Molecular Characterization of Local Walnut (Juglans regia) Genotypes in the North-East Parnon Mountain Region of Greece
Abstract
:1. Introduction
2. Results
2.1. Polymorphism of SSR Primer Pairs
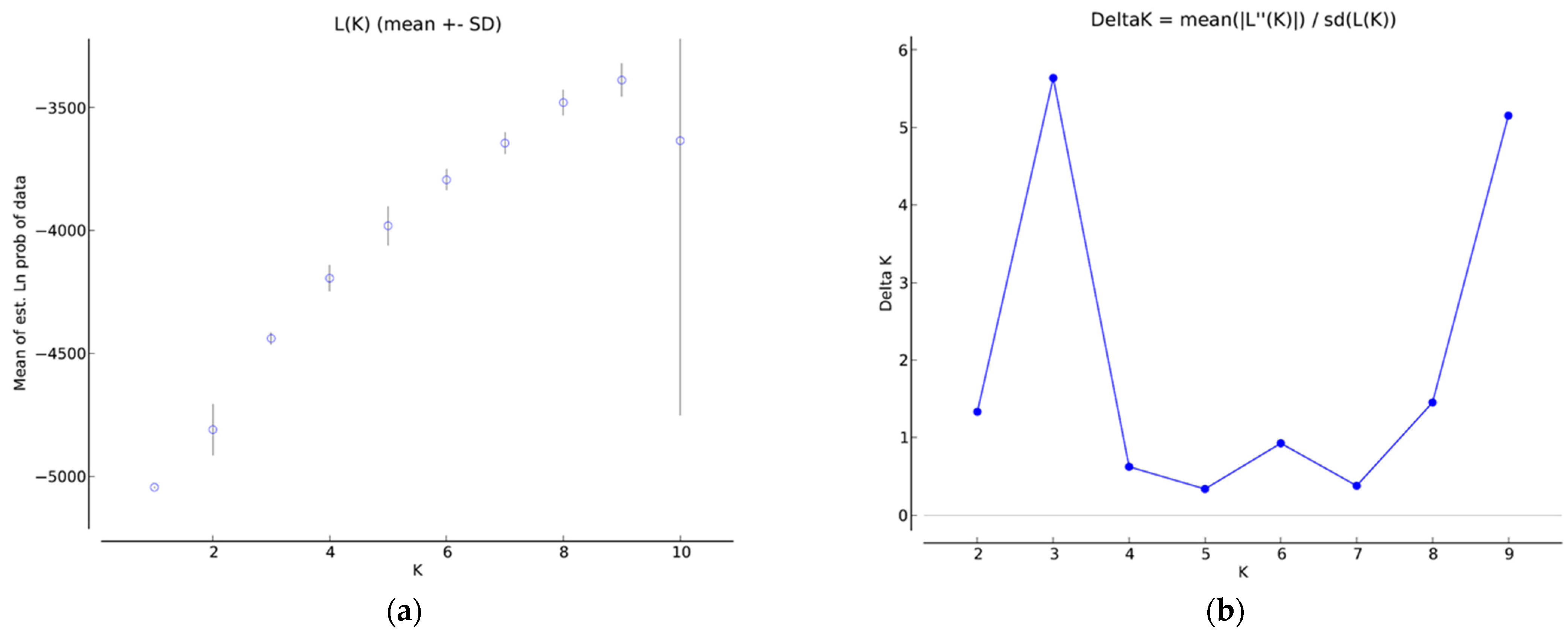
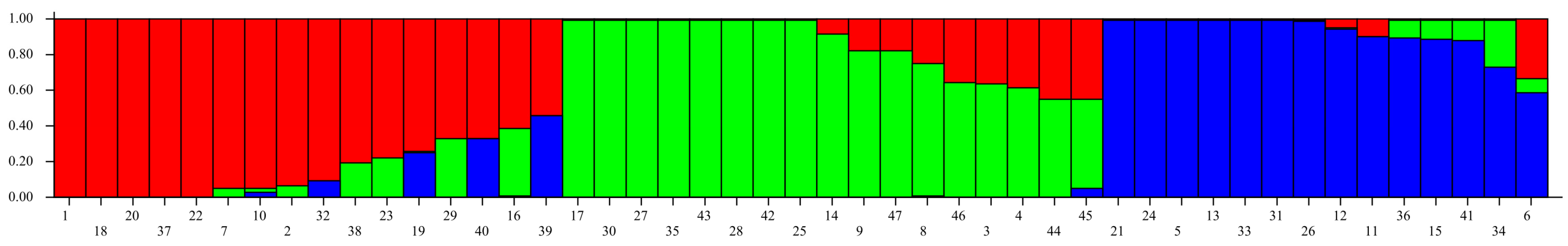
2.2. Population Structure Analysis
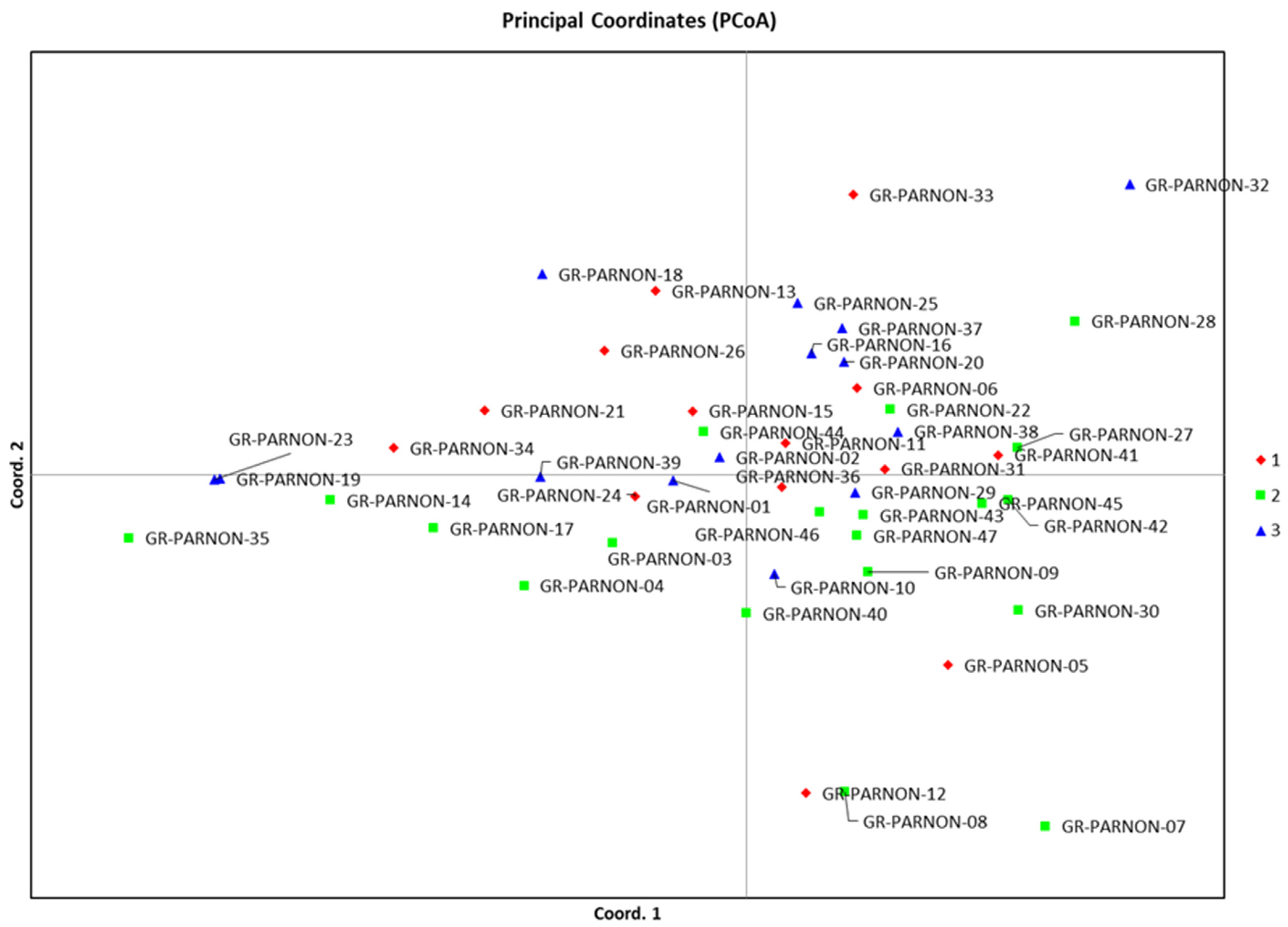
2.3. Principal Coordinate Analysis
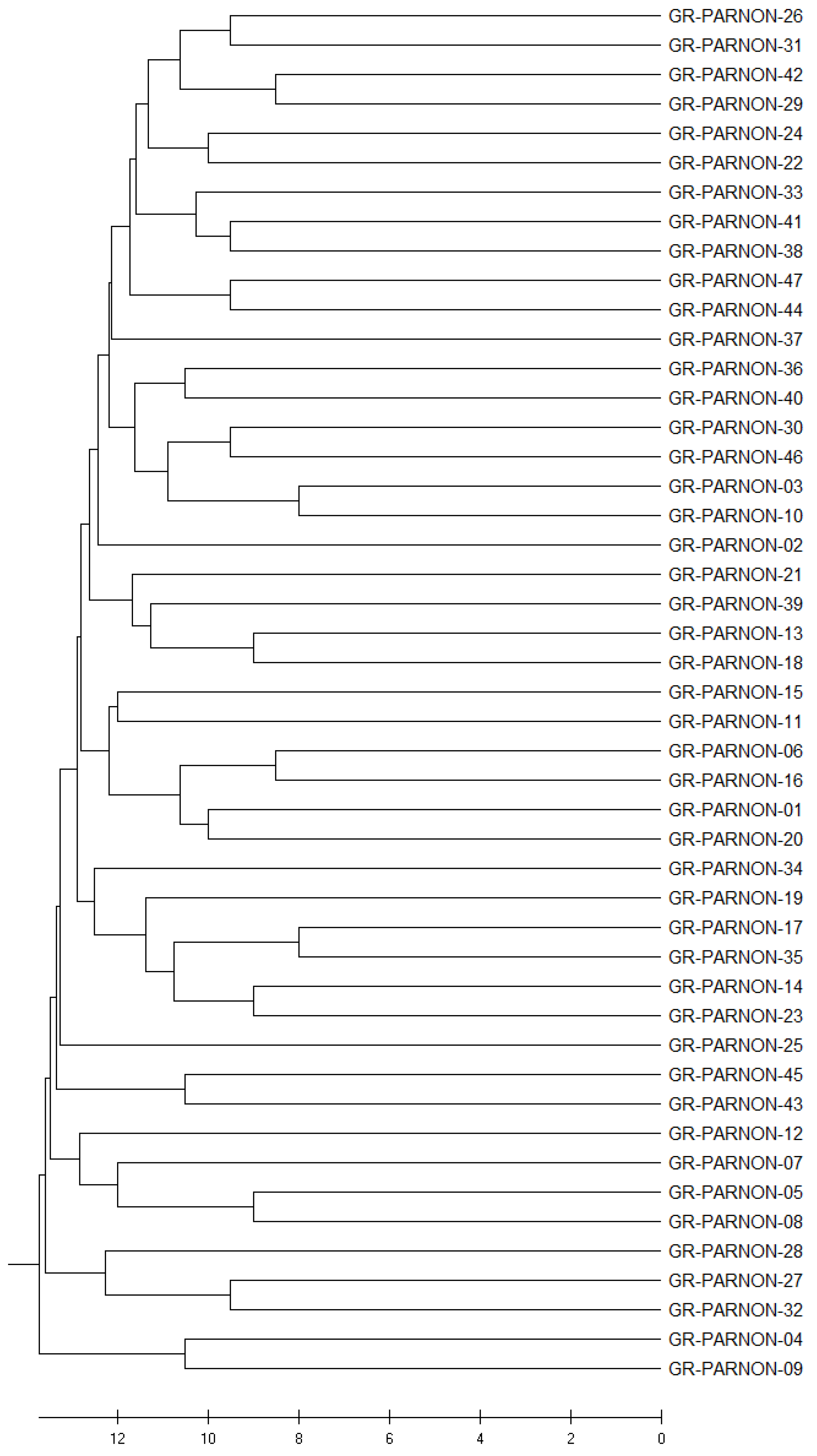
