M.; Balog, E.R.; Rubin, S.M.; Morgan, D.O.; Loog, M. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature 2011, 480, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Schneider, B.L.; Yang, Q.H.; Futcher, A.B. Linkage of replication to start by the Cdk inhibitor Sic1. Science 1996, 272, 560–562. [Google Scholar] [CrossRef] [PubMed]
- Koivomagi, M.; Valk, E.; Venta, R.; Iofik, A.; Lepiku, M.; Morgan, D.O.; Loog, M. Dynamics of Cdk1 substrate specificity during the cell cycle. Mol. Cell 2011, 42, 610–623. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Bhaduri, S.; Ord, M.; Davey, N.E.; Loog, M.; Pryciak, P.M. Comprehensive Analysis of G1 Cyclin Docking Motif Sequences that Control CDK Regulatory Potency In Vivo. Curr. Biol. 2020, 30, 4454–4466.e4455. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Annan, R.S.; Huddleston, M.J.; Carr, S.A.; Reynard, G.; Deshaies, R.J. Phosphorylation of Sic1p by G1 Cdk Required for Its Degradation and Entry into S Phase. Science 1997, 278, 455–460. [Google Scholar] [CrossRef]
- Nash, P.; Tang, X.; Orlicky, S.; Chen, Q.; Gertler, F.B.; Mendenhall, M.D.; Sicheri, F.; Pawson, T.; Tyers, M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 2001, 414, 514–521. [Google Scholar] [CrossRef]
- Feldman, R.M.; Correll, C.C.; Kaplan, K.B.; Deshaies, R.J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 1997, 91, 221–230. [Google Scholar] [CrossRef]
- Bai, C.; Sen, P.; Hofmann, K.; Ma, L.; Goebl, M.; Harper, J.W.; Elledge, S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996, 86, 263–274. [Google Scholar] [CrossRef]
- Skowyra, D.; Craig, K.L.; Tyers, M.; Elledge, S.J.; Harper, J.W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 1997, 91, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Escote, X.; Zapater, M.; Clotet, J.; Posas, F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 2004, 6, 997–1002. [Google Scholar] [CrossRef]
- Moreno-Torres, M.; Jaquenoud, M.; De Virgilio, C. TORC1 controls G1-S cell cycle transition in yeast via Mpk1 and the greatwall kinase pathway. Nat. Commun. 2015, 6, 8256. [Google Scholar] [CrossRef]
- Moreno-Torres, M.; Jaquenoud, M.; Peli-Gulli, M.P.; Nicastro, R.; De Virgilio, C. TORC1 coordinates the conversion of Sic1 from a target to an inhibitor of cyclin-CDK-Cks1. Cell Discov. 2017, 3, 17012. [Google Scholar] [CrossRef]
- Venta, R.; Valk, E.; Ord, M.; Kosik, O.; Paabo, K.; Maljavin, A.; Kivi, R.; Faustova, I.; Shtaida, N.; Lepiku, M.; et al. A processive phosphorylation circuit with multiple kinase inputs and mutually diversional routes controls G1/S decision. Nat. Commun. 2020, 11, 1836. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Gutierrez, E.; Alegria-Carrasco, E.; Sellers-Moya, A.; Molina, M.; Martin, H. Not just the wall: The other ways to turn the yeast CWI pathway on. Int. Microbiol. 2020, 23, 107–119. [Google Scholar] [CrossRef]
- Torres, J.; Di Como, C.J.; Herrero, E.; De La Torre-Ruiz, M.A. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 2002, 277, 43495–43504. [Google Scholar] [CrossRef]
- Sun, S.; Gresham, D. Cellular quiescence in budding yeast. Yeast 2021, 38, 12–29. [Google Scholar] [CrossRef]
