Unraveling the Janus-Faced Role of Autophagy in Hepatocellular Carcinoma: Implications for Therapeutic Interventions
Abstract
1. Introduction
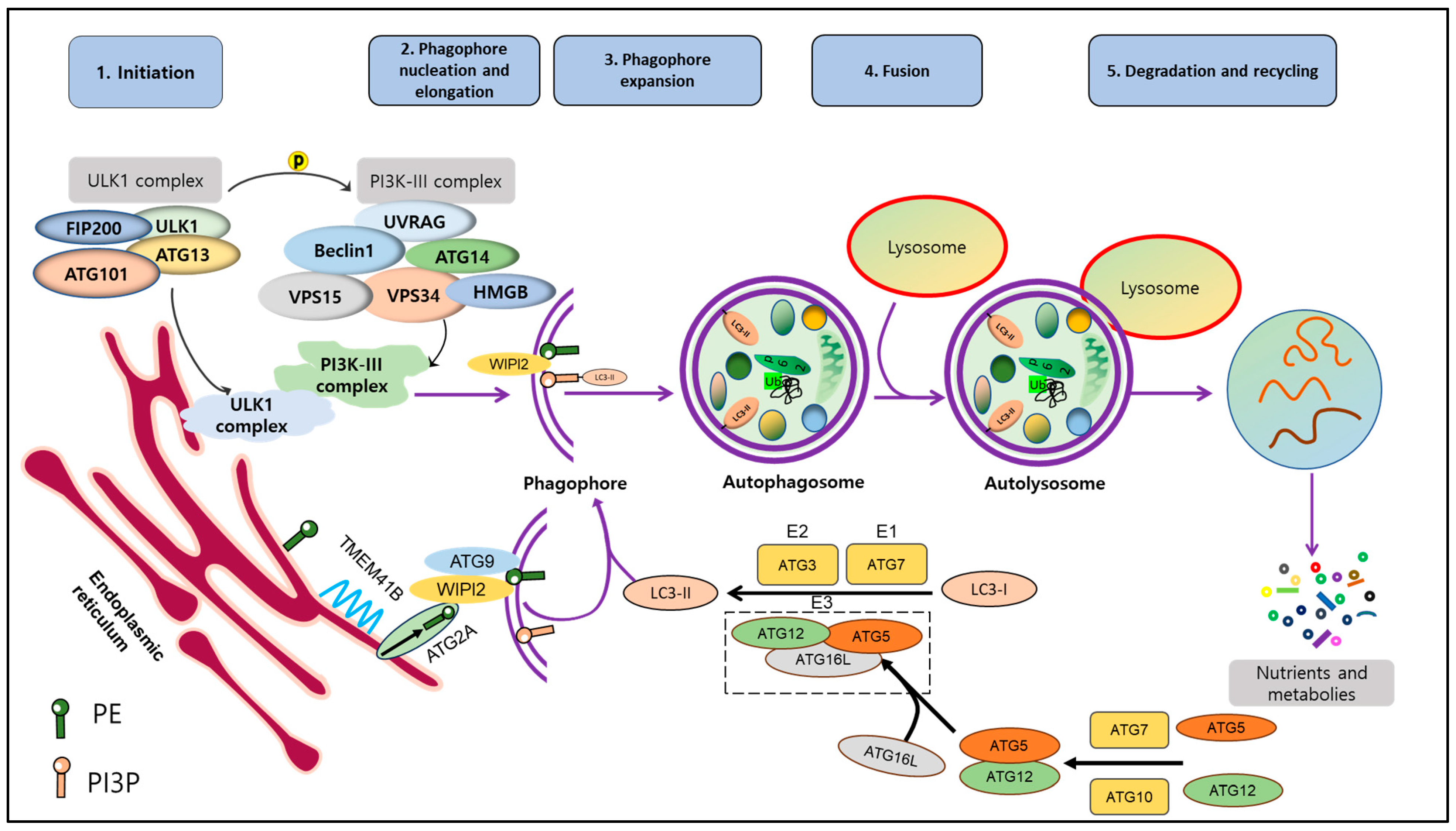
2. Mechanism of Autophagy
3. Autophagy-Regulating Signaling Pathways in HCC
3.1. PI3K/AKT/mTOR
3.2. AMPK/mTOR
3.3. The MAPK (JNK, p38 and ERK)
3.4. Nrf2-p62 Pathway
3.5. Crosstalks between These Pathways
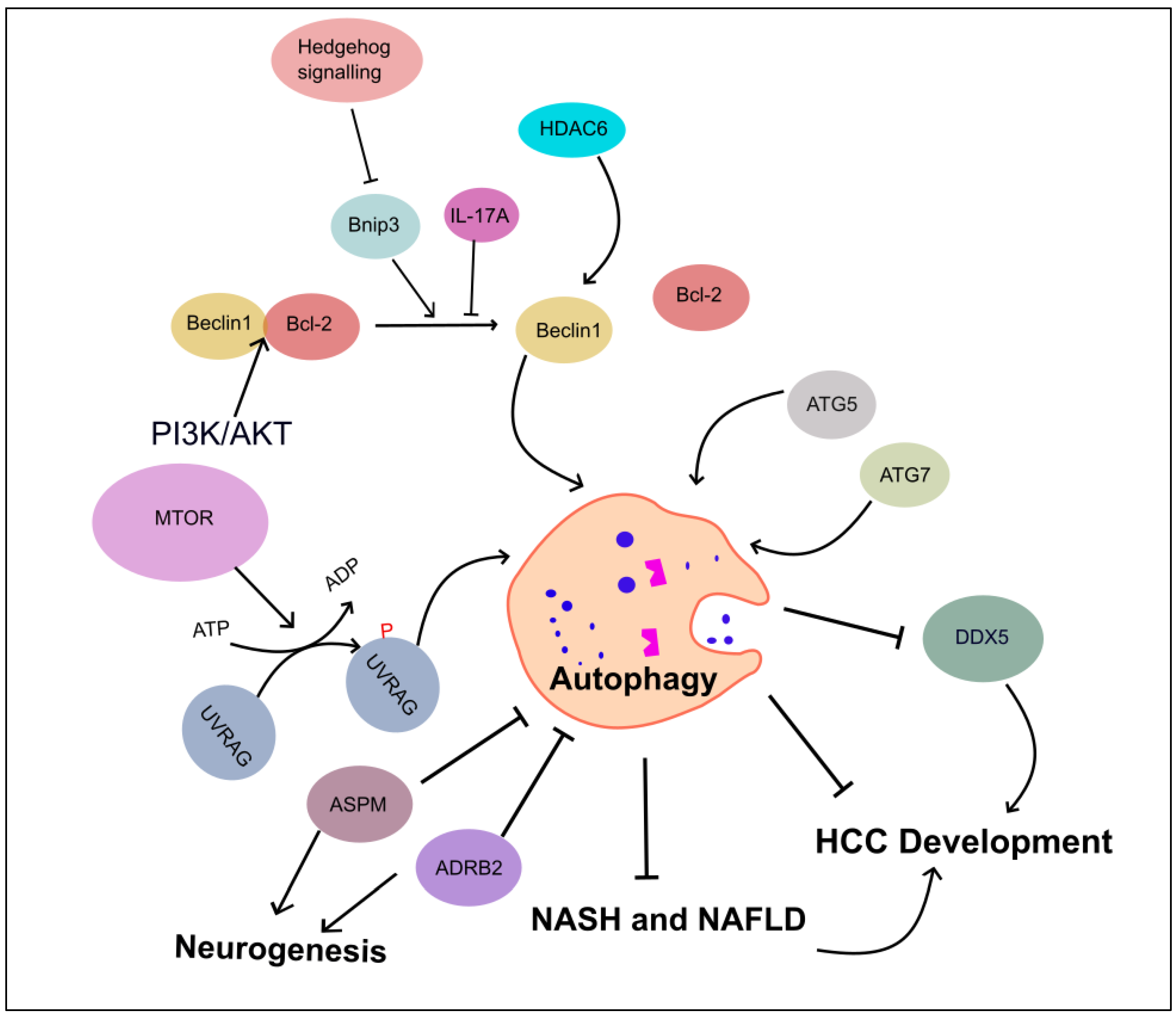
4. Dual Role of Autophagy in HCC
4.1. Autophagy Inhibits HCC Progression
4.2. Autophagy Promotes HCC Progression
4.3. Autophagy in Drug Resistance
5. Selective Autophagy in HCC
5.1. Mitophagy in HCC
5.2. Other Selective Autophagy in HCC
6. The Agents That Target Autophagy in HCC Treatment
6.1. Autophagy Inhibitors
6.2. Autophagy Activators
| Compounds | Autophagy Modulators | Experimental Objects | Mechanism |
|---|---|---|---|
| CQ | Inhibitor | HCC cell line: HepG2 [241] | Lysosomal Inhibitor |
| HCQ | Inhibitor | HCC cell lines: HepG2 and Huh7 [242] | |
| BafA1 | Inhibitor | Rat liver cell line H-4-II-E [203] | V-ATPase Inhibitor |
| HCC cell lines: (BEL7402, LO2, SMMC-7721, Huh7, and HepG2) [204] | |||
| 3-methyladenine plus delphinidin | Inhibitor | HCC cell line: SMMC7721 [209] | PI3K Inhibitors |
| LY294002 | Inhibitor | Normal liver cell line: HL-7702 [210] HCC cell lines: SMMC-7721, BEL-7401, SK-Hep-1 and HEP-G2 [210] | |
| VPS34-IN-1 | Inhibitor | HCC cell line: Huh7, MHCC-97H [217] | VPS34 Inhibitor |
| MRT68921 plus Cinobufagin | Inhibitor | HCC cell line: HepG2 and Huh7 [221] | ULK1/2 Inhibitor |
| SBI-0206965 | Inhibitor | Normal human liver cells (L02) [227] Nutrient-starved human HCC [227] | |
| DC-ATG4in | Inhibitor | HCC cell lines: Huh7 and SK-HEP-1 [230] | ATG4B Inhibitor |
| Temsirolimus plus Sorafenib | Activator | HCC patients [238] | mTOR inhibitor |
| Everolimus plus Sorafenib | Activator | HCC patients [239] |
7. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rumgay, H.; Arnold, M.; Ferlay, J.; Lesi, O.; Cabasag, C.J.; Vignat, J.; Laversanne, M.; McGlynn, K.A.; Soerjomataram, I. Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 2022, 77, 1598–1606. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Arnold, M.; Abnet, C.C.; Neale, R.E.; Vignat, J.; Giovannucci, E.L.; McGlynn, K.A.; Bray, F. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology 2020, 159, 335–349.e15. [Google Scholar] [CrossRef]
- Chon, Y.E.; Jeong, S.W.; Jun, D.W. Hepatocellular carcinoma statistics in South Korea. Clin. Mol. Hepatol. 2021, 27, 512. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef]
- Debnath, J.; Gammoh, N.; Ryan, K.M. Autophagy and autophagy-related pathways in cancer. Nat. Rev. Mol. Cell Biol. 2023, 24, 560–575. [Google Scholar] [CrossRef]
- Yamamoto, H.; Matsui, T. Molecular mechanisms of macroautophagy, microautophagy, and chaperone-mediated autophagy. J. Nippon Med. Sch. 2023, JNMS.2024_2091-2102. [Google Scholar] [CrossRef]
- Vargas, J.N.S.; Hamasaki, M.; Kawabata, T.; Youle, R.J.; Yoshimori, T. The mechanisms and roles of selective autophagy in mammals. Nat. Rev. Mol. Cell Biol. 2023, 24, 167–185. [Google Scholar] [CrossRef]
- Chun, Y.; Kim, J. Autophagy: An essential degradation program for cellular homeostasis and life. Cells 2018, 7, 278. [Google Scholar] [CrossRef]
- Bi, T.; Lu, Q.; Pan, X.; Dong, F.; Hu, Y.; Xu, Z.; Xiu, P.; Liu, Z.; Li, J. circFAM134B is a key factor regulating reticulophagy-mediated ferroptosis in hepatocellular carcinoma. Cell Cycle 2023, 22, 1900–1920. [Google Scholar] [CrossRef]
- Mehta, P.; Shende, P. Dual role of autophagy for advancements from conventional to new delivery systems in cancer. Biochim. Biophys. Acta Gen. Subj. 2023, 1867, 130430. [Google Scholar] [CrossRef]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Nadafzadeh, N.; Imani, M.H.; Rajabi, R.; Ziaolhagh, S.; Bayanzadeh, S.D.; Norouzi, R.; Rafiei, R.; Koohpar, Z.K.; Raei, B. Targeting and regulation of autophagy in hepatocellular carcinoma: Revisiting the molecular interactions and mechanisms for new therapy approaches. Cell Commun. Signal. 2023, 21, 32. [Google Scholar] [CrossRef] [PubMed]
- Hama, Y.; Ogasawara, Y.; Noda, N.N. Autophagy and cancer: Basic mechanisms and inhibitor development. Cancer Sci. 2023, 114, 2699–2708. [Google Scholar] [CrossRef] [PubMed]
- Phung, H.H.; Lee, C.H. Mouse models of nonalcoholic steatohepatitis and their application to new drug development. Arch. Pharm. Res. 2022, 45, 761–794. [Google Scholar] [CrossRef]
- Wu, W.K.; Zhang, L.; Chan, M.T. Autophagy, nafld and nafld-related hcc. Adv. Exp. Med. Biol. 2018, 1061, 127–138. [Google Scholar] [CrossRef]
- Huang, F.; Wang, B.-R.; Wang, Y.-G. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J. Gastroenterol. 2018, 24, 4643. [Google Scholar] [CrossRef]
- Yamamoto, H.; Zhang, S.; Mizushima, N. Autophagy genes in biology and disease. Nat. Rev. Genet. 2023, 24, 382–400. [Google Scholar] [CrossRef]
- Metur, S.P.; Lei, Y.; Zhang, Z.; Klionsky, D.J. Regulation of autophagy gene expression and its implications in cancer. J. Cell Sci. 2023, 136, jcs260631. [Google Scholar] [CrossRef]
- Mizushima, N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010, 22, 132–139. [Google Scholar] [CrossRef]
- Jean, S.; Kiger, A.A. Classes of phosphoinositide 3-kinases at a glance. J. Cell Sci. 2014, 127, 923–928. [Google Scholar] [CrossRef]
- Moretti, F.; Bergman, P.; Dodgson, S.; Marcellin, D.; Claerr, I.; Goodwin, J.M.; DeJesus, R.; Kang, Z.; Antczak, C.; Begue, D. TMEM 41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 2018, 19, e45889. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Hama, Y.; Izume, T.; Tamura, N.; Ueno, T.; Yamashita, Y.; Sakamaki, Y.; Mimura, K.; Morishita, H.; Shihoya, W. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 2018, 217, 3817–3828. [Google Scholar] [CrossRef]
- van Vliet, A.R.; Chiduza, G.N.; Maslen, S.L.; Pye, V.E.; Joshi, D.; De Tito, S.; Jefferies, H.B.; Christodoulou, E.; Roustan, C.; Punch, E. ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation. Mol. Cell 2022, 82, 4324–4339.e8. [Google Scholar] [CrossRef] [PubMed]
- Jain, V.; Singh, M.P.; Amaravadi, R.K. Recent advances in targeting autophagy in cancer. Trends Pharmacol. Sci. 2023, 44, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Amaya, C.; Fader, C.M.; Colombo, M.I. Autophagy and proteins involved in vesicular trafficking. FEBS Lett. 2015, 589, 3343–3353. [Google Scholar] [CrossRef]
- Lu, X.; Paliogiannis, P.; Calvisi, D.F.; Chen, X. Role of the mammalian target of rapamycin pathway in liver cancer: From molecular genetics to targeted therapies. Hepatology 2021, 73, 49–61. [Google Scholar] [CrossRef]
- Boyault, S.; Rickman, D.S.; De Reyniès, A.; Balabaud, C.; Rebouissou, S.; Jeannot, E.; Hérault, A.; Saric, J.; Belghiti, J.; Franco, D. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007, 45, 42–52. [Google Scholar] [CrossRef]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.-C.; Llovet, J.M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015, 149, 1226–1239.e4. [Google Scholar] [CrossRef]
- Hoshida, Y.; Nijman, S.M.; Kobayashi, M.; Chan, J.A.; Brunet, J.-P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Venkatesh, S.K.; Torbenson, M.S.; Roberts, L.R. Clinical implications of basic research in hepatocellular carcinoma. J. Hepatol. 2016, 64, 736–745. [Google Scholar] [CrossRef]
