Co-Expression Network Analysis of the Transcriptome Identified Hub Genes and Pathways Responding to Saline–Alkaline Stress in Sorghum bicolor L.
Abstract
:1. Introduction
2. Results
2.1. Phenotype of Sorghum under NaHCO3 Stress
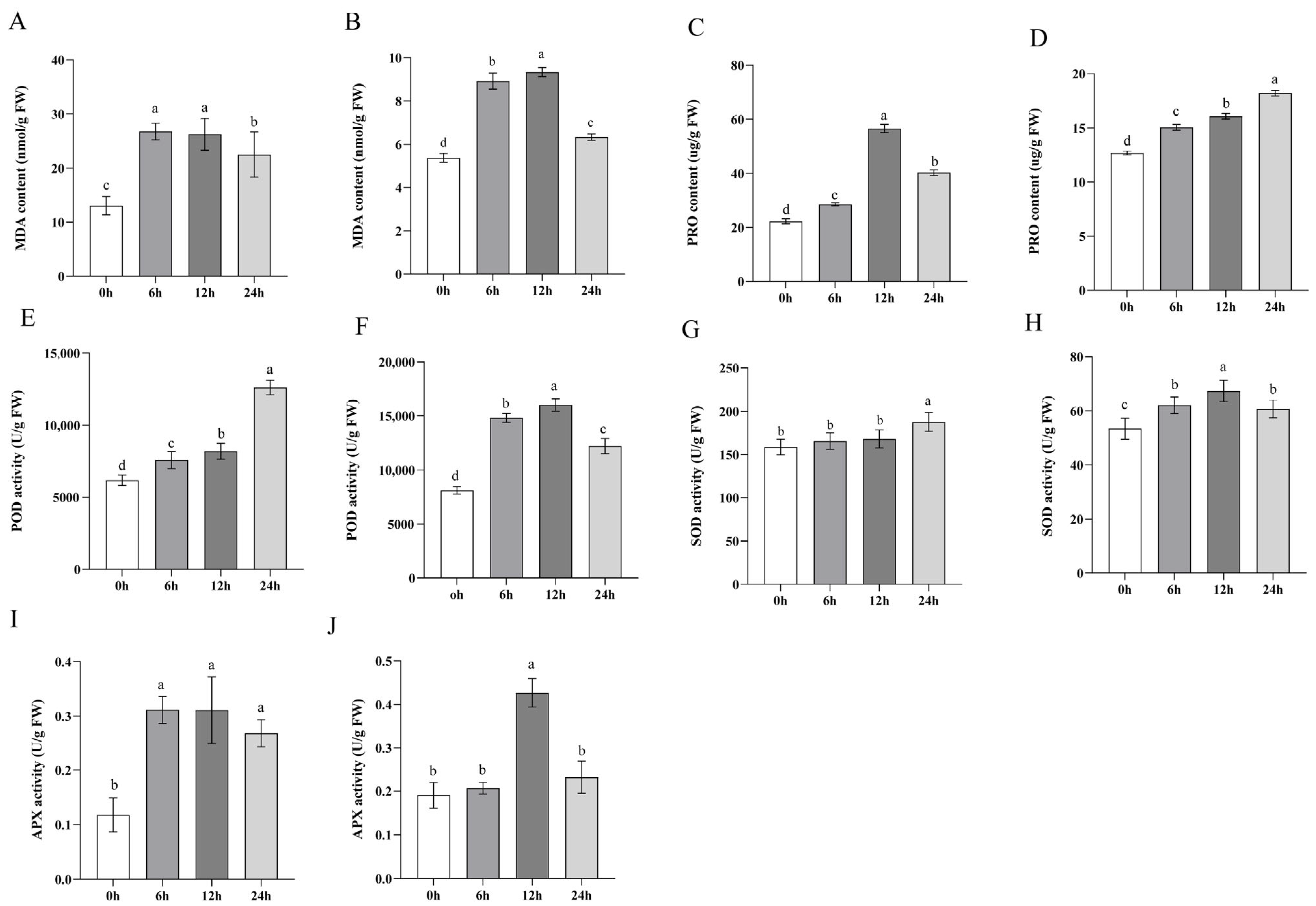
2.2. Antioxidant System Responds to Saline–Alkali Stress
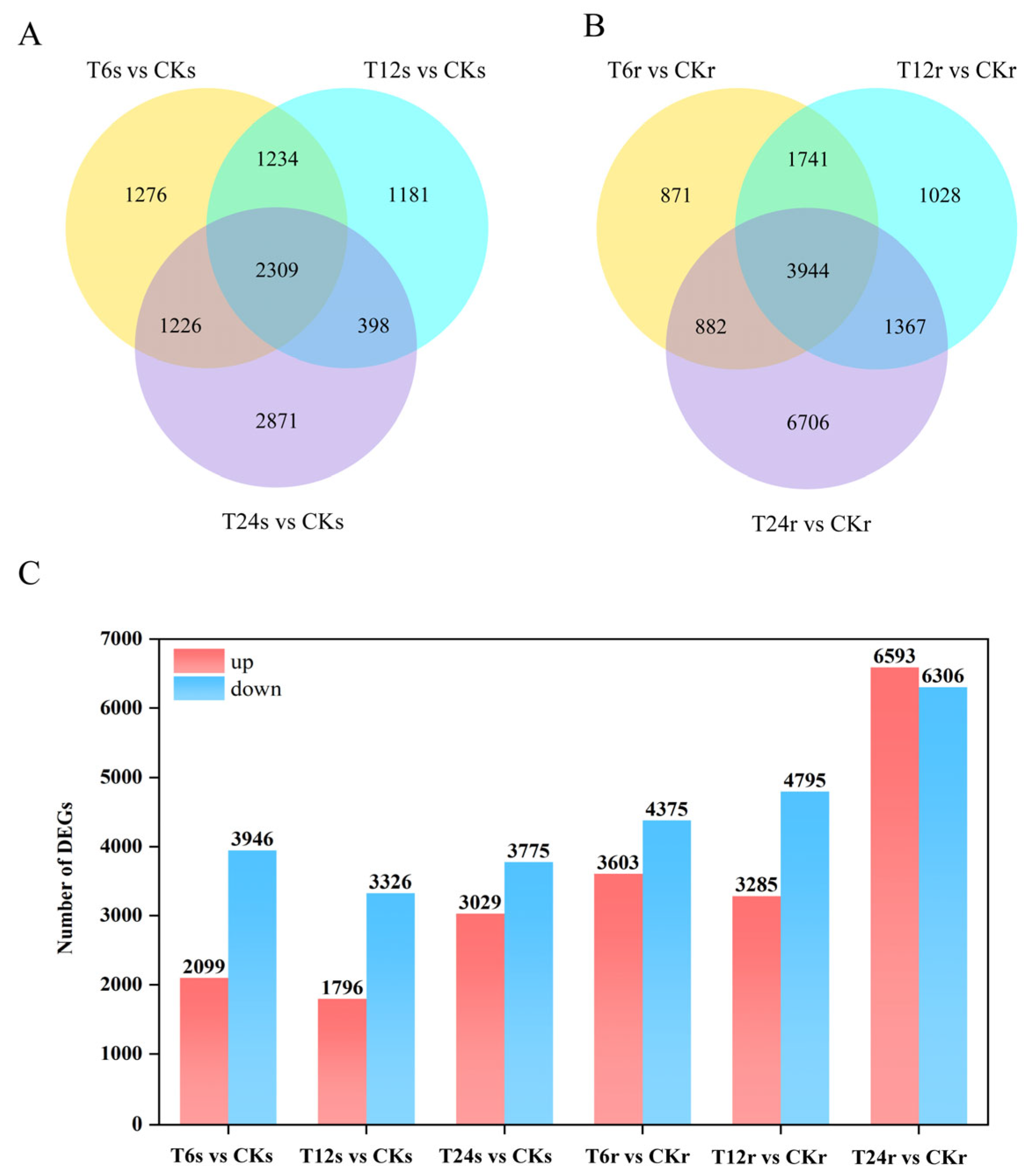
2.3. Statistics of DEGs from RNA-Seq Data
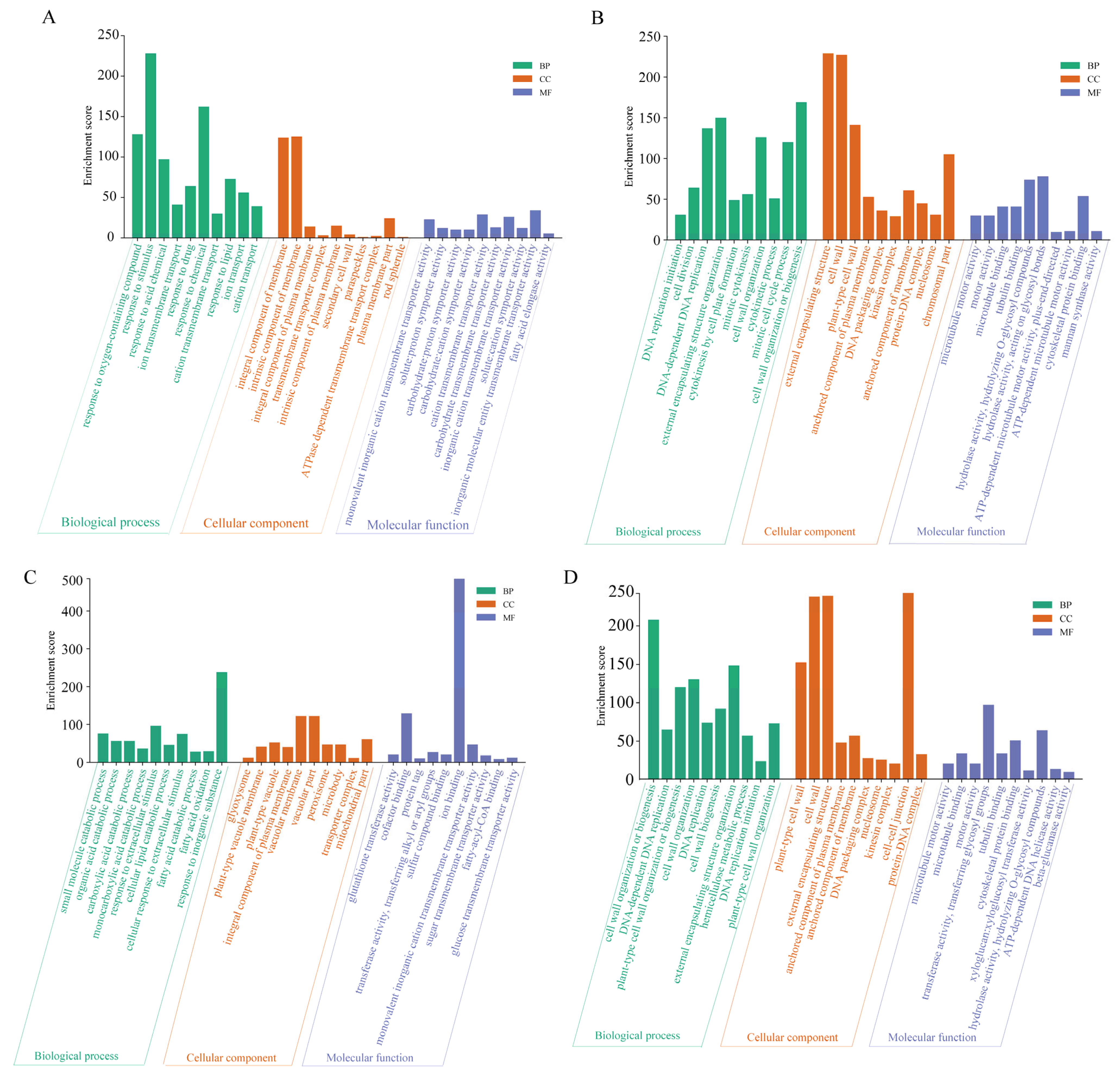
2.4. Biological Function Analysis of DEGs Corresponding to Saline–Alkaline Stress
2.5. Construction of WGCNA and Biological Function Analysis
2.6. PPI Network Validated the Key Hub Genes
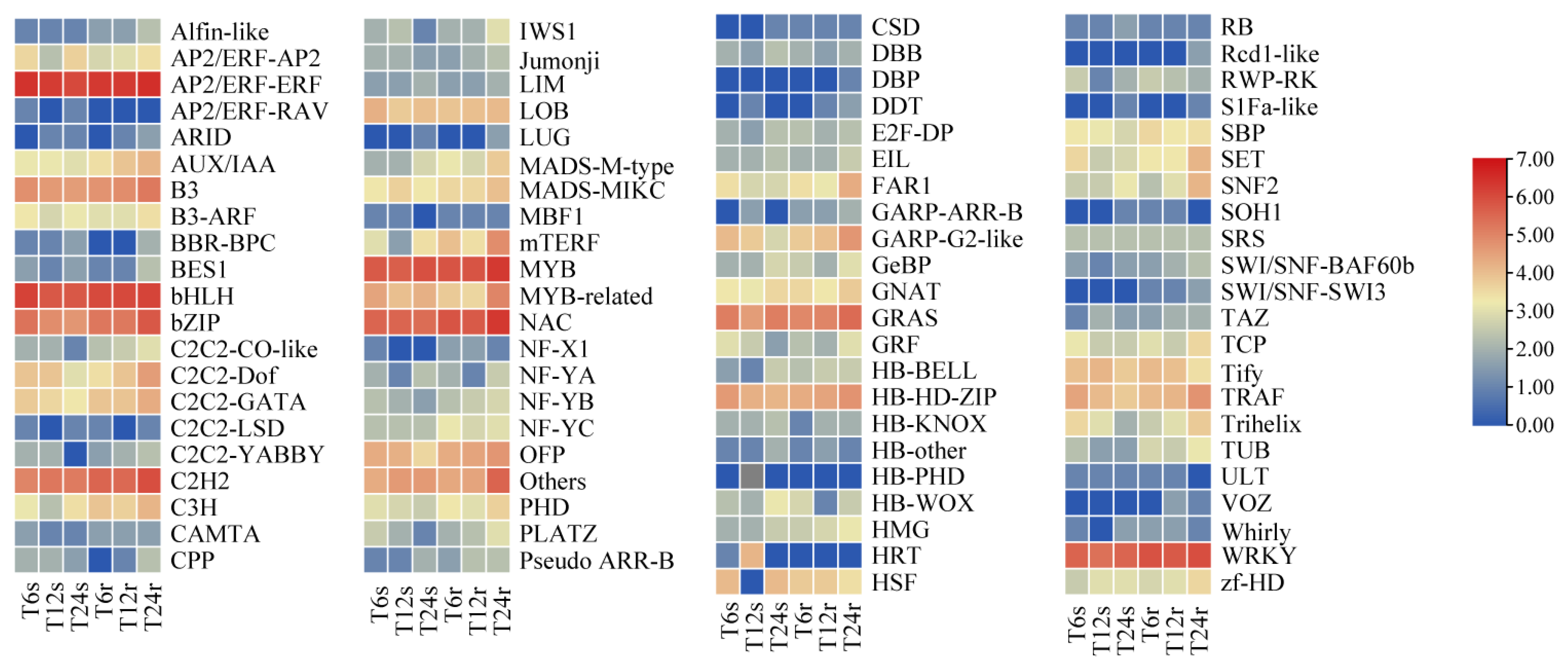
2.7. Identification of DEGs Encoding Transcription Factors
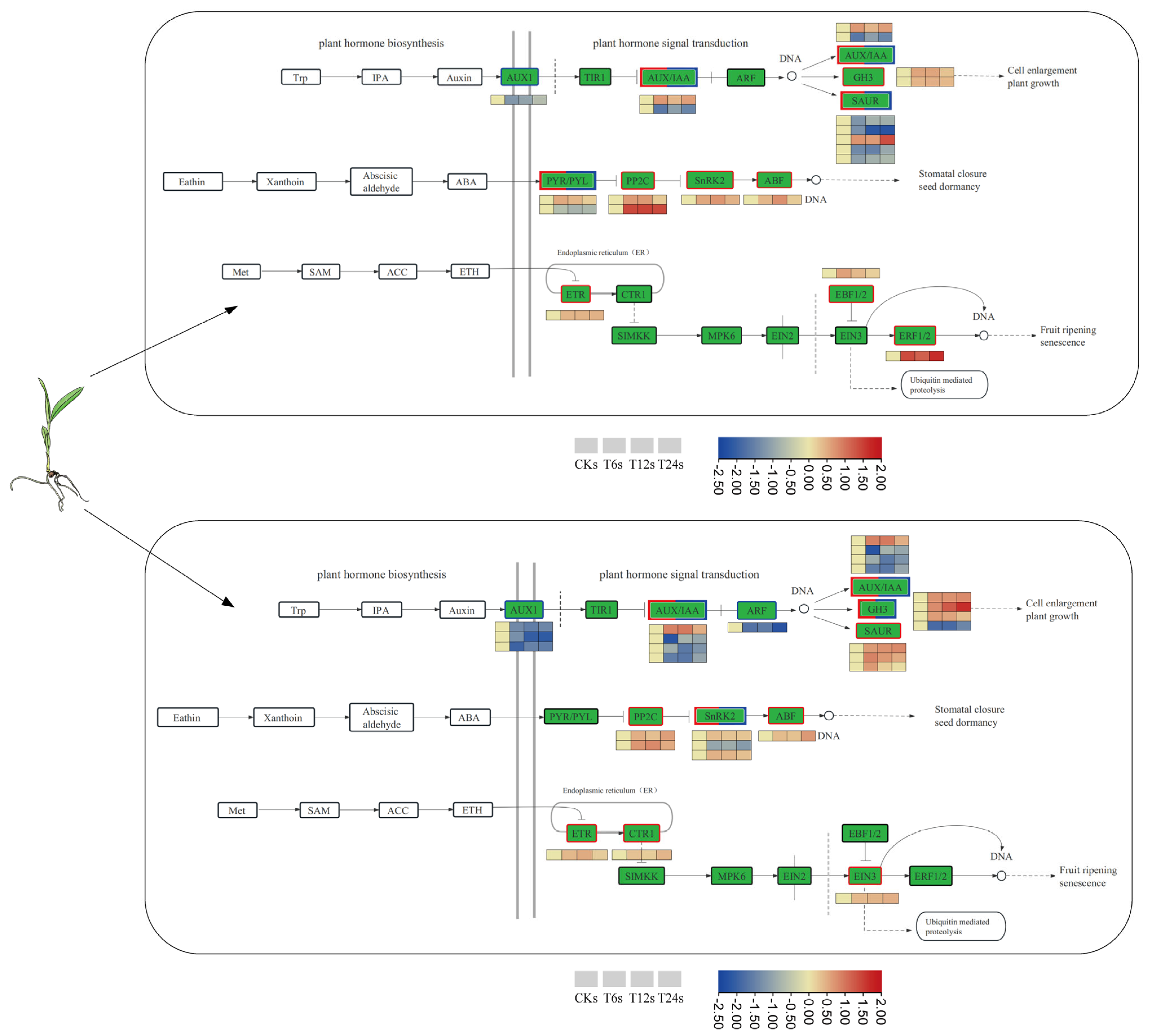
2.8. Analysis of DEGs Involved in Phytohormone Biosynthesis and Signal Transduction
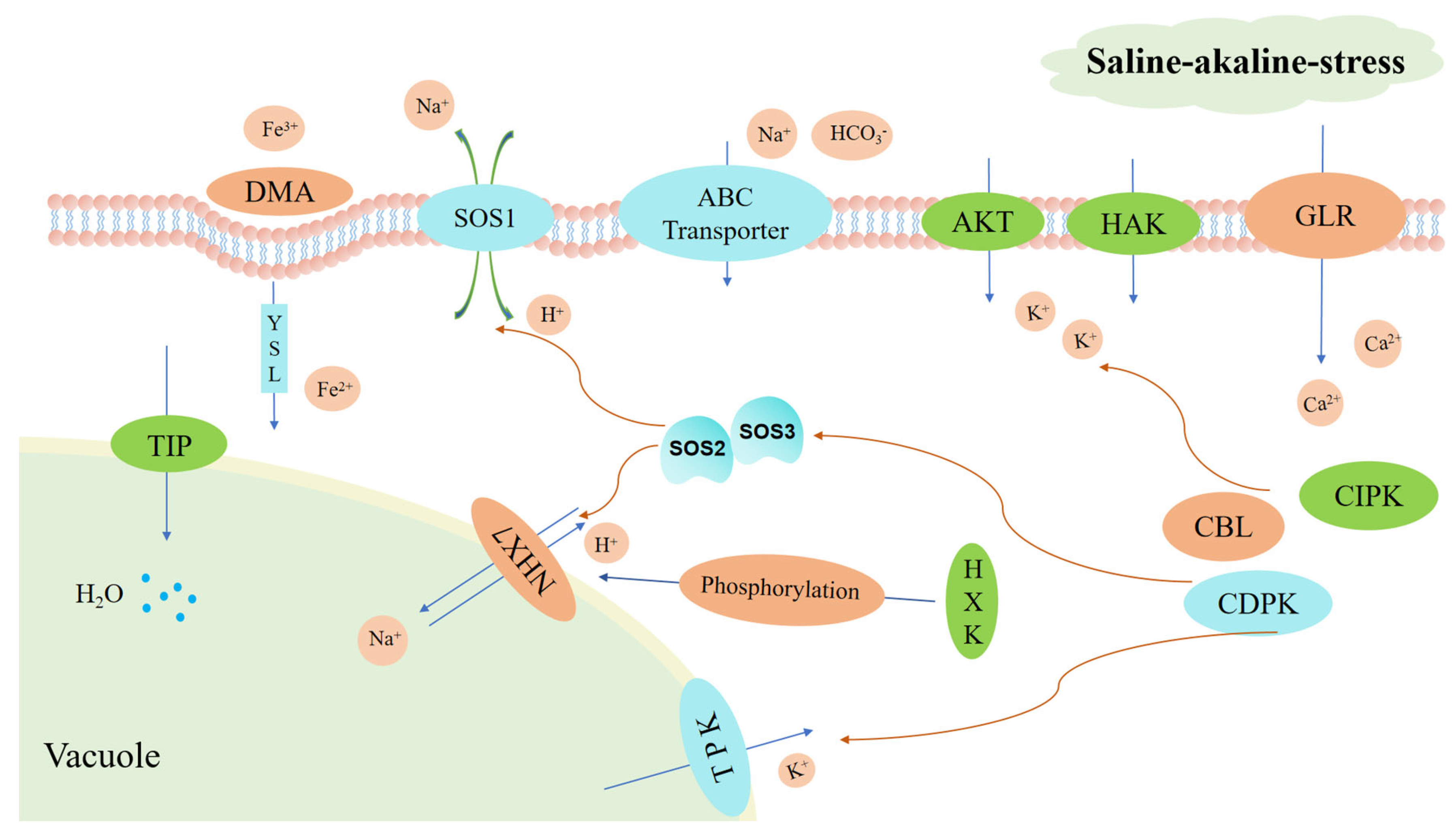
2.9. Analysis of DEGs Involved in Intracellular Ion Transport
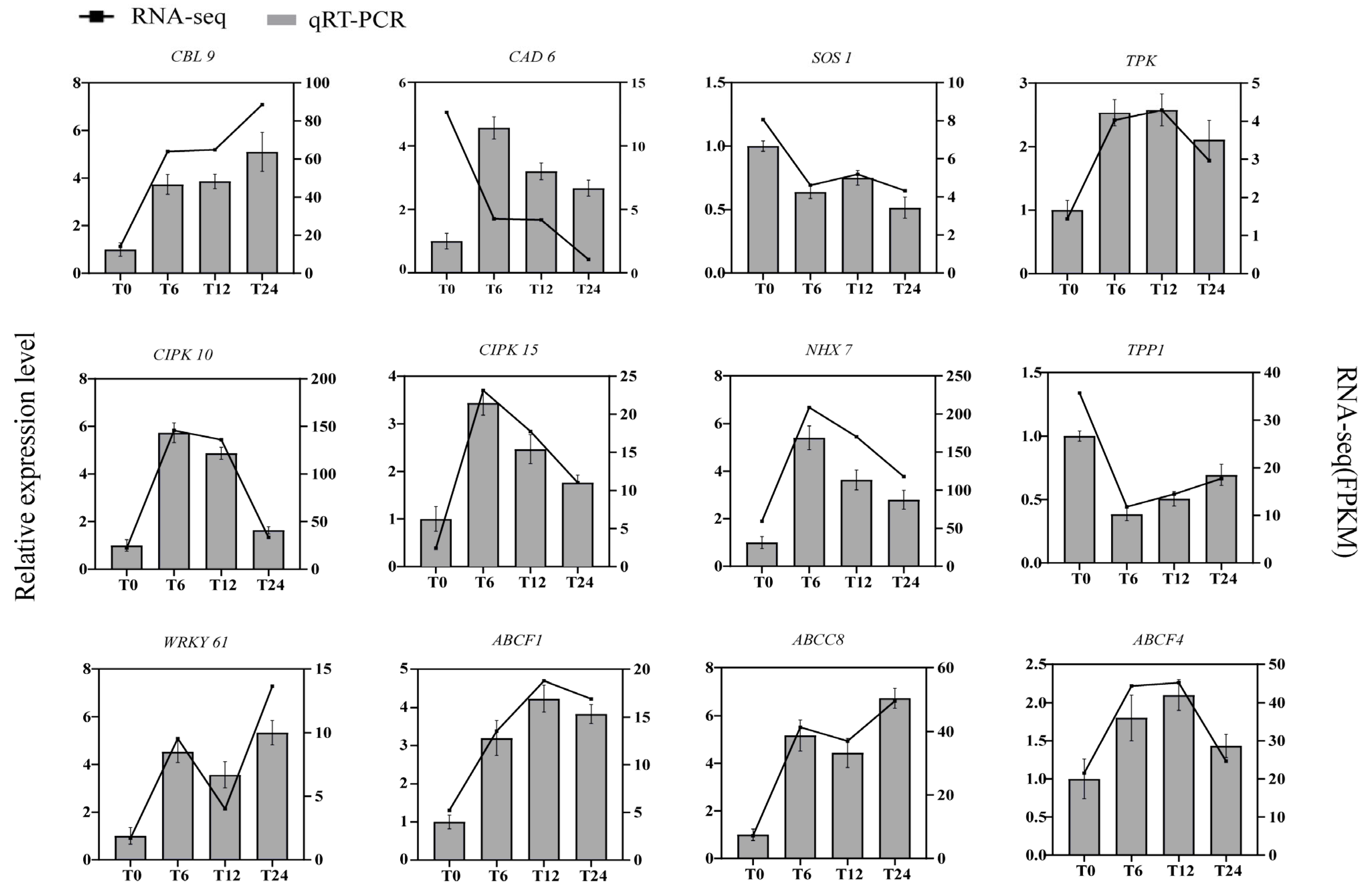
2.10. Validation of RNA-Seq Via RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Plant Culture and Treatments
4.2. Determination of Antioxidant Enzyme Activities, Proline and Malondialdehyde Concentrations
4.3. RNA Extraction and Transcriptome Sequencing
4.4. Transcriptome Data Quality Control and Assembly
4.5. Analysis of Functional Enrichment of DEGs
4.6. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.7. Construction of the Protein–Protein Interaction (PPI) Network
4.8. Identification of Transcription Factors
4.9. Validation using Real-Time Quantitative PCR (RT-qPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CDPK | Calcium-dependent protein kinase |
| CBL | Calcineurin B-like protein |
| CIPK | CBL-interacting protein kinase |
| KT | Potassium transporter |
| NHX | Na+/H+ antiporter |
| AMT | Ammonium transporter |
| NRT | Nitrate transporter |
| WGCNA | Weighted gene co-expression network analysis |
| PPI | Protein–protein interaction |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Feng, C.; Gao, H.; Zhou, Y.; Jing, Y.; Li, S.; Yan, Z.; Xu, K.; Zhou, F.; Zhang, W.; Yang, X.; et al. Unfolding Molecular Switches for Salt Stress Resilience in Soybean: Recent Advances and Prospects for Salt-Tolerant Smart Plant Production. Front. Plant Sci. 2023, 14, 1162014. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to Increasing the Salt Tolerance of Wheat and Other Cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-Tolerant Plant Growth Promoting Rhizobacteria for Enhancing Crop Productivity of Saline Soils. Front. Microbiol. 2019, 10, 2791. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, P.; Kumar, R. Soil Salinity: A Serious Environmental Issue and Plant Growth Promoting Bacteria as One of the Tools for Its Alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.-L.; Gao, Z.-W.; Gao, Y.; Liu, G.-F.; Sheng, L.-X.; Wang, D.-L. Eco-Physiological Characteristics of Alfalfa Seedlings in Response to Various Mixed Salt-Alkaline Stresses. J. Integr. Plant Biol. 2008, 50, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S. Learning from Halophytes: Physiological Basis and Strategies to Improve Abiotic Stress Tolerance in Crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Wei, Y.; Xu, Y.; Lu, P.; Wang, X.; Li, Z.; Cai, X.; Zhou, Z.; Wang, Y.; Zhang, Z.; Lin, Z.; et al. Salt Stress Responsiveness of a Wild Cotton Species (Gossypium klotzschianum) Based on Transcriptomic Analysis. PLoS ONE 2017, 12, e0178313. [Google Scholar] [CrossRef]