2.4. UPGMA Cluster Analysis
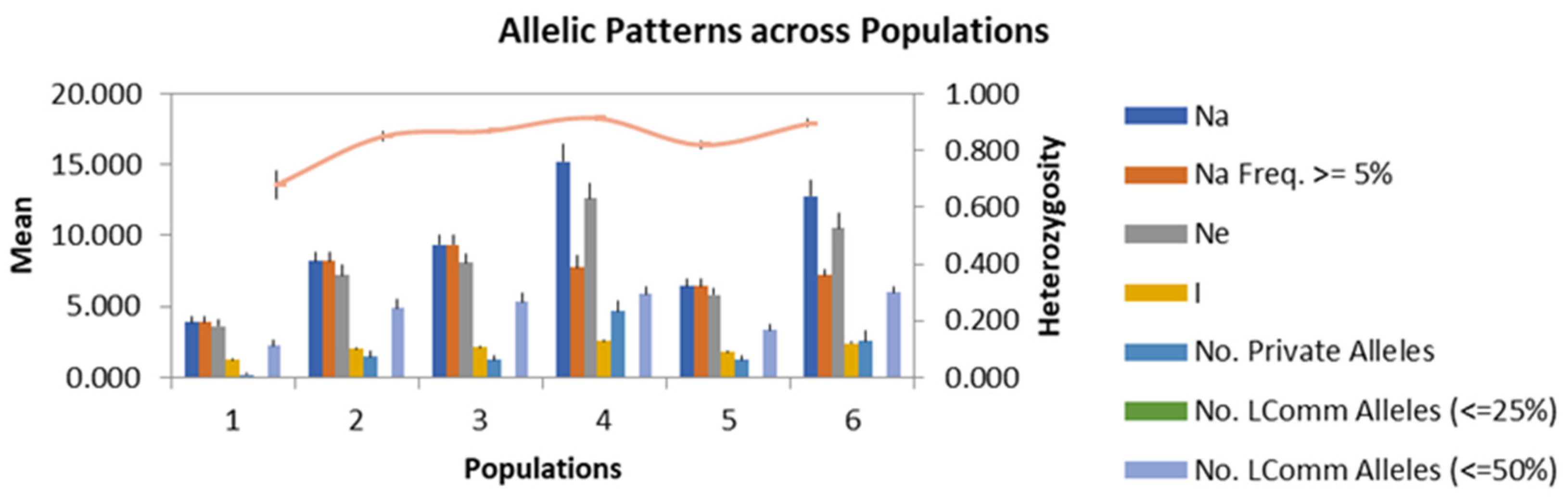
2.5. Genetic Diversity Analysis
3. Discussion
4. Materials and Methods
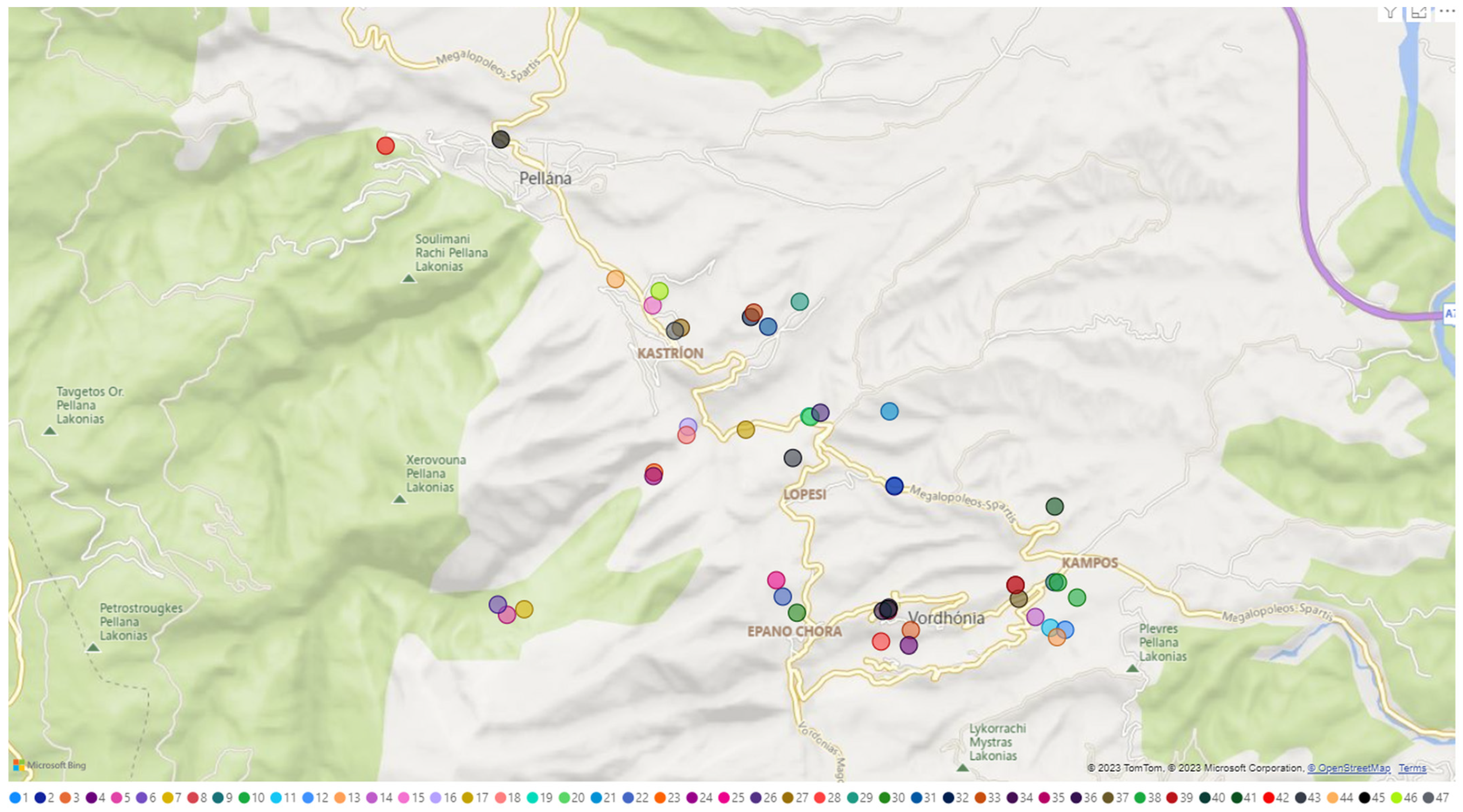
4.1. Plant Material
4.2. Molecular Analysis Using SSR Markers
4.3. Molecular Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Available online: https://www.fao.org/faostat/en/ (accessed on 1 October 2023).