- Bhaskar, P.T.; Hay, N. The two TORCs and AKT. Dev. Cell 2007, 12, 487–502. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Hay, N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell 2005, 8, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Cuyàs, E.; Corominas-Faja, B.; Joven, J.; Menendez, J.A. Cell cycle regulation by the nutrient-sensing mammalian target of rapamycin (mTOR) pathway. Methods Mol. Biol. 2014, 1170, 113–144. [Google Scholar] [CrossRef]
- Noda, T.; Ohsumi, Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998, 273, 3963–3966. [Google Scholar] [CrossRef]
- Schmelzle, T.; Hall, M.N. TOR, a central controller of cell growth. Cell 2000, 103, 253–262. [Google Scholar] [CrossRef]
- Bortolami, M.; Comparato, A.; Benna, C.; Errico, A.; Maretto, I.; Pucciarelli, S.; Cillo, U.; Farinati, F. Gene and protein expression of mTOR and LC3 in hepatocellular carcinoma, colorectal liver metastasis and “normal” liver tissues. PLoS ONE 2020, 15, e0244356. [Google Scholar] [CrossRef]
- Chen, N.; Karantza-Wadsworth, V. Role and regulation of autophagy in cancer. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1793, 1516–1523. [Google Scholar] [CrossRef]
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the integrated stress response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Heras-Sandoval, D.; Pérez-Rojas, J.M.; Hernández-Damián, J.; Pedraza-Chaverri, J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell. Signal. 2014, 26, 2694–2701. [Google Scholar] [CrossRef]
- Antonioli, M.; Albiero, F.; Nazio, F.; Vescovo, T.; Perdomo, A.B.; Corazzari, M.; Marsella, C.; Piselli, P.; Gretzmeier, C.; Dengjel, J. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev. Cell 2014, 31, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Cianfanelli, V.; D’Orazio, M.; Cecconi, F. AMBRA1 and BECLIN 1 interplay in the crosstalk between autophagy and cell proliferation. Cell Cycle 2015, 14, 959–963. [Google Scholar] [CrossRef]
- Xiang, H.; Zhang, J.; Lin, C.; Zhang, L.; Liu, B.; Ouyang, L. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm. Sin. B 2020, 10, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Ruf, S.; Heberle, A.M.; Langelaar-Makkinje, M.; Gelino, S.; Wilkinson, D.; Gerbeth, C.; Schwarz, J.J.; Holzwarth, B.; Warscheid, B.; Meisinger, C. PLK1 (polo like kinase 1) inhibits MTOR complex 1 and promotes autophagy. Autophagy 2017, 13, 486–505. [Google Scholar] [CrossRef]
- Cao, Y.-Y.; Qiao, Y.; Wang, Z.-H.; Chen, Q.; Qi, Y.-P.; Lu, Z.-M.; Wang, Z.; Lu, W.-H. The Polo-Like Kinase 1–Mammalian Target of Rapamycin Axis Regulates Autophagy to Prevent Intestinal Barrier Dysfunction during Sepsis. Am. J. Pathol. 2023, 193, 296–312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, S.; Chua, M.-S.; Li, H.; Luo, D.; Wang, S.; Zhang, S.; Han, B.; Sun, C. SOCS5 inhibition induces autophagy to impair metastasis in hepatocellular carcinoma cells via the PI3K/Akt/mTOR pathway. Cell Death Dis. 2019, 10, 612. [Google Scholar] [CrossRef]
- Li, J.; Zhai, D.-S.; Huang, Q.; Chen, H.-L.; Zhang, Z.; Tan, Q.-F. LncRNA DCST1-AS1 accelerates the proliferation, metastasis and autophagy of hepatocellular carcinoma cell by AKT/mTOR signaling pathways. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6091–6104. [Google Scholar] [CrossRef]
- Wang, S.; Zhu, M.; Wang, Q.; Hou, Y.; Li, L.; Weng, H.; Zhao, Y.; Chen, D.; Ding, H.; Guo, J. Alpha-fetoprotein inhibits autophagy to promote malignant behaviour in hepatocellular carcinoma cells by activating PI3K/AKT/mTOR signalling. Cell Death Dis. 2018, 9, 1027. [Google Scholar] [CrossRef]
- Chen, J.S.; Wang, Q.; Fu, X.H.; Huang, X.H.; Chen, X.L.; Cao, L.Q.; Chen, L.Z.; Tan, H.X.; Li, W.; Bi, J. Involvement of PI3K/PTEN/AKT/mTOR pathway in invasion and metastasis in hepatocellular carcinoma: Association with MMP-9. Hepatol. Res. 2009, 39, 177–186. [Google Scholar] [CrossRef]
- Niu, Y.; Sun, W.; Lu, J.-J.; Ma, D.-L.; Leung, C.-H.; Pei, L.; Chen, X. PTEN activation by DNA damage induces protective autophagy in response to cucurbitacin B in hepatocellular carcinoma cells. Oxid. Med. Cell. Longev. 2016, 2016, 4313204. [Google Scholar] [CrossRef]
- Sun, E.J.; Wankell, M.; Palamuthusingam, P.; McFarlane, C.; Hebbard, L. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Biomedicines 2021, 9, 1639. [Google Scholar] [CrossRef] [PubMed]
- Alers, S.; Löffler, A.S.; Wesselborg, S.; Stork, B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: Cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 2012, 32, 2–11. [Google Scholar] [CrossRef]
- Endo, H.; Owada, S.; Inagaki, Y.; Shida, Y.; Tatemichi, M. Glucose starvation induces LKB1-AMPK-mediated MMP-9 expression in cancer cells. Sci. Rep. 2018, 8, 10122. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Shen, H.-M.; Lim, L.H.K. The Role of Autophagy in Liver Cancer: Crosstalk in Signaling Pathways and Potential Therapeutic Targets. Pharmaceuticals 2020, 13, 432. [Google Scholar] [CrossRef] [PubMed]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Weerasekara, V.K.; Panek, D.J.; Broadbent, D.G.; Mortenson, J.B.; Mathis, A.D.; Logan, G.N.; Prince, J.T.; Thomson, D.M.; Thompson, J.W.; Andersen, J.L. Metabolic-stress-induced rearrangement of the 14-3-3ζ interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3ζ interaction with phosphorylated Atg9. Mol. Cell. Biol. 2014, 34, 4379–4388. [Google Scholar] [CrossRef]
- Kim, J.; Kim, Y.C.; Fang, C.; Russell, R.C.; Kim, J.H.; Fan, W.; Liu, R.; Zhong, Q.; Guan, K.L. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013, 152, 290–303. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, W.; Sun, X.; Xu, D.; Wang, C.; Zhang, Q.; Wang, H.; Luo, W.; Chen, Y.; Chen, H.; et al. AMPK regulates autophagy by phosphorylating BECN1 at threonine 388. Autophagy 2016, 12, 1447–1459. [Google Scholar] [CrossRef]
- Kim, Y.W.; Jang, E.J.; Kim, C.H.; Lee, J.H. Sauchinone exerts anticancer effects by targeting AMPK signaling in hepatocellular carcinoma cells. Chem. Biol. Interact. 2017, 261, 108–117. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Z.X.; Huang, F.; Yuan, X.W.; Deng, L.; Tang, D. MicroRNA-1271 functions as a potential tumor suppressor in hepatitis B virus-associated hepatocellular carcinoma through the AMPK signaling pathway by binding to CCNA1. J. Cell. Physiol. 2019, 234, 3555–3569. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, Y.; Li, M.; Wu, H.; Wang, Y.; You, Y.; Li, P.; Ding, X.; Liu, C.; Gong, J. Predictive and preventive significance of AMPK activation on hepatocarcinogenesis in patients with liver cirrhosis. Cell Death Dis. 2018, 9, 264. [Google Scholar] [CrossRef]
- Sun, Y.; Lei, B.; Huang, Q. SOX18 Affects Cell Viability, Migration, Invasiveness, and Apoptosis in Hepatocellular Carcinoma (HCC) Cells by Participating in Epithelial-to-Mesenchymal Transition (EMT) Progression and Adenosine Monophosphate Activated Protein Kinase (AMPK)/Mammalian Target of Rapamycin (mTOR). Med. Sci. Monit. 2019, 25, 6244–6254. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Lv, G.; Li, R.; Liu, W.T.; Zong, C.; Ye, F.; Li, X.Y.; Yang, X.; Jiang, J.H.; Hou, X.J.; et al. Glycochenodeoxycholate promotes hepatocellular carcinoma invasion and migration by AMPK/mTOR dependent autophagy activation. Cancer Lett. 2019, 454, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, W.; Mao, Q.; Gao, D.; Xiong, L.; Hu, X.; Zheng, Y.; Xu, X. HMGB1 Promotes Resistance to Doxorubicin in Human Hepatocellular Carcinoma Cells by Inducing Autophagy via the AMPK/mTOR Signaling Pathway. Front. Oncol. 2021, 11, 739145. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.Y.; Tsai, H.H.; Yen, C.J.; Hung, L.Y.; Yang, C.C.; Ho, C.H.; Liang, H.Y.; Chen, F.W.; Li, C.F.; Wang, J.M. Metformin Resensitizes Sorafenib-Resistant HCC Cells through AMPK-Dependent Autophagy Activation. Front. Cell Dev. Biol. 2020, 8, 596655. [Google Scholar] [CrossRef]