- Romano-Armada, N.; Yañez-Yazlle, M.F.; Irazusta, V.P.; Rajal, V.B.; Moraga, N.B. Potential of Bioremediation and PGP Traits in Streptomyces as Strategies for Bio-Reclamation of Salt-Affected Soils for Agriculture. Pathogens 2020, 9, 117. [Google Scholar] [CrossRef]
- Wang, J.; Lin, C.; Han, Z.; Fu, C.; Huang, D.; Cheng, H. Dissolved Nitrogen in Salt-Affected Soils Reclaimed by Planting Rice: How Is It Influenced by Soil Physicochemical Properties? Sci. Total Environ. 2022, 824, 153863. [Google Scholar] [CrossRef]
- Roy, S.J.; Negrão, S.; Tester, M. Salt Resistant Crop Plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Al-Harrasi, I.; Jana, G.A.; Patankar, H.V.; Al-Yahyai, R.; Rajappa, S.; Kumar, P.P.; Yaish, M.W. A Novel Tonoplast Na+/H+ Antiporter Gene from Date Palm (PdNHX6) Confers Enhanced Salt Tolerance Response in Arabidopsis. Plant Cell Rep. 2020, 39, 1079–1093. [Google Scholar] [CrossRef]
- Brini, F.; Hanin, M.; Mezghani, I.; Berkowitz, G.A.; Masmoudi, K. Overexpression of Wheat Na+/H+ Antiporter TNHX1 and H+-Pyrophosphatase TVP1 Improve Salt- and Drought-Stress Tolerance in Arabidopsis Thaliana Plants. J. Exp. Bot. 2006, 58, 301–308. [Google Scholar] [CrossRef]
- Penton, D.; Czogalla, J.; Wengi, A.; Himmerkus, N.; Loffing-Cueni, D.; Carrel, M.; Rajaram, R.D.; Staub, O.; Bleich, M.; Schweda, F.; et al. Extracellular K + Rapidly Controls NaCl Cotransporter Phosphorylation in the Native Distal Convoluted Tubule by Cl−-dependent and Independent Mechanisms. J. Physiol. 2016, 594, 6319–6331. [Google Scholar] [CrossRef]
- Feria, A.B.; Bosch, N.; Sánchez, A.; Nieto-Ingelmo, A.I.; De La Osa, C.; Echevarría, C.; García-Mauriño, S.; Monreal, J.A. Phosphoenolpyruvate Carboxylase (PEPC) and PEPC-Kinase (PEPC-k) Isoenzymes in Arabidopsis Thaliana: Role in Control and Abiotic Stress Conditions. Planta 2016, 244, 901–913. [Google Scholar] [CrossRef]
- Maathuis, F. K+ Nutrition and Na+Toxicity: The Basis of Cellular K+/Na+ Ratios. Ann. Bot. 1999, 84, 123–133. [Google Scholar] [CrossRef]
- Rohilla, P.; Yadav, J.P. Acute Salt Stress Differentially Modulates Nitrate Reductase Expression in Contrasting Salt Responsive Rice Cultivars. Protoplasma 2019, 256, 1267–1278. [Google Scholar] [CrossRef] [PubMed]
- Baghour, M.; Gálvez, F.J.; Sánchez, M.E.; Aranda, M.N.; Venema, K.; Rodríguez-Rosales, M.P. Overexpression of LeNHX2 and SlSOS2 Increases Salt Tolerance and Fruit Production in Double Transgenic Tomato Plants. Plant Physiol. Biochem. 2019, 135, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Hussain, S.; Ali, B.; Ren, X.; Chen, X.; Li, Q.; Saqib, M.; Ahmad, N. Recent Progress in Understanding Salinity Tolerance in Plants: Story of Na+/K+ Balance and Beyond. Plant Physiol. Biochem. 2021, 160, 239–256. [Google Scholar] [CrossRef] [PubMed]
- Talbi Zribi, O.; Abdelly, C.; Debez, A. Interactive Effects of Salinity and Phosphorus Availability on Growth, Water Relations, Nutritional Status and Photosynthetic Activity of Barley (Hordeum vulgare L.): Effects of Salinity and Phosphorus Interaction on Barley. Plant Biol. 2011, 13, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Freyria, N.J.; Kuo, A.; Chovatia, M.; Johnson, J.; Lipzen, A.; Barry, K.W.; Grigoriev, I.V.; Lovejoy, C. Salinity Tolerance Mechanisms of an Arctic Pelagophyte Using Comparative Transcriptomic and Gene Expression Analysis. Commun. Biol. 2022, 5, 500. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Chaki, M.; Begara-Morales, J.C.; Barroso, J.B. Oxidative Stress in Plants. Antioxidants 2020, 9, 481. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Salehi, H.; Rahman, M.A.; Zahid, Z.; Madadkar Haghjou, M.; Najafi-Kakavand, S.; Charagh, S.; Osman, H.S.; Albaqami, M.; Zhuang, Y.; et al. Plant Hormones and Neurotransmitter Interactions Mediate Antioxidant Defenses under Induced Oxidative Stress in Plants. Front. Plant Sci. 2022, 13, 961872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Y.; Deng, C.; Deng, S.; Li, N.; Zhao, C.; Zhao, R.; Liang, S.; Chen, S. The Arabidopsis Ca2+-Dependent Protein Kinase CPK12 Is Involved in Plant Response to Salt Stress. Int. J. Mol. Sci. 2018, 19, 4062. [Google Scholar] [CrossRef]
- Guo, R.; Yang, Z.; Li, F.; Yan, C.; Zhong, X.; Liu, Q.; Xia, X.; Li, H.; Zhao, L. Comparative Metabolic Responses and Adaptive Strategies of Wheat (Triticum aestivum) to Salt and Alkali Stress. BMC Plant Biol. 2015, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Bartels, D.; Dinakar, C. Balancing Salinity Stress Responses in Halophytes and Non-Halophytes: A Comparison between Thellungiella and Arabidopsis Thaliana. Funct. Plant Biol. 2013, 40, 819. [Google Scholar] [CrossRef] [PubMed]
- Faried, H.N.; Ayyub, C.M.; Amjad, M.; Ahmed, R.; Wattoo, F.M.; Butt, M.; Bashir, M.; Shaheen, M.R.; Waqas, M.A. Salicylic Acid Confers Salt Tolerance in Potato Plants by Improving Water Relations, Gaseous Exchange, Antioxidant Activities and Osmoregulation: Salt Stress Alleviation in Potato by Foliar Application of Salicylic Acid. J. Sci. Food Agric. 2017, 97, 1868–1875. [Google Scholar] [CrossRef]
- Meng, X.; Zhou, J.; Sui, N. Mechanisms of Salt Tolerance in Halophytes: Current Understanding and Recent Advances. Open Life Sci. 2018, 13, 149–154. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Yu, W.; Wu, W.; Zhang, N.; Wang, L.; Wang, Y.; Wang, B.; Lan, Q.; Wang, Y. Research Advances on Molecular Mechanism of Salt Tolerance in Suaeda. Biology 2022, 11, 1273. [Google Scholar] [CrossRef]
- Sicilia, A.; Testa, G.; Santoro, D.F.; Cosentino, S.L.; Lo Piero, A.R. RNASeq Analysis of Giant Cane Reveals the Leaf Transcriptome Dynamics under Long-Term Salt Stress. BMC Plant Biol. 2019, 19, 355. [Google Scholar] [CrossRef]
- Sicilia, A.; Santoro, D.F.; Testa, G.; Cosentino, S.L.; Lo Piero, A.R. Transcriptional Response of Giant Reed (Arundo donax L.) Low Ecotype to Long-Term Salt Stress by Unigene-Based RNAseq. Phytochemistry 2020, 177, 112436. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Cao, M.; Wang, Q.; Xu, J.; Liu, C.; Ullah, N.; Li, J.; Hou, Z.; Liang, Z.; Zhou, W.; et al. Insights into the Plateau Adaptation of Salvia Castanea by Comparative Genomic and WGCNA Analyses. J. Adv. Res. 2022, 42, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pu, L.; Han, M.; Zhu, M.; Zhang, R.; Xiang, Y. Soil Salinization Research in China: Advances and Prospects. J. Geogr. Sci. 2014, 24, 943–960. [Google Scholar] [CrossRef]
- Kerchev, P.I.; Van Breusegem, F. Improving Oxidative Stress Resilience in Plants. Plant J. 2022, 109, 359–372. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X. Response Mechanisms of Plants Under Saline-Alkali Stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef] [PubMed]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional Factors Regulate Plant Stress Responses Through Mediating Secondary Metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef]