- Bontron, S.; Jaquenoud, M.; Vaga, S.; Talarek, N.; Bodenmiller, B.; Aebersold, R.; De Virgilio, C. Yeast endosulfines control entry into quiescence and chronological life span by inhibiting protein phosphatase 2A. Cell Rep. 2013, 3, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Dokladal, L.; Stumpe, M.; Hu, Z.; Jaquenoud, M.; Dengjel, J.; De Virgilio, C. Phosphoproteomic responses of TORC1 target kinases reveal discrete and convergent mechanisms that orchestrate the quiescence program in yeast. Cell Rep. 2021, 37, 110149. [Google Scholar] [CrossRef]
- Wanke, V.; Cameroni, E.; Uotila, A.; Piccolis, M.; Urban, J.; Loewith, R.; De Virgilio, C. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 2008, 69, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Reinders, A.; Burckert, N.; Boller, T.; Wiemken, A.; De Virgilio, C. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 1998, 12, 2943–2955. [Google Scholar] [CrossRef] [PubMed]
- Soulard, A.; Cremonesi, A.; Moes, S.; Schutz, F.; Jeno, P.; Hall, M.N. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 2010, 21, 3475–3486. [Google Scholar] [CrossRef] [PubMed]
- Deprez, M.A.; Caligaris, M.; Rosseels, J.; Hatakeyama, R.; Ghillebert, R.; Sampaio-Marques, B.; Mudholkar, K.; Eskes, E.; Meert, E.; Ungermann, C.; et al. The nutrient-responsive CDK Pho85 primes the Sch9 kinase for its activation by TORC1. PLoS Genet. 2023, 19, e1010641. [Google Scholar] [CrossRef]
- Wanke, V.; Pedruzzi, I.; Cameroni, E.; Dubouloz, F.; De Virgilio, C. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 2005, 24, 4271–4278. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, J.; Ricco, N.; Grijota-Martinez, C.; Fado, R.; Clotet, J. Redundancy or specificity? The role of the CDK Pho85 in cell cycle control. Int. J. Biochem. Mol. Biol. 2013, 4, 140–149. [Google Scholar]
- Pedruzzi, I.; Burckert, N.; Egger, P.; De Virgilio, C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 2000, 19, 2569–2579. [Google Scholar] [CrossRef]
- Lee, P.; Paik, S.M.; Shin, C.S.; Huh, W.K.; Hahn, J.S. Regulation of yeast Yak1 kinase by PKA and autophosphorylation-dependent 14-3-3 binding. Mol. Microbiol. 2011, 79, 633–646. [Google Scholar] [CrossRef]
- Cai, T.; Aulds, J.; Gill, T.; Cerio, M.; Schmitt, M.E. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics 2002, 161, 1029–1042. [Google Scholar] [CrossRef]
- Gill, T.; Cai, T.; Aulds, J.; Wierzbicki, S.; Schmitt, M.E. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: Novel method of mRNA degradation. Mol. Cell. Biol. 2004, 24, 945–953. [Google Scholar] [CrossRef]
- Trcek, T.; Larson, D.R.; Moldon, A.; Query, C.C.; Singer, R.H. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell 2011, 147, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Messier, V.; Zenklusen, D.; Michnick, S.W. A nutrient-responsive pathway that determines M phase timing through control of B-cyclin mRNA stability. Cell 2013, 153, 1080–1093. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, A.; Maruki, Y.; Imamura, Y.; Kondo, C.; Kawamata, T.; Kawanishi, I.; Takata, H.; Matsuura, A.; Lee, K.S.; Kikkawa, U.; et al. The yeast Tor signaling pathway is involved in G2/M transition via polo-kinase. PLoS ONE 2008, 3, e2223. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.J.; Ewald, J.C.; Skotheim, J.M. Cell size control in yeast. Curr. Biol. 2012, 22, R350–R359. [Google Scholar] [CrossRef]