- Ma, X.; Qiu, Y.; Sun, Y.; Zhu, L.; Zhao, Y.; Li, T.; Lin, Y.; Ma, D.; Qin, Z.; Sun, C.; et al. NOD2 inhibits tumorigenesis and increases chemosensitivity of hepatocellular carcinoma by targeting AMPK pathway. Cell Death Dis. 2020, 11, 174. [Google Scholar] [CrossRef]
- Wang, L.; Li, H.; Zhen, Z.; Ma, X.; Yu, W.; Zeng, H.; Li, L. CXCL17 promotes cell metastasis and inhibits autophagy via the LKB1-AMPK pathway in hepatocellular carcinoma. Gene 2019, 690, 129–136. [Google Scholar] [CrossRef]
- Jiang, X.; Tan, H.Y.; Teng, S.; Chan, Y.T.; Wang, D.; Wang, N. The Role of AMP-Activated Protein Kinase as a Potential Target of Treatment of Hepatocellular Carcinoma. Cancers 2019, 11, 647. [Google Scholar] [CrossRef]
- Qin, Q.F.; Li, X.J.; Li, Y.S.; Zhang, W.K.; Tian, G.H.; Shang, H.C.; Tang, H.B. AMPK-ERK/CARM1 Signaling Pathways Affect Autophagy of Hepatic Cells in Samples of Liver Cancer Patients. Front. Oncol. 2019, 9, 1247. [Google Scholar] [CrossRef]
- Wei, X.; Li, X.; Yan, W.; Zhang, X.; Sun, Y.; Zhang, F. SKP2 Promotes Hepatocellular Carcinoma Progression through Nuclear AMPK-SKP2-CARM1 Signaling Transcriptionally Regulating Nutrient-Deprived Autophagy Induction. Cell. Physiol. Biochem. 2018, 47, 2484–2497. [Google Scholar] [CrossRef]
- Davis, R.J. MAPKs: New JNK expands the group. Trends Biochem. Sci. 1994, 19, 470–473. [Google Scholar] [CrossRef]
- Ito, Y.; Sasaki, Y.; Horimoto, M.; Wada, S.; Tanaka, Y.; Kasahara, A.; Ueki, T.; Hirano, T.; Yamamoto, H.; Fujimoto, J.; et al. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology 1998, 27, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Kempf, R.C.; Long, J.; Laidler, P.; Mijatovic, S.; Maksimovic-Ivanic, D.; Stivala, F.; Mazzarino, M.C.; et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging 2011, 3, 192–222. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, K.J.; Wohlschlaeger, J.; Lang, H.; Sotiropoulos, G.C.; Malago, M.; Steveling, K.; Reis, H.; Cicinnati, V.R.; Schmid, K.W.; Baba, H.A. Activation of the ERK and AKT signalling pathway predicts poor prognosis in hepatocellular carcinoma and ERK activation in cancer tissue is associated with hepatitis C virus infection. J. Hepatol. 2008, 48, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Liu, D.; Chen, L.; He, Y.; Tian, X.; Qi, L.; Chen, L.; Luo, Y.; Chen, Z.; Hu, X.; et al. PNO1 regulates autophagy and apoptosis of hepatocellular carcinoma via the MAPK signaling pathway. Cell Death Dis. 2021, 12, 552. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xia, C.; Lv, Y.; Li, C.; Mei, Q.; Li, H.; Wang, H.; Li, S. Crosstalk Influence between P38MAPK and Autophagy on Mitochondria-Mediated Apoptosis Induced by Anti-Fas Antibody/Actinomycin D in Human Hepatoma Bel-7402 Cells. Molecules 2017, 22, 1705. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, P.; Chen, Y.; Yao, H.; Wang, S. Novel 2-phenyloxypyrimidine derivative induces apoptosis and autophagy via inhibiting PI3K pathway and activating MAPK/ERK signaling in hepatocellular carcinoma cells. Sci. Rep. 2018, 8, 10923. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Y.; Li, Y.; Jiang, W.Q.; Zhou, L.F. MAPK/JNK signalling: A potential autophagy regulation pathway. Biosci. Rep. 2015, 35, e00199. [Google Scholar] [CrossRef]
- Xie, C.M.; Chan, W.Y.; Yu, S.; Zhao, J.; Cheng, C.H. Bufalin induces autophagy-mediated cell death in human colon cancer cells through reactive oxygen species generation and JNK activation. Free Radic. Biol. Med. 2011, 51, 1365–1375. [Google Scholar] [CrossRef]
- Yang, W.; Su, J.; Li, M.; Li, T.; Wang, X.; Zhao, M.; Hu, X. Myricetin Induces Autophagy and Cell Cycle Arrest of HCC by Inhibiting MARCH1-Regulated Stat3 and p38 MAPK Signaling Pathways. Front. Pharmacol. 2021, 12, 709526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; He, J.; Ye, X.; Zhu, J.; Hu, X.; Shen, M.; Ma, Y.; Mao, Z.; Song, H.; Chen, F. β-Thujaplicin induces autophagic cell death, apoptosis, and cell cycle arrest through ROS-mediated Akt and p38/ERK MAPK signaling in human hepatocellular carcinoma. Cell Death Dis. 2019, 10, 255. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Copple, I.M. Advances and challenges in therapeutic targeting of NRF2. Trends Pharmacol. Sci. 2023, 44, 137–149. [Google Scholar] [CrossRef]
- Kageyama, S.; Gudmundsson, S.R.; Sou, Y.-S.; Ichimura, Y.; Tamura, N.; Kazuno, S.; Ueno, T.; Miura, Y.; Noshiro, D.; Abe, M. p62/SQSTM1-droplet serves as a platform for autophagosome formation and anti-oxidative stress response. Nat. Commun. 2021, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, R.; Noshiro, D.; Morishita, H.; Takada, S.; Kageyama, S.; Fujioka, Y.; Funakoshi, T.; Komatsu-Hirota, S.; Arai, R.; Ryzhii, E. Phosphorylation of phase-separated p62 bodies by ULK1 activates a redox-independent stress response. EMBO J. 2023, 42, e113349. [Google Scholar] [CrossRef]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.-S.; Ueno, I.; Sakamoto, A.; Tong, K.I. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Geng, X.; Dong, Q.; Li, X.; Ye, P.; Lin, M.; Xu, B.; Jiang, H. Crosstalk between autophagy and the Keap1-Nrf2-ARE pathway regulates realgar-induced neurotoxicity. J. Ethnopharmacol. 2023, 301, 115776. [Google Scholar] [CrossRef]
- Zhang, X.; Dai, M.; Li, S.; Li, M.; Cheng, B.; Ma, T.; Zhou, Z. The emerging potential role of p62 in cancer treatment by regulating metabolism. Trends Endocrinol. Metab. 2023, 34, 474–488. [Google Scholar] [CrossRef]
- Saito, T.; Ichimura, Y.; Taguchi, K.; Suzuki, T.; Mizushima, T.; Takagi, K.; Hirose, Y.; Nagahashi, M.; Iso, T.; Fukutomi, T.; et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat. Commun. 2016, 7, 12030. [Google Scholar] [CrossRef]
- Kahroba, H.; Shirmohamadi, M.; Hejazi, M.S.; Samadi, N. The Role of Nrf2 signaling in cancer stem cells: From stemness and self-renewal to tumorigenesis and chemoresistance. Life Sci. 2019, 239, 116986. [Google Scholar] [CrossRef]
- Robertson, H.; Dinkova-Kostova, A.T.; Hayes, J.D. NRF2 and the Ambiguous Consequences of Its Activation during Initiation and the Subsequent Stages of Tumourigenesis. Cancers 2020, 12, 3609. [Google Scholar] [CrossRef] [PubMed]
- Gureev, A.P.; Popov, V.N.; Starkov, A.A. Crosstalk between the mTOR and Nrf2/ARE signaling pathways as a target in the improvement of long-term potentiation. Exp. Neurol. 2020, 328, 113285. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-H.; Yen, T.-L.; Hsu, C.-Y.; Thomas, P.-A.; Sheu, J.-R.; Jayakumar, T. Multi-targeting andrographolide, a novel NF-κB inhibitor, as a potential therapeutic agent for stroke. Int. J. Mol. Sci. 2017, 18, 1638. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.-L.; Mizushima, N.; Ohsumi, Y. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef]
- Jung, K.H.; Noh, J.H.; Kim, J.K.; Eun, J.W.; Bae, H.J.; Chang, Y.G.; Kim, M.G.; Park, W.S.; Lee, J.Y.; Lee, S.-Y. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology 2012, 56, 644–657. [Google Scholar] [CrossRef]