- Chuamnakthong, S.; Nampei, M.; Ueda, A. Characterization of Na+ Exclusion Mechanism in Rice under Saline-Alkaline Stress Conditions. Plant Sci. 2019, 287, 110171. [Google Scholar] [CrossRef]
- Tian, Q.; Shen, L.; Luan, J.; Zhou, Z.; Guo, D.; Shen, Y.; Jing, W.; Zhang, B.; Zhang, Q.; Zhang, W. Rice Shaker Potassium Channel OsAKT2 Positively Regulates Salt Tolerance and Grain Yield by Mediating K + Redistribution. Plant Cell Environ. 2021, 44, 2951–2965. [Google Scholar] [CrossRef]
- Haj-Amor, Z.; Araya, T.; Kim, D.-G.; Bouri, S.; Lee, J.; Ghiloufi, W.; Yang, Y.; Kang, H.; Jhariya, M.K.; Banerjee, A.; et al. Soil Salinity and Its Associated Effects on Soil Microorganisms, Greenhouse Gas Emissions, Crop Yield, Biodiversity and Desertification: A Review. Sci. Total Environ. 2022, 843, 156946. [Google Scholar] [CrossRef]
- Yang, H.; Chen, W.; Chen, Y.; Zhang, F.; Yang, X. Assessing the Impact of Shallow Subsurface Pipe Drainage on Soil Salinity and Crop Yield in Arid Zone. PeerJ 2021, 9, e12622. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Du, C.; Yu, B.; Wang, G.; Deng, Y.; Wang, Y.; Chen, M.; Chang, J.; Yang, G.; He, G.; et al. Current Advances in the Molecular Regulation of Abiotic Stress Tolerance in Sorghum via Transcriptomic, Proteomic, and Metabolomic Approaches. Front. Plant Sci. 2023, 14, 1147328. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Ma, H.; Liu, E.; Jiang, T.; Feng, S.; Gong, S.; Wang, J. Transcriptome Sequencing of Dianthus Spiculifolius and Analysis of the Genes Involved in Responses to Combined Cold and Drought Stress. Int. J. Mol. Sci. 2017, 18, 849. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Y.; Zhu, J.-K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef]
- Markham, K.K.; Greenham, K. Abiotic Stress through Time. New Phytol. 2021, 231, 40–46. [Google Scholar] [CrossRef]
- Zhan, X.; Chen, Z.; Chen, R.; Shen, C. Environmental and Genetic Factors Involved in Plant Protection-Associated Secondary Metabolite Biosynthesis Pathways. Front. Plant Sci. 2022, 13, 877304. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, B.; Li, J.; Song, Z.; Lu, B.; Chi, M.; Yang, B.; Liu, J.; Lam, Y.-W.; Li, J.; et al. Salt-Response Analysis in Two Rice Cultivars at Seedling Stage. Acta Physiol. Plant. 2017, 39, 215. [Google Scholar] [CrossRef]
- Kaneko, T.; Horie, T.; Nakahara, Y.; Tsuji, N.; Shibasaka, M.; Katsuhara, M. Dynamic Regulation of the Root Hydraulic Conductivity of Barley Plants in Response to Salinity/Osmotic Stress. Plant Cell Physiol. 2015, 56, 875–882. [Google Scholar] [CrossRef]
- Zhao, C.; Zayed, O.; Zeng, F.; Liu, C.; Zhang, L.; Zhu, P.; Hsu, C.; Tuncil, Y.E.; Tao, W.A.; Carpita, N.C.; et al. Arabinose Biosynthesis Is Critical for Salt Stress Tolerance in Arabidopsis. New Phytol. 2019, 224, 274–290. [Google Scholar] [CrossRef]
- Buer, C.S.; Wasteneys, G.O.; Masle, J. Ethylene Modulates Root-Wave Responses in Arabidopsis. Plant Physiol. 2003, 132, 1085–1096. [Google Scholar] [CrossRef]
- Hong, J.H.; Seah, S.W.; Xu, J. The Root of ABA Action in Environmental Stress Response. Plant Cell Rep. 2013, 32, 971–983. [Google Scholar] [CrossRef]