- Halvorsen, B.L.; Carlsen, M.H.; Phillips, K.M.; Bøhn, S.K.; Holte, K.; Jacobs, D.R.; Blomhoff, R. Content of Redox-Active Compounds (Ie, Antioxidants) in Foods Consumed in the United States. Am. J. Clin. Nutr. 2006, 84, 95–135. [Google Scholar] [CrossRef]
- Kornsteiner, M.; Wagner, K.-H.; Elmadfa, I. Tocopherols and Total Phenolics in 10 Different Nut Types. Food Chem. 2006, 98, 381–387. [Google Scholar] [CrossRef]
- Hayes, D.; Angove, M.J.; Tucci, J.; Dennis, C. Walnuts (Juglans regia) Chemical Composition and Research in Human Health. Crit. Rev. Food Sci. Nutr. 2016, 56, 1231–1241. [Google Scholar] [CrossRef]
- Ros, E.; Izquierdo-Pulido, M.; Sala-Vila, A. Beneficial Effects of Walnut Consumption on Human Health: Role of Micronutrients. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Guasch-Ferré, M.; Tobias, D.K.; Li, Y. Association of Walnut Consumption with Total and Cause-Specific Mortality and Life Expectancy in U.S. Adults . Nutrients 2021, 13, 2699. [Google Scholar] [CrossRef]
- Germain, E.; Aletà, N.; Ninot, A.; Rouskas, D.; Zakinthinos, G.; Pereira, G.; Monastra, F. Prospections réalisées dansles populations de semis de noyer d’Espagne, de Grèce, d’Italie etdu Portugal: Caractérisation des populations et description en collections d’études des présélections issues deces prospections. Options Méditerr 1997, 16, 7–40. [Google Scholar]
- Rouskas, D.; Katranis, N.; Zakynthinos, G.; Isaakidis, R. Walnuts (Junglans regia L.) Seedlings Selection in Greece. Acta Hortic. 1997, 442, 109–116. [Google Scholar] [CrossRef]
- Rouskas, D.; Zakynthinos, G. Preliminary Evaluation of Seventy Walnut (Juglans regia L.) Seedlings Selections in Greece. Acta Hortic. 2001, 544, 61–72. [Google Scholar] [CrossRef]
- Christopoulos, M.V.; Rouskas, D.; Tsantili, E.; Bebeli, P.J. Germplasm Diversity and Genetic Relationships among Walnut (Juglans regia L.) Cultivars and Greek Local Selections Revealed by Inter-Simple Sequence Repeat (ISSR) Markers. Sci. Hortic. 2010, 125, 584–592. [Google Scholar] [CrossRef]
- Avanzato, D. Following Walnut Footprints (Juglans regia L.): Cultivation and Culture, Folklore and History, Traditions and Uses; Scripta Horticulturae; ISHS: Leuven, Belgium, 2014; ISBN 978-94-6261-003-3. [Google Scholar]
- Hellenic Statistical Authority–ELSTAT. Available online: https://www.statistics.gr/el/statistics/ (accessed on 1 October 2023).