- Takamura, A.; Komatsu, M.; Hara, T.; Sakamoto, A.; Kishi, C.; Waguri, S.; Eishi, Y.; Hino, O.; Tanaka, K.; Mizushima, N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011, 25, 795–800. [Google Scholar] [CrossRef]
- Liang, C.; Feng, P.; Ku, B.; Dotan, I.; Canaani, D.; Oh, B.-H.; Jung, J.U. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006, 8, 688–698. [Google Scholar] [CrossRef]
- Feng, X.; Jia, Y.; Zhang, Y.; Ma, F.; Zhu, Y.; Hong, X.; Zhou, Q.; He, R.; Zhang, H.; Jin, J. Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy 2019, 15, 1130–1149. [Google Scholar] [CrossRef]
- Zhou, B.; Yang, C.; Yan, X.; Shi, Z.; Xiao, H.; Wei, X.; Jiang, N.; Wu, Z. LETM1 knockdown promotes autophagy and apoptosis through AMP-activated protein kinase phosphorylation-mediated Beclin-1/Bcl-2 complex dissociation in hepatocellular carcinoma. Front. Oncol. 2021, 10, 606790. [Google Scholar] [CrossRef]
- Li, S.; Lin, Z.; Zheng, W.; Zheng, L.; Chen, X.; Yan, Z.; Cheng, Z.; Yan, H.; Zheng, C.; Guo, P. IL-17A inhibits autophagic activity of HCC cells by inhibiting the degradation of Bcl2. Biochem. Biophys. Res. Commun. 2019, 509, 194–200. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Deng, L.; Liu, H.; Chen, T.; Chen, S.; Xia, S.; Liu, Y. BCL2L10/BECN1 modulates hepatoma cells autophagy by regulating PI3K/AKT signaling pathway. Aging 2019, 11, 350. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, C.; Lu, L.; Magliato, S.; Wu, T. Hedgehog signaling pathway regulates autophagy in human hepatocellular carcinoma cells. Hepatology 2013, 58, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yang, X.; Xu, W.; Wang, J.; Wu, D.; Hong, Z.; Yuan, S.; Zeng, Z.; Jia, X.; Lu, S. Heat shock protein 90 promotes RNA helicase DDX5 accumulation and exacerbates hepatocellular carcinoma by inhibiting autophagy. Cancer Biol. Med. 2021, 18, 693. [Google Scholar] [CrossRef]
- Zhang, W.; Hou, J.; Wang, X.; Jiang, R.; Yin, Y.; Ji, J.; Deng, L.; Huang, X.; Wang, K.; Sun, B. PTPRO-mediated autophagy prevents hepatosteatosis and tumorigenesis. Oncotarget 2015, 6, 9420. [Google Scholar] [CrossRef][Green Version]
- Inokuchi-Shimizu, S.; Park, E.J.; Roh, Y.S.; Yang, L.; Zhang, B.; Song, J.; Liang, S.; Pimienta, M.; Taniguchi, K.; Wu, X. TAK1-mediated autophagy and fatty acid oxidation prevent hepatosteatosis and tumorigenesis. J. Clin. Investig. 2014, 124, 3566–3578. [Google Scholar] [CrossRef]
- Kakehashi, A.; Chariyakornkul, A.; Suzuki, S.; Khuanphram, N.; Tatsumi, K.; Yamano, S.; Fujioka, M.; Gi, M.; Wongpoomchai, R.; Wanibuchi, H. Cache Domain Containing 1 Is a Novel Marker of Non-Alcoholic Steatohepatitis-Associated Hepatocarcinogenesis. Cancers 2021, 13, 1216. [Google Scholar] [CrossRef]
- Winkler, F.; Venkatesh, H.S.; Amit, M.; Batchelor, T.; Demir, I.E.; Deneen, B.; Gutmann, D.H.; Hervey-Jumper, S.; Kuner, T.; Mabbott, D. Cancer neuroscience: State of the field, emerging directions. Cell 2023, 186, 1689–1707. [Google Scholar] [CrossRef]
- Lee, C.H.; Cho, J.; Lee, K. Tumour regression via integrative regulation of neurological, inflammatory, and hypoxic tumour microenvironment. Biomol. Ther. 2020, 28, 119. [Google Scholar] [CrossRef]
- Wu, F.-Q.; Fang, T.; Yu, L.-X.; Lv, G.-S.; Lv, H.-W.; Liang, D.; Li, T.; Wang, C.-Z.; Tan, Y.-X.; Ding, J. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1α. J. Hepatol. 2016, 65, 314–324. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, X.; Zhu, L.; Li, Z.; Zuo, P.; Wang, P.; Feng, J.; Mi, Y.; Zhang, C.; Xu, Y. ASPM promotes hepatocellular carcinoma progression by activating Wnt/β-catenin signaling through antagonizing autophagy-mediated Dvl2 degradation. FEBS Open Bio 2021, 11, 2784–2799. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Xia, S.L.; Liao, C.; Li, Y.L.; Wang, Y.F.; Li, T.P.; Zhao, M.J. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J. Gastroenterol. 2004, 10, 509–513. [Google Scholar] [CrossRef]
- Wang, X.K.; Liao, X.W.; Zhou, X.; Han, C.Y.; Chen, Z.J.; Yang, C.K.; Huang, J.L.; Wang, J.Y.; Liu, J.Q.; Huang, H.S.; et al. Oncogene UBE2I enhances cellular invasion, migration and proliferation abilities via autophagy-related pathway resulting in poor prognosis in hepatocellular carcinoma. Am. J. Cancer Res. 2020, 10, 4178–4197. [Google Scholar] [PubMed]
- Deng, G.; Zeng, S.; Qu, Y.; Luo, Q.; Guo, C.; Yin, L.; Han, Y.; Li, Y.; Cai, C.; Fu, Y.; et al. BMP4 promotes hepatocellular carcinoma proliferation by autophagy activation through JNK1-mediated Bcl-2 phosphorylation. J. Exp. Clin. Cancer Res. 2018, 37, 156. [Google Scholar] [CrossRef]
- Fu, N.; Du, H.; Li, D.; Lu, Y.; Li, W.; Wang, Y.; Kong, L.; Du, J.; Zhao, S.; Ren, W.; et al. Clusterin contributes to hepatitis C virus-related hepatocellular carcinoma by regulating autophagy. Life Sci. 2020, 256, 117911. [Google Scholar] [CrossRef] [PubMed]
- Osman, N.A.; Abd El-Rehim, D.M.; Kamal, I.M. Defective Beclin-1 and elevated hypoxia-inducible factor (HIF)-1α expression are closely linked to tumorigenesis, differentiation, and progression of hepatocellular carcinoma. Tumour Biol. 2015, 36, 4293–4299. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, X.; Xu, Y.; He, G.; Liu, Q.; Zhu, J.; Zhang, C. Elevated histone H3 citrullination is associated with increased Beclin1 expression in HBV-related hepatocellular carcinoma. J. Med. Virol. 2020, 92, 1221–1230. [Google Scholar] [CrossRef]
- Matsui, C.; Deng, L.; Minami, N.; Abe, T.; Koike, K.; Shoji, I. Hepatitis C Virus NS5A Protein Promotes the Lysosomal Degradation of Hepatocyte Nuclear Factor 1α via Chaperone-Mediated Autophagy. J. Virol. 2018, 92, 13. [Google Scholar] [CrossRef]
- Kimkong, I.; Kunanopparat, A. Autophagy related protein 9A increase in hepatitis B virus-associated hepatocellular carcinoma and the role in apoptosis. World J. Hepatol. 2020, 12, 1367–1371. [Google Scholar] [CrossRef]
- Khabir, M.; Aliche, A.Z.; Sureau, C.; Blanchet, M.; Labonté, P. Hepatitis Delta Virus Alters the Autophagy Process to Promote Its Genome Replication. J. Virol. 2020, 94, 4. [Google Scholar] [CrossRef]
- Xu, W.L.; Wang, S.H.; Sun, W.B.; Gao, J.; Ding, X.M.; Kong, J.; Xu, L.; Ke, S. Insufficient radiofrequency ablation-induced autophagy contributes to the rapid progression of residual hepatocellular carcinoma through the HIF-1α/BNIP3 signaling pathway. BMB Rep. 2019, 52, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Song, F.; Wang, B.; Li, K.; Tian, M.; Yu, M.; Pan, X.; Shi, B.; Liu, J.; Gu, J.; et al. The Effect of and Mechanism Underlying Autophagy in Hepatocellular Carcinoma Induced by CH12, a Monoclonal Antibody Directed against Epidermal Growth Factor Receptor Variant III. Cell. Physiol. Biochem. 2018, 46, 226–237. [Google Scholar] [CrossRef]
- Wu, J.Y.; Wang, Z.X.; Zhang, G.; Lu, X.; Qiang, G.H.; Hu, W.; Ji, A.L.; Wu, J.H.; Jiang, C.P. Targeted co-delivery of Beclin 1 siRNA and FTY720 to hepatocellular carcinoma by calcium phosphate nanoparticles for enhanced anticancer efficacy. Int. J. Nanomed. 2018, 13, 1265–1280. [Google Scholar] [CrossRef] [PubMed]
- Han, L.L.; Jia, L.; Wu, F.; Huang, C. Sirtuin6 (SIRT6) Promotes the EMT of Hepatocellular Carcinoma by Stimulating Autophagic Degradation of E-Cadherin. Mol. Cancer Res. 2019, 17, 2267–2280. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ma, Y.; He, H.W.; Zhao, W.L.; Shao, R.G. SPHK1 (sphingosine kinase 1) induces epithelial-mesenchymal transition by promoting the autophagy-linked lysosomal degradation of CDH1/E-cadherin in hepatoma cells. Autophagy 2017, 13, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ni, Y.; Zhang, L.; Jiang, R.; Xu, J.; Yang, H.; Hu, Y.; Qiu, J.; Pu, L.; Tang, J.; et al. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct. Target. Ther. 2021, 6, 76. [Google Scholar] [CrossRef]