- De Zelicourt, A.; Colcombet, J.; Hirt, H. The Role of MAPK Modules and ABA during Abiotic Stress Signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In Vitro Reconstitution of an Abscisic Acid Signalling Pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF Family Transcription Factors in Plant Abiotic Stress Responses. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Jain, M.; Khurana, J.P. Transcript Profiling Reveals Diverse Roles of Auxin-Responsive Genes during Reproductive Development and Abiotic Stress in Rice. FEBS J. 2009, 276, 3148–3162. [Google Scholar] [CrossRef]
- Lee, D.J.; Park, J.W.; Lee, H.W.; Kim, J. Genome-Wide Analysis of the Auxin-Responsive Transcriptome Downstream of Iaa1 and Its Expression Analysis Reveal the Diversity and Complexity of Auxin-Regulated Gene Expression. J. Exp. Bot. 2009, 60, 3935–3957. [Google Scholar] [CrossRef]
- Song, Y.; Wang, L.; Xiong, L. Comprehensive Expression Profiling Analysis of OsIAA Gene Family in Developmental Processes and in Response to Phytohormone and Stress Treatments. Planta 2009, 229, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Zolla, G.; Heimer, Y.M.; Barak, S. Mild Salinity Stimulates a Stress-Induced Morphogenic Response in Arabidopsis Thaliana Roots. J. Exp. Bot. 2010, 61, 211–224. [Google Scholar] [CrossRef]
- Xu, Y.-X.; Mao, J.; Chen, W.; Qian, T.-T.; Liu, S.-C.; Hao, W.-J.; Li, C.-F.; Chen, L. Identification and Expression Profiling of the Auxin Response Factors (ARFs) in the Tea Plant (Camellia sinensis (L.) O. Kuntze) under Various Abiotic Stresses. Plant Physiol. Biochem. 2016, 98, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhang, H.; Tian, Y.; Li, F.; Zhang, Z.; Lu, X.; Chen, X.; Huang, R. Expression of TERF1 in Rice Regulates Expression of Stress-Responsive Genes and Enhances Tolerance to Drought and High-Salinity. Plant Cell Rep. 2008, 27, 1787–1795. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhang, Z.; Zhang, X.; Zhang, H.; Huang, D.; Huang, R. Tomato TERF1 Modulates Ethylene Response and Enhances Osmotic Stress Tolerance by Activating Expression of Downstream Genes. FEBS Lett. 2004, 573, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Z.; Quan, R.; Li, G.; Wang, R.; Huang, R. An AP2 Domain-Containing Gene, ESE1, Targeted by the Ethylene Signaling Component EIN3 Is Important for the Salt Response in Arabidopsis. Plant Physiol. 2011, 157, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Qari, S.H.; Tarbiyyah, I. The Genetic Regulation of Secondary Metabolic Pathways in Response to Salinity and Drought as Abiotic Stresses. Appl. Sci. 2021, 11, 6668. [Google Scholar] [CrossRef]
- Dorion, S.; Ouellet, J.C.; Rivoal, J. Glutathione Metabolism in Plants under Stress: Beyond Reactive Oxygen Species Detoxification. Metabolites 2021, 11, 641. [Google Scholar] [CrossRef]
- Hernández Estévez, I.; Rodríguez Hernández, M. Plant Glutathione S-Transferases: An Overview. Plant Gene 2020, 23, 100233. [Google Scholar] [CrossRef]
- Vaish, S.; Gupta, D.; Mehrotra, R.; Mehrotra, S.; Basantani, M.K. Glutathione S-Transferase: A Versatile Protein Family. 3 Biotech 2020, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.-T.; Ma, S.-L.; Bai, L.-P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.-F. Signal Transduction during Cold, Salt, and Drought Stresses in Plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef]
- Yen, W.-J.; Chyau, C.-C.; Lee, C.-P.; Chu, H.-L.; Chang, L.-W.; Duh, P.-D. Cytoprotective Effect of White Tea against H2O2-Induced Oxidative Stress in Vitro. Food Chem. 2013, 141, 4107–4114. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, Z.; Wang, T.; Mantri, N.; Huang, H.; Li, H.; Tao, Z.; Guo, Q. Physiological and Transcriptomic Analyses of Cadmium Stress Response in Dendrobium Officinale Seedling. Plant Physiol. Biochem. 2020, 148, 152–165. [Google Scholar] [CrossRef]