- Leitao, R.M.; Kellogg, D.R. The duration of mitosis and daughter cell size are modulated by nutrients in budding yeast. J. Cell Biol. 2017, 216, 3463–3470. [Google Scholar] [CrossRef]
- Foltman, M.; Mendez, I.; Bech-Serra, J.J.; de la Torre, C.; Brace, J.L.; Weiss, E.L.; Lucas, M.; Queralt, E.; Sanchez-Diaz, A. TOR complex 1 negatively regulates NDR kinase Cbk1 to control cell separation in budding yeast. PLoS Biol. 2023, 21, e3002263. [Google Scholar] [CrossRef]
- Yamada, C.; Morooka, A.; Miyazaki, S.; Nagai, M.; Mase, S.; Iemura, K.; Tasnin, M.N.; Takuma, T.; Nakamura, S.; Morshed, S.; et al. TORC1 inactivation promotes APC/C-dependent mitotic slippage in yeast and human cells. iScience 2022, 25, 103675. [Google Scholar] [CrossRef]
- Odle, R.I.; Walker, S.A.; Oxley, D.; Kidger, A.M.; Balmanno, K.; Gilley, R.; Okkenhaug, H.; Florey, O.; Ktistakis, N.T.; Cook, S.J. An mTORC1-to-CDK1 Switch Maintains Autophagy Suppression during Mitosis. Mol. Cell 2020, 77, 228–240.e227. [Google Scholar] [CrossRef]
- Choi, J.H.; Adames, N.R.; Chan, T.F.; Zeng, C.; Cooper, J.A.; Zheng, X.F. TOR signaling regulates microtubule structure and function. Curr. Biol. 2000, 10, 861–864. [Google Scholar] [CrossRef]
- Tran, L.T.; Wang’ondu, R.W.; Weng, J.B.; Wanjiku, G.W.; Fong, C.M.; Kile, A.C.; Koepp, D.M.; Hood-DeGrenier, J.K. TORC1 kinase and the S-phase cyclin Clb5 collaborate to promote mitotic spindle assembly and DNA replication in S. cerevisiae. Curr. Genet. 2010, 56, 479–493. [Google Scholar] [CrossRef]
- Chen, X.; Portran, D.; Widmer, L.A.; Stangier, M.M.; Czub, M.P.; Liakopoulos, D.; Stelling, J.; Steinmetz, M.O.; Barral, Y. The motor domain of the kinesin Kip2 promotes microtubule polymerization at microtubule tips. J. Cell Biol. 2023, 222, e202110126. [Google Scholar] [CrossRef]
- van der Vaart, B.; Fischbock, J.; Mieck, C.; Pichler, P.; Mechtler, K.; Medema, R.H.; Westermann, S. TORC1 signaling exerts spatial control over microtubule dynamics by promoting nuclear export of Stu2. J. Cell Biol. 2017, 216, 3471–3484. [Google Scholar] [CrossRef]
- Ruchaud, S.; Carmena, M.; Earnshaw, W.C. Chromosomal passengers: Conducting cell division. Nat. Rev. Mol. Cell Biol. 2007, 8, 798–812. [Google Scholar] [CrossRef]
- Tatchell, K.; Makrantoni, V.; Stark, M.J.; Robinson, L.C. Temperature-sensitive ipl1-2/Aurora B mutation is suppressed by mutations in TOR complex 1 via the Glc7/PP1 phosphatase. Proc. Natl. Acad. Sci. USA 2011, 108, 3994–3999. [Google Scholar] [CrossRef] [PubMed]
- Queralt, E.; Uhlmann, F. Cdk-counteracting phosphatases unlock mitotic exit. Curr. Opin. Cell Biol. 2008, 20, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Foltman, M.; Molist, I.; Arcones, I.; Sacristan, C.; Filali-Mouncef, Y.; Roncero, C.; Sanchez-Diaz, A. Ingression Progression Complexes Control Extracellular Matrix Remodelling during Cytokinesis in Budding Yeast. PLoS Genet. 2016, 12, e1005864. [Google Scholar] [CrossRef] [PubMed]
- Bhavsar-Jog, Y.P.; Bi, E. Mechanics and regulation of cytokinesis in budding yeast. Semin. Cell Dev. Biol. 2017, 66, 107–118. [Google Scholar] [CrossRef]
- Okada, H.; MacTaggart, B.; Ohya, Y.; Bi, E. The kinetic landscape and interplay of protein networks in cytokinesis. iScience 2021, 24, 101917. [Google Scholar] [CrossRef]