- Che, N.; Ng, K.Y.; Wong, T.L.; Tong, M.; Kau, P.W.; Chan, L.H.; Lee, T.K.; Huen, M.S.; Yun, J.P.; Ma, S. PRMT6 deficiency induces autophagy in hostile microenvironments of hepatocellular carcinoma tumors by regulating BAG5-associated HSC70 stability. Cancer Lett. 2021, 501, 247–262. [Google Scholar] [CrossRef]
- Zhang, J.Q.; Li, Y.M.; Liu, T.; He, W.T.; Chen, Y.T.; Chen, X.H.; Li, X.; Zhou, W.C.; Yi, J.F.; Ren, Z.J. Antitumor effect of matrine in human hepatoma G2 cells by inducing apoptosis and autophagy. World J. Gastroenterol. 2010, 16, 4281–4290. [Google Scholar] [CrossRef]
- Yoo, S.H.; Yoon, Y.G.; Lee, J.S.; Song, Y.S.; Oh, J.S.; Park, B.S.; Kwon, T.K.; Park, C.; Choi, Y.H.; Yoo, Y.H. Etoposide induces a mixed type of programmed cell death and overcomes the resistance conferred by Bcl-2 in Hep3B hepatoma cells. Int. J. Oncol. 2012, 41, 1443–1454. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, E.; Zhang, L.; Zhou, B. PSPH induces cell autophagy and promotes cell proliferation and invasion in the hepatocellular carcinoma cell line Huh7 via the AMPK/mTOR/ULK1 signaling pathway. Cell Biol. Int. 2021, 45, 305–319. [Google Scholar] [CrossRef]
- Zhao, M.; Ying, L.; Wang, R.; Yao, J.; Zhu, L.; Zheng, M.; Chen, Z.; Yang, Z. DHX15 Inhibits Autophagy and the Proliferation of Hepatoma Cells. Front. Med. 2020, 7, 591736. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. CCND1 silencing suppresses liver cancer stem cell differentiation through inhibiting autophagy. Hum. Cell 2020, 33, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Do, H.Q.; Luong, A.B.; Bonazza, D.; Bottin, C.; Doan, T.P.; Tran, L.D.; Truong, N.H.; Tell, G.; Pham, H.L.; Tiribelli, C.; et al. Differential capacity of CD90+ cells in autophagy activation following chemotherapy in hepatocellular carcinoma. Ann. Hepatol. 2020, 19, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Alawyia, B.; Constantinou, C. Hepatocellular Carcinoma: A Narrative Review on Current Knowledge and Future Prospects. Curr. Treat. Options Oncol. 2023, 24, 711–724. [Google Scholar] [CrossRef]
- Jindal, A.; Thadi, A.; Shailubhai, K. Hepatocellular carcinoma: Etiology and current and future drugs. J. Clin. Exp. Hepatol. 2019, 9, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Ladd, A.; Duarte, S.; Sahin, I.; Zarrinpar, A. Mechanisms of drug resistance in hepatocellular carcinoma. Hepatology 2023. [Google Scholar] [CrossRef]
- Marin, J.J.; Macias, R.I.; Monte, M.J.; Romero, M.R.; Asensio, M.; Sanchez-Martin, A.; Cives-Losada, C.; Temprano, A.G.; Espinosa-Escudero, R.; Reviejo, M. Molecular bases of drug resistance in hepatocellular carcinoma. Cancers 2020, 12, 1663. [Google Scholar] [CrossRef]
- Mele, L.; Del Vecchio, V.; Liccardo, D.; Prisco, C.; Schwerdtfeger, M.; Robinson, N.; Desiderio, V.; Tirino, V.; Papaccio, G.; La Noce, M. The role of autophagy in resistance to targeted therapies. Cancer Treat. Rev. 2020, 88, 102043. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, Y.; Liu, S.; Sun, Z.; Wang, C.; Li, L.; Zhou, W.; Cheng, S. 14-3-3ζ binds to and stabilizes phospho-beclin 1(S295) and induces autophagy in hepatocellular carcinoma cells. J. Cell. Mol. Med. 2020, 24, 954–964. [Google Scholar] [CrossRef]
- Yang, Y.; Liao, Y.; Gui, Y.P.; Zhao, L.; Guo, L.B. GL-V9 reverses adriamycin resistance in hepatocellular carcinoma cells by affecting JNK2-related autophagy. Chin. J. Nat. Med. 2020, 18, 491–499. [Google Scholar] [CrossRef]
- Ni, Z.; Wang, B.; Dai, X.; Ding, W.; Yang, T.; Li, X.; Lewin, S.; Xu, L.; Lian, J.; He, F. HCC cells with high levels of Bcl-2 are resistant to ABT-737 via activation of the ROS-JNK-autophagy pathway. Free Radic. Biol. Med. 2014, 70, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Li, X.; Zhang, D.; Xie, Y.; Sun, B.; Li, H.; Sun, L.; Zhang, X. B-cell lymphoma 2 inhibitor ABT-737 induces Beclin1- and reactive oxygen species-dependent autophagy in Adriamycin-resistant human hepatocellular carcinoma cells. Tumour Biol. 2017, 39, 1010428317695965. [Google Scholar] [CrossRef]
- Cheng, P.; Ni, Z.; Dai, X.; Wang, B.; Ding, W.; Rae Smith, A.; Xu, L.; Wu, D.; He, F.; Lian, J. The novel BH-3 mimetic apogossypolone induces Beclin-1- and ROS-mediated autophagy in human hepatocellular carcinoma [corrected] cells. Cell Death Dis. 2013, 4, e489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yang, M.; Qu, Z.; Zhou, J.; Jiang, Q. Autophagy prevention sensitizes AKTi-1/2-induced anti-hepatocellular carcinoma cell activity in vitro and in vivo. Biochem. Biophys. Res. Commun. 2016, 480, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wang, H.; Liang, B.; Liu, G.; Tang, M.; Jia, R.; Fan, X.; Jing, W.; Zhou, X.; Wang, H.; et al. Downregulation of ASPP2 improves hepatocellular carcinoma cells survival via promoting BECN1-dependent autophagy initiation. Cell Death Dis. 2016, 7, e2512. [Google Scholar] [CrossRef]
- Tong, Y.; Huang, H.; Pan, H. Inhibition of MEK/ERK activation attenuates autophagy and potentiates pemetrexed-induced activity against HepG2 hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2015, 456, 86–91. [Google Scholar] [CrossRef]
- Zai, W.; Chen, W.; Han, Y.; Wu, Z.; Fan, J.; Zhang, X.; Luan, J.; Tang, S.; Jin, X.; Fu, X.; et al. Targeting PARP and autophagy evoked synergistic lethality in hepatocellular carcinoma. Carcinogenesis 2020, 41, 345–357. [Google Scholar] [CrossRef]
- Han, R.; Li, S. Regorafenib delays the proliferation of hepatocellular carcinoma by inducing autophagy. Pharmazie 2018, 73, 218–222. [Google Scholar] [CrossRef]
- Niture, S.; Gyamfi, M.A.; Lin, M.; Chimeh, U.; Dong, X.; Zheng, W.; Moore, J.; Kumar, D. TNFAIP8 regulates autophagy, cell steatosis, and promotes hepatocellular carcinoma cell proliferation. Cell Death Dis. 2020, 11, 178. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, H.; Xing, X.; Ma, Y.; Ji, S.; Xu, X.; Zhao, X.; Wang, S.; Jiang, W.; Fang, C.; et al. CD13 Induces Autophagy to Promote Hepatocellular Carcinoma Cell Chemoresistance through the P38/Hsp27/CREB/ATG7 Pathway. J. Pharmacol. Exp. Ther. 2020, 374, 512–520. [Google Scholar] [CrossRef]
- Yuan, H.; Li, A.J.; Ma, S.L.; Cui, L.J.; Wu, B.; Yin, L.; Wu, M.C. Inhibition of autophagy significantly enhances combination therapy with sorafenib and HDAC inhibitors for human hepatoma cells. World J. Gastroenterol. 2014, 20, 4953–4962. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Dong, X.; Zhai, B.; Jiang, X.; Dong, D.; Li, B.; Jiang, H.; Xu, S.; Sun, X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget 2015, 6, 28867–28881. [Google Scholar] [CrossRef] [PubMed]
- Dai, N.; Ye, R.; He, Q.; Guo, P.; Chen, H.; Zhang, Q. Capsaicin and sorafenib combination treatment exerts synergistic anti-hepatocellular carcinoma activity by suppressing EGFR and PI3K/Akt/mTOR signaling. Oncol. Rep. 2018, 40, 3235–3248. [Google Scholar] [PubMed]
- Pan, J.; Zhang, M.; Dong, L.; Ji, S.; Zhang, J.; Zhang, S.; Lin, Y.; Wang, X.; Ding, Z.; Qiu, S.; et al. Genome-Scale CRISPR screen identifies LAPTM5 driving lenvatinib resistance in hepatocellular carcinoma. Autophagy 2023, 19, 1184–1198. [Google Scholar] [CrossRef]