- Rentel, M.C.; Lecourieux, D.; Ouaked, F.; Usher, S.L.; Petersen, L.; Okamoto, H.; Knight, H.; Peck, S.C.; Grierson, C.S.; Hirt, H.; et al. OXI1 Kinase Is Necessary for Oxidative Burst-Mediated Signalling in Arabidopsis. Nature 2004, 427, 858–861. [Google Scholar] [CrossRef]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef]
- Islam, M.O.; Kato, H.; Shima, S.; Tezuka, D.; Matsui, H.; Imai, R. Functional Identification of a Rice Trehalase Gene Involved in Salt Stress Tolerance. Gene 2019, 685, 42–49. [Google Scholar] [CrossRef]
- Golldack, D.; Lüking, I.; Yang, O. Plant Tolerance to Drought and Salinity: Stress Regulating Transcription Factors and Their Functional Significance in the Cellular Transcriptional Network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Mieulet, D.; Hubberten, H.-M.; Obata, T.; Hoefgen, R.; Fernie, A.R.; Fisahn, J.; San Segundo, B.; Guiderdoni, E.; Schippers, J.H.M.; et al. SALT-RESPONSIVE ERF1 Regulates Reactive Oxygen Species-Dependent Signaling during the Initial Response to Salt Stress in Rice. Plant Cell 2013, 25, 2115–2131. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Ma, X.; Fan, W.; Su, M.; Cheng, L.; Alam, I.; Lee, B.-H.; Dongmei, Q.; Shihua, S.; Gongshe, L. Improved Drought and Salt Tolerance of Arabidopsis Thaliana by Transgenic Expression of a Novel DREB Gene from Leymus chinensis. Plant Cell Rep. 2011, 30, 1493–1502. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the Soybean GmERF3 Gene, an AP2/ERF Type Transcription Factor for Increased Tolerances to Salt, Drought, and Diseases in Transgenic Tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef]
- Zhai, Y.; Wang, Y.; Li, Y.; Lei, T.; Yan, F.; Su, L.; Li, X.; Zhao, Y.; Sun, X.; Li, J.; et al. Isolation and Molecular Characterization of GmERF7, a Soybean Ethylene-Response Factor That Increases Salt Stress Tolerance in Tobacco. Gene 2013, 513, 174–183. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Y.; Liu, J.; Zhao, T.; Yang, C.; Ding, Q.; Zhang, Y.; Mu, J.; Wang, D. Identification of WRKY Transcription Factors Responding to Abiotic Stresses in Brassica napus L. Planta 2022, 255, 3. [Google Scholar] [CrossRef] [PubMed]
- Van Aken, O.; Zhang, B.; Law, S.; Narsai, R.; Whelan, J. AtWRKY40 and AtWRKY63 Modulate the Expression of Stress-Responsive Nuclear Genes Encoding Mitochondrial and Chloroplast Proteins. Plant Physiol. 2013, 162, 254–271. [Google Scholar] [CrossRef]
- Wu, Q.-W.; Wei, M.; Feng, L.-F.; Ding, L.; Wei, W.-K.; Yang, J.-F.; Lin, X.-J.; Liang, H.-L.; Zhan, R.-T.; Ma, D.-M. Rhamnosyltransferases Involved in the Biosynthesis of Flavone Rutinosides in Chrysanthemum Species. Plant Physiol. 2022, 190, 2122–2136. [Google Scholar] [CrossRef]