- Bayazit, S.; Kazan, K.; Gülbitti, S.; Çevik, V.; Ayanoğlu, H.; Ergül, A. AFLP Analysis of Genetic Diversity in Low Chill Requiring Walnut (Juglans regia L.) Genotypes from Hatay, Turkey. Sci. Hortic. 2007, 111, 394–398. [Google Scholar] [CrossRef]
- Ahmed, N.; Mir, J.I.; Mir, R.R.; Rather, N.A.; Rashid, R.; Wani, S.H.; Shafi, W.; Mir, H.; Sheikh, M.A. SSR and RAPD Analysis of Genetic Diversity in Walnut (Juglans regia L.) Genotypes from Jammu and Kashmir, India. Physiol. Mol. Biol. Plants 2012, 18, 149–160. [Google Scholar] [CrossRef]
- Nickravesh, M.H.; Vahdati, K.; Amini, F.; Amiri, R.; Woeste, K. Application of Microsatellite and Inter Simple Sequence Markers for Certifying Trueness to Type of Grafted Ramets and Clustering of Persian Walnut Varieties. Res. Sq. 2021. in review. [Google Scholar] [CrossRef]
- Ji, A.; Wang, Y.; Wu, G.; Wu, W.; Yang, H.; Wang, Q. Genetic Diversity and Population Structure of North China Mountain Walnut Revealed by ISSR. Am. J. Plant Sci. 2014, 05, 3194–3202. [Google Scholar] [CrossRef]
- Kabiri, G.; Bouda, S.; Haddioui, A. Evaluation of Genetic Diversity and Structuration across Altitude of Walnut (Juglans regia L.) Accessions from Morocco Using SSR Markers. Bulg. J. Agric. Sci. 2022, 28, 451–458. [Google Scholar]
- Bernard, A.; Marrano, A.; Donkpegan, A.; Brown, P.J.; Leslie, C.A.; Neale, D.B.; Lheureux, F.; Dirlewanger, E. Association and Linkage Mapping to Unravel Genetic Architecture of Phenological Traits and Lateral Bearing in Persian Walnut (Juglans regia L.). BMC Genom. 2020, 21, 203. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.A.; Baksi, P.; Jasrotia, A.; Bhat, D.J.; Gupta, R.; Bakshi, M. Genetic Diversity of Walnut (Juglans regia L.) Seedlings through SSR Markers in North-Western Himalayan Region of Jammu. Bangladesh J. Bot. 2020, 49, 1003–1012. [Google Scholar] [CrossRef]
- Balapanov, I.; Suprun, I.; Stepanov, I.; Tokmakov, S.; Lugovskoy, A. Comparative Analysis Crimean, Moldavian and Kuban Persian Walnut Collections Genetic Variability by SSR-Markers. Sci. Hortic. 2019, 253, 322–326. [Google Scholar] [CrossRef]
- Dangl, G.S.; Woeste, K.; Aradhya, M.K.; Koehmstedt, A.; Simon, C.; Potter, D.; Leslie, C.A.; McGranahan, G. Characterization of 14 Microsatellite Markers for Genetic Analysis and Cultivar Identification of Walnut. J. Am. Soc. Hortic. Sci. 2005, 130, 348–354. [Google Scholar] [CrossRef]
- Mohsenipoor, S.; Vahdati, K.; Amiri, R.; Mozaffari, M.R. Study of the Genetic Structure and Gene Flow in Persian Walnut (Juglans regia L.) Using SSR Markers. Acta Hortic. 2010, 861, 133–142. [Google Scholar] [CrossRef]
- Bükücü, Ş.B.; Sütyemez, M.; Kefayati, S.; Paizila, A.; Jighly, A.; Kafkas, S. Major QTL with Pleiotropic Effects Controlling Time of Leaf Budburst and Flowering-Related Traits in Walnut (Juglans regia L.). Sci. Rep. 2020, 10, 15207. [Google Scholar] [CrossRef]
- Aradhya, M.K.; Velasco, D.; Wang, J.; Ramasamy, R.; You, F.M.; Leslie, C.; Dandekar, A.; Luo, M.; Dvorak, J. A Fine-scale Genetic Linkage Map Reveals Genomic Regions Associated with Economic Traits in Walnut (Juglans regia). Plant Breed. 2019, 138, 635–646. [Google Scholar] [CrossRef]
- Foroni, I.; Rao, R.; Woeste, K.; Gallitelli, M. Characterisation of Juglans regia L. with SSR Markers and Evaluation of Genetic Relationships among Cultivars and the ‘Sorrento’ Landrace. J. Hortic. Sci. Biotechnol. 2005, 80, 49–53. [Google Scholar] [CrossRef]