- Li, D.; Yao, Y.; Rao, Y.; Huang, X.; Wei, L.; You, Z.; Zheng, G.; Hou, X.; Su, Y.; Varghese, Z.; et al. Cholesterol sensor SCAP contributes to sorafenib resistance by regulating autophagy in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2022, 41, 116. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y. The role and mechanism of action of mitophagy in various liver diseases. Antioxid. Redox Signal. 2023, 38, 529–549. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Chao, X.; Ding, W.X. A PINK1-mediated mitophagy pathway decides the fate of tumors-to be benign or malignant? Autophagy 2018, 14, 563–566. [Google Scholar] [CrossRef]
- Lu, Y.; Li, Z.; Zhang, S.; Zhang, T.; Liu, Y.; Zhang, L. Cellular mitophagy: Mechanism, roles in diseases and small molecule pharmacological regulation. Theranostics 2023, 13, 736–766. [Google Scholar] [CrossRef]
- Feng, J.; Zhou, J.; Wu, Y.; Shen, H.M.; Peng, T.; Lu, G.D. Targeting mitophagy as a novel therapeutic approach in liver cancer. Autophagy 2023, 19, 2164–2165. [Google Scholar] [CrossRef]
- Kawajiri, S.; Saiki, S.; Sato, S.; Sato, F.; Hatano, T.; Eguchi, H.; Hattori, N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010, 584, 1073–1079. [Google Scholar] [CrossRef]
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.S.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010, 189, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; McKeen, T.; Zhang, J.; Ding, W.X. Role and Mechanisms of Mitophagy in Liver Diseases. Cells 2020, 9, 837. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Y.; Yang, B.; Zhou, L.; Ren, F.; Duan, Z.P.; Ma, Y.J. Promotion of mitochondrial energy metabolism during hepatocyte apoptosis in a rat model of acute liver failure. Mol. Med. Rep. 2015, 12, 5035–5041. [Google Scholar] [CrossRef]
- Chi, H.C.; Chen, S.L.; Lin, S.L.; Tsai, C.Y.; Chuang, W.Y.; Lin, Y.H.; Huang, Y.H.; Tsai, M.M.; Yeh, C.T.; Lin, K.H. Thyroid hormone protects hepatocytes from HBx-induced carcinogenesis by enhancing mitochondrial turnover. Oncogene 2017, 36, 5274–5284. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Siraj, S.; Jin, H.; Fan, Y.; Yang, X.; Huang, X.; Wang, X.; Wang, J.; Liu, L.; et al. FUN14 Domain-Containing 1-Mediated Mitophagy Suppresses Hepatocarcinogenesis by Inhibition of Inflammasome Activation in Mice. Hepatology 2019, 69, 604–621. [Google Scholar] [CrossRef]
- Prieto-Domínguez, N.; Ordóñez, R.; Fernández, A.; García-Palomo, A.; Muntané, J.; González-Gallego, J.; Mauriz, J.L. Modulation of autophagy by sorafenib: Effects on treatment response. Front. Pharmacol. 2016, 7, 151. [Google Scholar] [CrossRef]
- Prieto-Domínguez, N.; Méndez-Blanco, C.; Carbajo-Pescador, S.; Fondevila, F.; García-Palomo, A.; González-Gallego, J.; Mauriz, J.L. Melatonin enhances sorafenib actions in human hepatocarcinoma cells by inhibiting mTORC1/p70S6K/HIF-1α and hypoxia-mediated mitophagy. Oncotarget 2017, 8, 91402. [Google Scholar] [CrossRef]
- Wu, H.; Wang, T.; Liu, Y.; Li, X.; Xu, S.; Wu, C.; Zou, H.; Cao, M.; Jin, G.; Lang, J. Mitophagy promotes sorafenib resistance through hypoxia-inducible ATAD3A dependent Axis. J. Exp. Clin. Cancer Res. 2020, 39, 274. [Google Scholar] [CrossRef]
- Liu, Z.; Ren, B.; Wang, Y.; Zou, C.; Qiao, Q.; Diao, Z.; Mi, Y.; Zhu, D.; Liu, X. Sesamol Induces Human Hepatocellular Carcinoma Cells Apoptosis by Impairing Mitochondrial Function and Suppressing Autophagy. Sci. Rep. 2017, 7, 45728. [Google Scholar] [CrossRef]
- Wei, R.; Cao, J.; Yao, S. Matrine promotes liver cancer cell apoptosis by inhibiting mitophagy and PINK1/Parkin pathways. Cell Stress Chaperones 2018, 23, 1295–1309. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Wang, H.; Li, Y.; Xiao, Y.; Zhao, L.; Zhang, T.; Zhou, S.; Zhou, X.; Li, Y.; Shou, Z.; et al. Alantolactone induces apoptosis through ROS-mediated AKT pathway and inhibition of PINK1-mediated mitophagy in human HepG2 cells. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1961–1970. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, H.N.; Wang, K.; Zhang, L.; Huang, Z.; Liu, J.; Zhang, Z.; Luo, M.; Lei, Y.; Peng, Y.; et al. Ketoconazole exacerbates mitophagy to induce apoptosis by downregulating cyclooxygenase-2 in hepatocellular carcinoma. J. Hepatol. 2019, 70, 66–77. [Google Scholar] [CrossRef]
- Chen, H.N.; Chen, Y.; Zhou, Z.G.; Wei, Y.; Huang, C. A novel role for ketoconazole in hepatocellular carcinoma treatment: Linking PTGS2 to mitophagy machinery. Autophagy 2019, 15, 733–734. [Google Scholar] [CrossRef]
- Liu, K.; Lee, J.; Kim, J.Y.; Wang, L.; Tian, Y.; Chan, S.T.; Cho, C.; Machida, K.; Chen, D.; Ou, J.J. Mitophagy Controls the Activities of Tumor Suppressor p53 to Regulate Hepatic Cancer Stem Cells. Mol. Cell 2017, 68, 281–292.e5. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Wang, J.; Liu, F.; Zhang, Y. Blocking Parkin/PINK1-mediated mitophagy sensitizes hepatocellular carcinoma cells to sanguinarine-induced mitochondrial apoptosis. Toxicol. In Vitro 2020, 66, 104840. [Google Scholar] [CrossRef]
- Yao, N.; Wang, C.; Hu, N.; Li, Y.; Liu, M.; Lei, Y.; Chen, M.; Chen, L.; Chen, C.; Lan, P.; et al. Inhibition of PINK1/Parkin-dependent mitophagy sensitizes multidrug-resistant cancer cells to B5G1, a new betulinic acid analog. Cell Death Dis. 2019, 10, 232. [Google Scholar] [CrossRef] [PubMed]
- Inokuchi, S.; Yoshizumi, T.; Toshima, T.; Itoh, S.; Yugawa, K.; Harada, N.; Mori, H.; Fukuhara, T.; Matsuura, Y.; Mori, M. Suppression of optineurin impairs the progression of hepatocellular carcinoma through regulating mitophagy. Cancer Med. 2021, 10, 1501–1514. [Google Scholar] [CrossRef]
- Zeng, Y.; Guo, Z.; Hu, Z.; Liu, M.; Chen, Y.; Chen, S.; Peng, B.; Zhang, P.; Wu, Z.; Luo, H.; et al. FGD1 exhibits oncogenic properties in hepatocellular carcinoma through regulating cell morphology, autophagy and mitochondrial function. Biomed. Pharmacother. 2020, 125, 110029. [Google Scholar] [CrossRef]
- Zhou, J.; Feng, J.; Wu, Y.; Dai, H.-Q.; Zhu, G.-Z.; Chen, P.-H.; Wang, L.-M.; Lu, G.; Liao, X.-W.; Lu, P.-Z.; et al. Simultaneous treatment with sorafenib and glucose restriction inhibits hepatocellular carcinoma in vitro and in vivo by impairing SIAH1-mediated mitophagy. Exp. Mol. Med. 2022, 54, 2007–2021. [Google Scholar] [CrossRef]
- Li, W.; He, P.; Huang, Y.; Li, Y.F.; Lu, J.; Li, M.; Kurihara, H.; Luo, Z.; Meng, T.; Onishi, M.; et al. Selective autophagy of intracellular organelles: Recent research advances. Theranostics 2021, 11, 222–256. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ni, H.M.; Guo, F.; Ding, Y.; Shi, Y.H.; Lahiri, P.; Fröhlich, L.F.; Rülicke, T.; Smole, C.; Schmidt, V.C.; et al. Sequestosome 1/p62 Protein Is Associated with Autophagic Removal of Excess Hepatic Endoplasmic Reticulum in Mice. J. Biol. Chem. 2016, 291, 18663–18674. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Luo, S.; Wang, X.; Li, C.; Yang, J.; Zhu, X.; Xiao, L.; Sun, L. ER-Phagy: A New Regulator of ER Homeostasis. Front. Cell Dev. Biol. 2021, 9, 684526. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Parson, C.; Ding, W.X. Regulation of the homeostasis of hepatic endoplasmic reticulum and cytochrome P450 enzymes by autophagy. Liver Res. 2018, 2, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Fan, Y.; Li, J.; Cheng, B.; Lin, W.; Li, X.; Du, J.; Ling, C. Inhibition of cFLIP overcomes acquired resistance to sorafenib via reducing ER stress-related autophagy in hepatocellular carcinoma. Oncol. Rep. 2018, 40, 2206–2214. [Google Scholar] [CrossRef] [PubMed]