- Jin, T.; An, J.; Xu, H.; Chen, J.; Pan, L.; Zhao, R.; Wang, N.; Gai, J.; Li, Y. A Soybean Sodium/Hydrogen Exchanger GmNHX6 Confers Plant Alkaline Salt Tolerance by Regulating Na+/K+ Homeostasis. Front. Plant Sci. 2022, 13, 938635. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Maathuis, F.; Voitsekhovskaja, O. Unravelling the Plant Signalling Machinery: An Update on the Cellular and Genetic Basis of Plant Signal Transduction. Funct. Plant Biol. 2018, 45, 1. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Kumar, P.; Bae, H. Genomics and Evolutionary Aspect of Calcium Signaling Event in Calmodulin and Calmodulin-like Proteins in Plants. BMC Plant Biol. 2017, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, G.; Alli, A.; Yu, L. Plant HAK/KUP/KT K+ Transporters: Function and Regulation. Semin. Cell Dev. Biol. 2018, 74, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Latz, A.; Mehlmer, N.; Zapf, S.; Mueller, T.D.; Wurzinger, B.; Pfister, B.; Csaszar, E.; Hedrich, R.; Teige, M.; Becker, D. Salt Stress Triggers Phosphorylation of the Arabidopsis Vacuolar K+ Channel TPK1 by Calcium-Dependent Protein Kinases (CDPKs). Mol. Plant 2013, 6, 1274–1289. [Google Scholar] [CrossRef]
- Maathuis, F.J.M. Vacuolar Two-pore K+ Channels Act as Vacuolar Osmosensors. New Phytol. 2011, 191, 84–91. [Google Scholar] [CrossRef]
- Gómez-Galera, S.; Sudhakar, D.; Pelacho, A.M.; Capell, T.; Christou, P. Constitutive Expression of a Barley Fe Phytosiderophore Transporter Increases Alkaline Soil Tolerance and Results in Iron Partitioning between Vegetative and Storage Tissues under Stress. Plant Physiol. Biochem. 2012, 53, 46–53. [Google Scholar] [CrossRef]
- Aoyama, T.; Kobayashi, T.; Takahashi, M.; Nagasaka, S.; Usuda, K.; Kakei, Y.; Ishimaru, Y.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL18 Is a Rice Iron(III)–Deoxymugineic Acid Transporter Specifically Expressed in Reproductive Organs and Phloem of Lamina Joints. Plant Mol. Biol. 2009, 70, 681–692. [Google Scholar] [CrossRef]
- Dolatabadian, A.; Modarres Sanavy, S.A.M.; Ghanati, F.; Gresshoff, P.M. Agrobacterium Rhizogenes Transformed Soybean Roots Differ in Their Nodulation and Nitrogen Fixation Response to Genistein and Salt Stress. World J. Microbiol. Biotechnol. 2013, 29, 1327–1339. [Google Scholar] [CrossRef]
- Wülser, J.; Ernst, C.; Vetsch, D.; Emmenegger, B.; Michel, A.; Lutz, S.; Ahrens, C.H.; Vorholt, J.A.; Ledermann, R.; Fischer, H.-M. Salt- and Osmo-Responsive Sensor Histidine Kinases Activate the Bradyrhizobium diazoefficiens General Stress Response to Initiate Functional Symbiosis. Mol. Plant-Microbe Interact. 2022, 35, 604–615. [Google Scholar] [CrossRef]
- Ali, A.; Petrov, V.; Yun, D.-J.; Gechev, T. Revisiting Plant Salt Tolerance: Novel Components of the SOS Pathway. Trends Plant Sci. 2023, 28, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ Homeostasis in Plants: Towards Improved Salt Stress Tolerance in Crop Plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.-K. The Putative Plasma Membrane Na+/H+ Antiporter SOS1 Controls Long-Distance Na+ Transport in Plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Lu, J.; Jiang, J.; Chen, S.; Guan, Z.; Fang, W.; Chen, F. Isolation and Characterisation of Chrysanthemum Crassum SOS1, Encoding a Putative Plasma Membrane Na+/H+ Antiporter: SOS1, a Putative Membrane Na+/H+ Antiporter in C. Crassum. Plant Biol. 2012, 14, 706–713. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, J.; Zhang, T.; Xu, T.; Yang, L.; Li, X.; Ji, F.; Gao, Y.; Ali, S.; Zhang, Q.; et al. A Putative Plasma Membrane Na+/H+ Antiporter GmSOS1 Is Critical for Salt Stress Tolerance in Glycine Max. Front. Plant Sci. 2022, 13, 870695. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, C.; Hou, L.; Wu, X.; Wang, D.; Zhang, L.; Liu, P. Exogenous SA Affects Rice Seed Germination under Salt Stress by Regulating Na+/K+ Balance and Endogenous GAs and ABA Homeostasis. Int. J. Mol. Sci. 2022, 23, 3293. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]