- Shah, R.A.; Bakshi, P.; Jasrotia, A.; Itoo, H.; Padder, B.A.; Gupta, R.; Kour, G.; Dolkar, D. Morphological to Molecular Markers: Plant Genetic Diversity Studies in Walnut (Juglans regia L.)—A Review. Erwerbs-Obstbau 2023, 65, 1499–1511. [Google Scholar] [CrossRef]
- Guichoux, E.; Lagache, L.; Wagner, S.; Chaumeil, P.; Léger, P.; Lepais, O.; Lepoittevin, C.; Malausa, T.; Revardel, E.; Salin, F.; et al. Current Trends in Microsatellite Genotyping. Mol. Ecol. Resour. 2011, 11, 591–611. [Google Scholar] [CrossRef] [PubMed]
- Boopathi, N.M. Genotyping of Mapping Population. In Genetic Mapping and Marker Assisted Selection; Springer: Singapore, 2020; pp. 107–178. ISBN 9789811529481. [Google Scholar]
- Szantai, E.; Ronai, Z.; Szilagyi, A.; Sasvari-Szekely, M.; Guttman, A. Haplotyping by Capillary Electrophoresis. J. Chromatogr. A 2005, 1079, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Shahi Shavvon, R.; Qi, H.-L.; Mafakheri, M.; Fan, P.-Z.; Wu, H.-Y.; Bazdid Vahdati, F.; Al-Shmgani, H.S.; Wang, Y.-H.; Liu, J. Unravelling the Genetic Diversity and Population Structure of Common Walnut in the Iranian Plateau. BMC Plant Biol. 2023, 23, 201. [Google Scholar] [CrossRef] [PubMed]
- Büyüksolak, Z.N.; Aşkın, M.A.; Kahramanoğlu, İ.; Okatan, V. Effects of Altitude on the Pomological Characteristics and Chemical Properties of ‘Chandler’ Walnuts: A Case Study in Uşak Province. Acta Agrobot. 2020, 73, 3. [Google Scholar] [CrossRef]
- Akça, Y.; Yuldaşulu, Y.B.; Murad, E.; Vahdati, K. Exploring of Walnut Genetic Resources in Kazakhstan and Evaluation of Promising Selections. Int. J. Hortic. Sci. Technol. 2020, 7, 93–102. [Google Scholar] [CrossRef]
- Pop, I.F.; Vicol, A.C.; Botu, M.; Raica, P.A.; Vahdati, K.; Pamfil, D. Relationships of Walnut Cultivars in a Germplasm Collection: Comparative Analysis of Phenotypic and Molecular Data. Sci. Hortic. 2013, 153, 124–135. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Khadivi-Khub, A.; Nosrati, Z.; Karimi, R. Identification of Superior Walnut (Juglans regia) Genotypes with Late Leafing and High Kernel Quality in Iran. Sci. Hortic. 2015, 193, 195–201. [Google Scholar] [CrossRef]
- Hassani, D.; Sarikhani, S.; Dastjerdi, R.; Mahmoudi, R.; Soleimani, A.; Vahdati, K. Situation and Recent Trends on Cultivation and Breeding of Persian Walnut in Iran. Sci. Hortic. 2020, 270, 109369. [Google Scholar] [CrossRef]
- Cervera, M.T.; Plomion, C.; Malpica, C. Molecular Markers and Genome Mapping in Woody Plants. In Molecular Biology of Woody Plants; Jain, S.M., Minocha, S.C., Eds.; Forestry Sciences; Springer: Dordrecht, The Netherlands, 2000; Volume 64, pp. 375–394. ISBN 978-90-481-5338-1. [Google Scholar]
- Crespel, L.; Chirollet, M.; Durel, C.; Zhang, D.; Meynet, J.; Gudin, S. Mapping of Qualitative and Quantitative Phenotypic Traits in Rosa Using AFLP Markers. Theor. Appl. Genet. 2002, 105, 1207–1214. [Google Scholar] [CrossRef]
- Priyanka, V.; Kumar, R.; Dhaliwal, I.; Kaushik, P. Germplasm Conservation: Instrumental in Agricultural Biodiversity—A Review. Sustainability 2021, 13, 6743. [Google Scholar] [CrossRef]
- De La Rosa, R.; Angiolillo, A.; Guerrero, C.; Pellegrini, M.; Rallo, L.; Besnard, G.; Bervillé, A.; Martin, A.; Baldoni, L. A First Linkage Map of Olive (Olea europaea L.) Cultivars Using RAPD, AFLP, RFLP and SSR Markers. Theor. Appl. Genet. 2003, 106, 1273–1282. [Google Scholar] [CrossRef]
- Woeste, K. Thirty Polymorphic Nuclear Microsatellite Loci from Black Walnut. J. Hered. 2002, 93, 58–60. [Google Scholar] [CrossRef]