- Minami, Y.; Hoshino, A.; Higuchi, Y.; Hamaguchi, M.; Kaneko, Y.; Kirita, Y.; Taminishi, S.; Nishiji, T.; Taruno, A.; Fukui, M.; et al. Liver lipophagy ameliorates nonalcoholic steatohepatitis through extracellular lipid secretion. Nat. Commun. 2023, 14, 4084. [Google Scholar] [CrossRef]
- Ding, W.X.; Li, M.; Yin, X.M. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy 2011, 7, 248–249. [Google Scholar] [CrossRef]
- Czaja, M.J. Function of Autophagy in Nonalcoholic Fatty Liver Disease. Dig. Dis. Sci. 2016, 61, 1304–1313. [Google Scholar] [CrossRef]
- Daher, D.; Dahan, K.S.E.; Singal, A.G. Non-alcoholic fatty liver disease-related hepatocellular carcinoma. J. Liver Cancer 2023, 23, 127–142. [Google Scholar] [CrossRef]
- Berardi, D.E.; Bock-Hughes, A.; Terry, A.R.; Drake, L.E.; Bozek, G.; Macleod, K.F. Lipid droplet turnover at the lysosome inhibits growth of hepatocellular carcinoma in a BNIP3-dependent manner. Sci. Adv. 2022, 8, eabo2510. [Google Scholar] [CrossRef] [PubMed]
- Dong-Hyung, C.; Yi Sak, K.; Doo Sin, J.; Seong-Kyu, C.; Eun-Kyeong, J. Pexophagy: Molecular Mechanisms and Implications for Health and Diseases. Mol. Cells 2018, 41, 55–64. [Google Scholar] [CrossRef]
- Terlecky, S.R.; Terlecky, L.J.; Giordano, C.R. Peroxisomes, oxidative stress, and inflammation. World J. Biol. Chem. 2012, 3, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Chung, F.L. Oxidative stress and hepatocarcinogenesis. Hepatoma Res. 2018, 4, 39. [Google Scholar] [CrossRef]
- Dutta, R.K.; Maharjan, Y.; Lee, J.N.; Park, C.; Ho, Y.S.; Park, R. Catalase deficiency induces reactive oxygen species mediated pexophagy and cell death in the liver during prolonged fasting. Biofactors 2021, 47, 112–125. [Google Scholar] [CrossRef]
- Yang, Y.P.; Hu, L.F.; Zheng, H.F.; Mao, C.J.; Hu, W.D.; Xiong, K.P.; Wang, F.; Liu, C.F. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol. Sin. 2013, 34, 625–635. [Google Scholar] [CrossRef]
- Zhao, X.G.; Sun, R.J.; Yang, X.Y.; Liu, D.Y.; Lei, D.P.; Jin, T.; Pan, X.L. Chloroquine-enhanced efficacy of cisplatin in the treatment of hypopharyngeal carcinoma in xenograft mice. PLoS ONE 2015, 10, e0126147. [Google Scholar] [CrossRef]
- Shi, Y.H.; Ding, Z.B.; Zhou, J.; Hui, B.; Shi, G.M.; Ke, A.W.; Wang, X.Y.; Dai, Z.; Peng, Y.F.; Gu, C.Y.; et al. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy 2011, 7, 1159–1172. [Google Scholar] [CrossRef]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Amaravadi, R.K.; Winkler, J.D. Lys05: A new lysosomal autophagy inhibitor. Autophagy 2012, 8, 1383–1384. [Google Scholar] [CrossRef]
- Gade, T.P.F.; Tucker, E.; Nakazawa, M.S.; Hunt, S.J.; Wong, W.; Krock, B.; Weber, C.N.; Nadolski, G.J.; Clark, T.W.I.; Soulen, M.C.; et al. Ischemia Induces Quiescence and Autophagy Dependence in Hepatocellular Carcinoma. Radiology 2017, 283, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Tagawa, Y.; Yoshimori, T.; Moriyama, Y.; Masaki, R.; Tashiro, Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998, 23, 33–42. [Google Scholar] [CrossRef]
- Yan, Y.; Jiang, K.; Liu, P.; Zhang, X.; Dong, X.; Gao, J.; Liu, Q.; Barr, M.P.; Zhang, Q.; Hou, X.; et al. Bafilomycin A1 induces caspase-independent cell death in hepatocellular carcinoma cells via targeting of autophagy and MAPK pathways. Sci. Rep. 2016, 6, 37052. [Google Scholar] [CrossRef]
- Dengjel, J.; Høyer-Hansen, M.; Nielsen, M.O.; Eisenberg, T.; Harder, L.M.; Schandorff, S.; Farkas, T.; Kirkegaard, T.; Becker, A.C.; Schroeder, S.; et al. Identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol. Cell. Proteom. 2012, 11, M111.014035. [Google Scholar] [CrossRef]
- Goodall, M.L.; Wang, T.; Martin, K.R.; Kortus, M.G.; Kauffman, A.L.; Trent, J.M.; Gately, S.; MacKeigan, J.P. Development of potent autophagy inhibitors that sensitize oncogenic BRAF V600E mutant melanoma tumor cells to vemurafenib. Autophagy 2014, 10, 1120–1136. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Lui, V.W.; Yeo, W. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol. 2011, 7, 1149–1167. [Google Scholar] [CrossRef] [PubMed]
- Michl, J.; Silverstein, S.C. Role of macrophage receptors in the ingestion phase of phagocytosis. Birth Defects Orig. Artic. Ser. 1978, 14, 99–117. [Google Scholar] [PubMed]
- Feng, R.; Wang, S.Y.; Shi, Y.H.; Fan, J.; Yin, X.M. Delphinidin induces necrosis in hepatocellular carcinoma cells in the presence of 3-methyladenine, an autophagy inhibitor. J. Agric. Food Chem. 2010, 58, 3957–3964. [Google Scholar] [CrossRef]
- Ma, J.; Xie, S.L.; Geng, Y.J.; Jin, S.; Wang, G.Y.; Lv, G.Y. In vitro regulation of hepatocellular carcinoma cell viability, apoptosis, invasion, and AEG-1 expression by LY294002. Clin. Res. Hepatol. Gastroenterol. 2014, 38, 73–80. [Google Scholar] [CrossRef]
- Jaber, N.; Dou, Z.; Chen, J.S.; Catanzaro, J.; Jiang, Y.P.; Ballou, L.M.; Selinger, E.; Ouyang, X.; Lin, R.Z.; Zhang, J.; et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. USA 2012, 109, 2003–2008. [Google Scholar] [CrossRef]
- Dyczynski, M.; Yu, Y.; Otrocka, M.; Parpal, S.; Braga, T.; Henley, A.B.; Zazzi, H.; Lerner, M.; Wennerberg, K.; Viklund, J.; et al. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer Lett. 2018, 435, 32–43. [Google Scholar] [CrossRef]
- Noman, M.Z.; Parpal, S.; Van Moer, K.; Xiao, M.; Yu, Y.; Viklund, J.; De Milito, A.; Hasmim, M.; Andersson, M.; Amaravadi, R.K.; et al. Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci. Adv. 2020, 6, eaax7881. [Google Scholar] [CrossRef]
- Janji, B.; Hasmim, M.; Parpal, S.; Berchem, G.; Noman, M.Z. Firing up the cold tumors by targeting Vps34. Oncoimmunology 2020, 9, 1809936. [Google Scholar] [CrossRef]
- Jaber, N.; Mohd-Naim, N.; Wang, Z.; DeLeon, J.L.; Kim, S.; Zhong, H.; Sheshadri, N.; Dou, Z.; Edinger, A.L.; Du, G.; et al. Vps34 regulates Rab7 and late endocytic trafficking through recruitment of the GTPase-activating protein Armus. J. Cell Sci. 2016, 129, 4424–4435. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Q.; Chen, S.; Li, Z.; Fu, L. Targeting VPS34 in autophagy: An update on pharmacological small-molecule compounds. Eur. J. Med. Chem. 2023, 256, 115467. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wu, X.; Qian, Y.; Jiang, X.; Wang, Y.; Gao, J. PIK3C3 regulates the expansion of liver CSCs and PIK3C3 inhibition counteracts liver cancer stem cell activity induced by PI3K inhibitor. Cell Death Dis. 2020, 11, 427. [Google Scholar] [CrossRef]
- Gao, Y.; Zhou, Z.; Zhang, T.; Xue, S.; Li, K.; Jiang, J. Structure-Based Virtual Screening towards the Discovery of Novel ULK1 Inhibitors with Anti-HCC Activities. Molecules 2022, 27, 2627. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.R.; Celano, S.L.; Solitro, A.R.; Gunaydin, H.; Scott, M.; O’Hagan, R.C.; Shumway, S.D.; Fuller, P.; MacKeigan, J.P. A Potent and Selective ULK1 Inhibitor Suppresses Autophagy and Sensitizes Cancer Cells to Nutrient Stress. iScience 2018, 8, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Petherick, K.J.; Conway, O.J.; Mpamhanga, C.; Osborne, S.A.; Kamal, A.; Saxty, B.; Ganley, I.G. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 2015, 290, 11376–11383. [Google Scholar] [CrossRef]