- Dohrmann, P.R.; Butler, G.; Tamai, K.; Dorland, S.; Greene, J.R.; Thiele, D.J.; Stillman, D.J. Parallel pathways of gene regulation: Homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 1992, 6, 93–104. [Google Scholar] [CrossRef]
- Colman-Lerner, A.; Chin, T.E.; Brent, R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 2001, 107, 739–750. [Google Scholar] [CrossRef]
- Baladron, V.; Ufano, S.; Duenas, E.; Martin-Cuadrado, A.B.; del Rey, F.; Vazquez de Aldana, C.R. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell 2002, 1, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Chica, N.; Rozalen, A.E.; Perez-Hidalgo, L.; Rubio, A.; Novak, B.; Moreno, S. Nutritional Control of Cell Size by the Greatwall-Endosulfine-PP2A.B55 Pathway. Curr. Biol. 2016, 26, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Tumaneng, K.; Russell, R.C.; Guan, K.L. Organ size control by Hippo and TOR pathways. Curr. Biol. 2012, 22, R368–R379. [Google Scholar] [CrossRef]
- Yu, F.X.; Guan, K.L. The Hippo pathway: Regulators and regulations. Genes Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.G.; Ng, Y.L.D.; Lam, W.L.M.; Plouffe, S.W.; Guan, K.L. The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res. 2015, 25, 1299–1313. [Google Scholar] [CrossRef]
- Gan, W.; Dai, X.; Dai, X.; Xie, J.; Yin, S.; Zhu, J.; Wang, C.; Liu, Y.; Guo, J.; Wang, M.; et al. LATS suppresses mTORC1 activity to directly coordinate Hippo and mTORC1 pathways in growth control. Nat. Cell Biol. 2020, 22, 246–256. [Google Scholar] [CrossRef]
- Zaman, A.; Wu, X.; Lemoff, A.; Yadavalli, S.; Lee, J.; Wang, C.; Cooper, J.; McMillan, E.A.; Yeaman, C.; Mirzaei, H.; et al. Exocyst protein subnetworks integrate Hippo and mTOR signaling to promote virus detection and cancer. Cell Rep. 2021, 36, 109491. [Google Scholar] [CrossRef]
- Honda, D.; Okumura, M.; Chihara, T. Crosstalk between the mTOR and Hippo pathways. Dev. Growth Differ. 2023, 65, 337–347. [Google Scholar] [CrossRef]
- Cornu, M.; Albert, V.; Hall, M.N. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 2013, 23, 53–62. [Google Scholar] [CrossRef]
- Kim, J.; Guan, K.L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019, 21, 63–71. [Google Scholar] [CrossRef]



| S. cerevisiae | Function |
|---|---|
| Tor1 or Tor2 | Serine/threonine protein kinases |
| Kog1 | Recruitment of substrates to the Tor kinase and regulation of Tor kinase function |
| Lst8 | Stabilize the complex |
| Tco89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foltman, M.; Sanchez-Diaz, A. TOR Complex 1: Orchestrating Nutrient Signaling and Cell Cycle Progression. Int. J. Mol. Sci. 2023, 24, 15745. https://doi.org/10.3390/ijms242115745
Foltman M, Sanchez-Diaz A. TOR Complex 1: Orchestrating Nutrient Signaling and Cell Cycle Progression. International Journal of Molecular Sciences. 2023; 24(21):15745. https://doi.org/10.3390/ijms242115745
Chicago/Turabian StyleFoltman, Magdalena, and Alberto Sanchez-Diaz. 2023. "TOR Complex 1: Orchestrating Nutrient Signaling and Cell Cycle Progression" International Journal of Molecular Sciences 24, no. 21: 15745. https://doi.org/10.3390/ijms242115745
APA StyleFoltman, M., & Sanchez-Diaz, A. (2023). TOR Complex 1: Orchestrating Nutrient Signaling and Cell Cycle Progression. International Journal of Molecular Sciences, 24(21), 15745. https://doi.org/10.3390/ijms242115745