| Gene Family | Gene Name | Average log2 (Fold Change) | TF Id |
|---|---|---|---|
| AP2/ERF-ERF | SORBI_3002G069400 | 5.628355581 | Sobic.002G069400.1.p |
| SORBI_3005G103000 | 5.185617358 | Sobic.005G103000.1.p | |
| SORBI_3007G205100 | 4.154163003 | Sobic.007G205100.1.p | |
| SORBI_3002G269600 | 3.383162754 | Sobic.002G269600.1.p | |
| SORBI_3007G162700 | 3.217188466 | Sobic.007G162700.1.p | |
| BHLH | SORBI_3009G057000 | 6.300233713 | Sobic.009G057000.3.p |
| SORBI_3003G077100 | 6.164343331 | Sobic.003G077100.1.p | |
| SORBI_3008G153801 | 6.129472986 | Sobic.008G153801.1.p | |
| SORBI_3002G267000 | 5.565088837 | Sobic.002G267000.1.p | |
| SORBI_3007G008500 | 5.507180988 | Sobic.007G008500.3.p | |
| MYB | SORBI_3001G056000 | 4.934458789 | Sobic.001G056000.1.p |
| SORBI_3001G158600 | 3.955308363 | Sobic.001G158600.2.p | |
| SORBI_3007G177100 | 2.598003641 | Sobic.007G177100.1.p | |
| SORBI_3003G034500 | 2.531600604 | Sobic.003G034500.1.p | |
| SORBI_3005G155900 | 2.451087788 | Sobic.005G155900.1.p | |
| NAC | SORBI_3006G141900 | 7.874140412 | Sobic.006G141900.1.p |
| SORBI_3001G018350 | 5.66101195 | Sobic.001G018350.1.p | |
| SORBI_3003G333300 | 5.361257257 | Sobic.003G333300.1.p | |
| SORBI_3005G028500 | 4.949789354 | Sobic.005G028500.1.p | |
| SORBI_3002G200300 | 4.380423241 | Sobic.002G200300.1.p | |
| WRKY | SORBI_3005G014200 | 7.17739049 | Sobic.005G014200.1.p |
| SORBI_3010G148800 | 5.826129024 | Sobic.010G148800.1.p | |
| SORBI_3008G029200 | 4.578702823 | Sobic.008G029200.1.p | |
| SORBI_3009G247700 | 2.276355842 | Sobic.009G247700.1.p | |
| SORBI_3007G217700 | 2.037259465 | Sobic.007G217700.6.p | |
| C2H2 | SORBI_3003G242300 | 7.142275339 | Sobic.003G242300.2.p |
| SORBI_3005G025100 | 6.269975035 | Sobic.005G025100.1.p | |
| SORBI_3003G242400 | 5.907308008 | Sobic.003G242400.2.p | |
| SORBI_3004G066700 | 3.077041905 | Sobic.004G066700.1.p | |
| SORBI_3005G225700 | 2.760863094 | Sobic.005G225700.1.p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Ye, L.; Zhou, L.; Yu, J.; Pang, B.; Zuo, D.; Gu, L.; Zhu, B.; Du, X.; Wang, H. Co-Expression Network Analysis of the Transcriptome Identified Hub Genes and Pathways Responding to Saline–Alkaline Stress in Sorghum bicolor L. Int. J. Mol. Sci. 2023, 24, 16831. https://doi.org/10.3390/ijms242316831
Wang H, Ye L, Zhou L, Yu J, Pang B, Zuo D, Gu L, Zhu B, Du X, Wang H. Co-Expression Network Analysis of the Transcriptome Identified Hub Genes and Pathways Responding to Saline–Alkaline Stress in Sorghum bicolor L. International Journal of Molecular Sciences. 2023; 24(23):16831. https://doi.org/10.3390/ijms242316831
Chicago/Turabian StyleWang, Hongcheng, Lvlan Ye, Lizhou Zhou, Junxing Yu, Biao Pang, Dan Zuo, Lei Gu, Bin Zhu, Xuye Du, and Huinan Wang. 2023. "Co-Expression Network Analysis of the Transcriptome Identified Hub Genes and Pathways Responding to Saline–Alkaline Stress in Sorghum bicolor L." International Journal of Molecular Sciences 24, no. 23: 16831. https://doi.org/10.3390/ijms242316831
APA StyleWang, H., Ye, L., Zhou, L., Yu, J., Pang, B., Zuo, D., Gu, L., Zhu, B., Du, X., & Wang, H. (2023). Co-Expression Network Analysis of the Transcriptome Identified Hub Genes and Pathways Responding to Saline–Alkaline Stress in Sorghum bicolor L. International Journal of Molecular Sciences, 24(23), 16831. https://doi.org/10.3390/ijms242316831