- Xu, Z.; Bao, J.; Jin, X.; Li, H.; Fan, K.; Wu, Z.; Yao, M.; Zhang, Y.; Liu, G.; Wang, D.; et al. The Effects of Cinobufagin on Hepatocellular Carcinoma Cells Enhanced by MRT68921, an Autophagy Inhibitor. Am. J. Chin. Med. 2023, 51, 1595–1611. [Google Scholar] [CrossRef]
- Smith, B.D.; Vogeti, L.; Gupta, A.; Singh, J.; Al-Ani, G.; Bulfer, S.L.; Caldwell, T.M.; Timson, M.J.; Vogeti, S.; Ahn, Y.M.; et al. Abstract B129: Preclinical studies with DCC-3116, an ULK kinase inhibitor designed to inhibit autophagy as a potential strategy to address mutant RAS cancers. Mol. Cancer Ther. 2019, 18, B129. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Hong, D.S.; Vandross, A.L.; Psoinos, C.M.; Brennan, D.M.; Sherman, M.L.; Ruiz-Soto, R.; Reu, F.J.; Weekes, C.D. A phase 1/2 study of DCC-3116 as a single agent and in combination with trametinib in patients with advanced or metastatic solid tumors with RAS or RAF mutations. J. Clin. Oncol. 2022, 40, TPS3178. [Google Scholar] [CrossRef]
- Egan, D.F.; Chun, M.G.; Vamos, M.; Zou, H.; Rong, J.; Miller, C.J.; Lou, H.J.; Raveendra-Panickar, D.; Yang, C.C.; Sheffler, D.J.; et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol. Cell 2015, 59, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Dower, C.M.; Bhat, N.; Gebru, M.T.; Chen, L.; Wills, C.A.; Miller, B.A.; Wang, H.G. Targeted Inhibition of ULK1 Promotes Apoptosis and Suppresses Tumor Growth and Metastasis in Neuroblastoma. Mol. Cancer Ther. 2018, 17, 2365–2376. [Google Scholar] [CrossRef]
- Qiu, L.; Zhou, G.; Cao, S. Targeted inhibition of ULK1 enhances daunorubicin sensitivity in acute myeloid leukemia. Life Sci. 2020, 243, 117234. [Google Scholar] [CrossRef]
- Xue, S.T.; Li, K.; Gao, Y.; Zhao, L.Y.; Gao, Y.; Yi, H.; Jiang, J.D.; Li, Z.R. The role of the key autophagy kinase ULK1 in hepatocellular carcinoma and its validation as a treatment target. Autophagy 2020, 16, 1823–1837. [Google Scholar] [CrossRef]
- Lee, M.J.; Park, J.S.; Jo, S.B.; Joe, Y.A. Enhancing Anti-Cancer Therapy with Selective Autophagy Inhibitors by Targeting Protective Autophagy. Biomol. Ther. 2023, 31, 1–15. [Google Scholar] [CrossRef]
- Fu, Y.; Huang, Z.; Hong, L.; Lu, J.H.; Feng, D.; Yin, X.M.; Li, M. Targeting ATG4 in Cancer Therapy. Cancers 2019, 11, 649. [Google Scholar] [CrossRef]
- Xie, Y.; Fan, S.; Ni, D.; Wan, W.; Xu, P.; Ding, Y.; Zhang, R.; Lu, J.; Zhang, N.; Zhang, Y.; et al. An ATG4B inhibitor blocks autophagy and sensitizes Sorafenib inhibition activities in HCC tumor cells. Bioorg. Med. Chem. 2023, 84, 117262. [Google Scholar] [CrossRef]
- Liu, P.F.; Tsai, K.L.; Hsu, C.J.; Tsai, W.L.; Cheng, J.S.; Chang, H.W.; Shiau, C.W.; Goan, Y.G.; Tseng, H.H.; Wu, C.H.; et al. Drug Repurposing Screening Identifies Tioconazole as an ATG4 Inhibitor that Suppresses Autophagy and Sensitizes Cancer Cells to Chemotherapy. Theranostics 2018, 8, 830–845. [Google Scholar] [CrossRef]
- Fu, Y.; Hong, L.; Xu, J.; Zhong, G.; Gu, Q.; Gu, Q.; Guan, Y.; Zheng, X.; Dai, Q.; Luo, X.; et al. Discovery of a small molecule targeting autophagy via ATG4B inhibition and cell death of colorectal cancer cells in vitro and in vivo. Autophagy 2019, 15, 295–311. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Kim, C.K.; Alvarez, A.A.; Pangeni, R.P.; Wan, X.; Song, X.; Shi, T.; Yang, Y.; Sastry, N.; Horbinski, C.M.; et al. MST4 Phosphorylation of ATG4B Regulates Autophagic Activity, Tumorigenicity, and Radioresistance in Glioblastoma. Cancer Cell 2017, 32, 840–855.e8. [Google Scholar] [CrossRef] [PubMed]
- Tanemura, M.; Ohmura, Y.; Deguchi, T.; Machida, T.; Tsukamoto, R.; Wada, H.; Kobayashi, S.; Marubashi, S.; Eguchi, H.; Ito, T.; et al. Rapamycin causes upregulation of autophagy and impairs islets function both in vitro and in vivo. Am. J. Transplant. 2012, 12, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Hartford, C.M.; Ratain, M.J. Rapamycin: Something old, something new, sometimes borrowed and now renewed. Clin. Pharmacol. Ther. 2007, 82, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Schnitzbauer, A.A.; Filmann, N.; Adam, R.; Bachellier, P.; Bechstein, W.O.; Becker, T.; Bhoori, S.; Bilbao, I.; Brockmann, J.; Burra, P.; et al. mTOR Inhibition Is Most Beneficial after Liver Transplantation for Hepatocellular Carcinoma in Patients with Active Tumors. Ann. Surg. 2020, 272, 855–862. [Google Scholar] [CrossRef]
- Geissler, E.K.; Schnitzbauer, A.A.; Zülke, C.; Lamby, P.E.; Proneth, A.; Duvoux, C.; Burra, P.; Jauch, K.W.; Rentsch, M.; Ganten, T.M.; et al. Sirolimus Use in Liver Transplant Recipients with Hepatocellular Carcinoma: A Randomized, Multicenter, Open-Label Phase 3 Trial. Transplantation 2016, 100, 116–125. [Google Scholar] [CrossRef]
- Kelley, R.K.; Nimeiri, H.S.; Munster, P.N.; Vergo, M.T.; Huang, Y.; Li, C.M.; Hwang, J.; Mulcahy, M.F.; Yeh, B.M.; Kuhn, P.; et al. Temsirolimus combined with sorafenib in hepatocellular carcinoma: A phase I dose-finding trial with pharmacokinetic and biomarker correlates. Ann. Oncol. 2013, 24, 1900–1907. [Google Scholar] [CrossRef]
- Koeberle, D.; Dufour, J.F.; Demeter, G.; Li, Q.; Ribi, K.; Samaras, P.; Saletti, P.; Roth, A.D.; Horber, D.; Buehlmann, M.; et al. Sorafenib with or without everolimus in patients with advanced hepatocellular carcinoma (HCC): A randomized multicenter, multinational phase II trial (SAKK 77/08 and SASL 29). Ann. Oncol. 2016, 27, 856–861. [Google Scholar] [CrossRef]
- Matter, M.S.; Decaens, T.; Andersen, J.B.; Thorgeirsson, S.S. Targeting the mTOR pathway in hepatocellular carcinoma: Current state and future trends. J. Hepatol. 2014, 60, 855–865. [Google Scholar] [CrossRef]
- Xu, F.; Tautenhahn, H.M.; Dirsch, O.; Dahmen, U. Blocking autophagy with chloroquine aggravates lipid accumulation and reduces intracellular energy synthesis in hepatocellular carcinoma cells, both contributing to its anti-proliferative effect. J. Cancer Res. Clin. Oncol. 2022, 148, 3243–3256. [Google Scholar] [CrossRef]
- Chen, M.Y.; Yadav, V.K.; Chu, Y.C.; Ong, J.R.; Huang, T.Y.; Lee, K.F.; Lee, K.H.; Yeh, C.T.; Lee, W.H. Hydroxychloroquine (HCQ) Modulates Autophagy and Oxidative DNA Damage Stress in Hepatocellular Carcinoma to Overcome Sorafenib Resistance via TLR9/SOD1/hsa-miR-30a-5p/Beclin-1 Axis. Cancers 2021, 13, 3227. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.H.; Nguyen, T.M.; Ngoc, D.T.M.; You, T.; Park, M.K.; Lee, C.H. Unraveling the Janus-Faced Role of Autophagy in Hepatocellular Carcinoma: Implications for Therapeutic Interventions. Int. J. Mol. Sci. 2023, 24, 16255. https://doi.org/10.3390/ijms242216255
Nguyen TH, Nguyen TM, Ngoc DTM, You T, Park MK, Lee CH. Unraveling the Janus-Faced Role of Autophagy in Hepatocellular Carcinoma: Implications for Therapeutic Interventions. International Journal of Molecular Sciences. 2023; 24(22):16255. https://doi.org/10.3390/ijms242216255
Chicago/Turabian StyleNguyen, Thi Ha, Tuan Minh Nguyen, Dinh Thi Minh Ngoc, Taesik You, Mi Kyung Park, and Chang Hoon Lee. 2023. "Unraveling the Janus-Faced Role of Autophagy in Hepatocellular Carcinoma: Implications for Therapeutic Interventions" International Journal of Molecular Sciences 24, no. 22: 16255. https://doi.org/10.3390/ijms242216255
APA StyleNguyen, T. H., Nguyen, T. M., Ngoc, D. T. M., You, T., Park, M. K., & Lee, C. H. (2023). Unraveling the Janus-Faced Role of Autophagy in Hepatocellular Carcinoma: Implications for Therapeutic Interventions. International Journal of Molecular Sciences, 24(22), 16255. https://doi.org/10.3390/ijms242216255