- Ruiz-Garcia, L.; Lopez-Ortega, G.; Fuentes Denia, A.; Frutos Tomas, D. Identification of a Walnut (Juglans regia L.) Germplasm Collection and Evaluation of Their Genetic Variability by Microsatellite Markers. Span. J. Agric. Res. 2011, 9, 179. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Zarei, A.; Lawson, S.; Woeste, K.E.; Smulders, M.J.M. Genetic Diversity and Genetic Structure of Persian Walnut (Juglans regia) Accessions from 14 European, African, and Asian Countries Using SSR Markers. Tree Genet. Genomes 2016, 12, 114. [Google Scholar] [CrossRef]
- Vischi, M.; Chiabà, C.; Raranciuc, S.; Poggetti, L.; Messina, R.; Ermacora, P.; Cipriani, G.; Paffetti, D.; Vettori, C.; Testolin, R. Genetic Diversity of Walnut (Juglans regia L.) in the Eastern Italian Alps. Forests 2017, 8, 81. [Google Scholar] [CrossRef]
- Victory, E.R.; Glaubitz, J.C.; Rhodes, O.E.; Woeste, K.E. Genetic Homogeneity in Juglans Nigra (Juglandaceae) at Nuclear Microsatellites. Am. J. Bot. 2006, 93, 118–126. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, A.; Wang, X.; Yu, J.; Zhang, H.; Gao, J.; Cheng, Y.; Deng, X. Development of Juglans regia SSR Markers by Data Mining of the EST Database. Plant Mol. Biol. Report. 2010, 28, 646–653. [Google Scholar] [CrossRef]
- Itoo, H.; Shah, R.A.; Qurat, S.; Jeelani, A.; Khursheed, S.; Bhat, Z.A.; Mir, M.A.; Rather, G.H.; Zargar, S.M.; Shah, M.D.; et al. Genome-Wide Characterization and Development of SSR Markers for Genetic Diversity Analysis in Northwestern Himalayas Walnut (Juglans regia L.). 3 Biotech 2023, 13, 136. [Google Scholar] [CrossRef]
- Orhan, E.; Eyduran, S.P.; Poljuha, D.; Akin, M.; Weber, T.; Ercisli, S. Genetic Diversity Detection of Seed-Propagated Walnut (Juglans regia L.) Germplasm from Eastern Anatolia Using SSR Markers. Folia Hortic. 2020, 32, 37–46. [Google Scholar] [CrossRef]
- Vahdati, K.; Mohseni Pourtaklu, S.; Karimi, R.; Barzehkar, R.; Amiri, R.; Mozaffari, M.; Woeste, K. Genetic Diversity and Gene Flow of Some Persian Walnut Populations in Southeast of Iran Revealed by SSR Markers. Plant Syst. Evol. 2015, 301, 691–699. [Google Scholar] [CrossRef]
- Mahmoodi, R.; Rahmani, F.; Rezaee, R. Genetic Diversity among Juglans regia L. Genotypes Assessed by Morphological Traits and Microsatellite Markers. Span. J. Agric. Res. 2013, 11, 431. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Fatahi, R.; Zamani, Z. Analysis of Genetic Diversity among Some Persian Walnut Genotypes (Juglans regia L.) Using Morphological Traits and SSRs Markers. Sci. Hortic. 2011, 130, 146–151. [Google Scholar] [CrossRef]
- Feng, X.; Zhou, H.; Zulfiqar, S.; Luo, X.; Hu, Y.; Feng, L.; Malvolti, M.E.; Woeste, K.; Zhao, P. The Phytogeographic History of Common Walnut in China. Front. Plant Sci. 2018, 9, 1399. [Google Scholar] [CrossRef]
- Eser, E.; Topçu, H.; Kefayati, S.; Sütyemez, M.; Islam, M.R.; Kafkas, S. Highly Polymorphic Novel Simple Sequence Repeat Markers from Class I Repeats in Walnut (Juglansregia L.). Turk. J. Agric. For. 2019, 43, 174–183. [Google Scholar] [CrossRef]
- Zhou, H.; Zhao, P.; Woeste, K.; Zhang, S. Gene Flow among Wild and Cultivated Common Walnut (Juglans regia) Trees in the Qinling Mountains Revealed by Microsatellite Markers. J. For. Res. 2021, 32, 2189–2201. [Google Scholar] [CrossRef]
- Bujdoso, G.; Cseke, K. The Persian (English) Walnut (Juglans regia L.) Assortment of Hungary: Nut Characteristics and Origin. Sci. Hortic. 2021, 283, 110035. [Google Scholar] [CrossRef]
- Aradhya, M.; Velasco, D.; Preece, J.; Kluepfel, D.A. Biogeographic and Glacial History of Walnut (Juglans regia L.). Acta Hortic. 2021, 1318, 9–16. [Google Scholar] [CrossRef]
- Pollegioni, P.; Woeste, K.; Chiocchini, F.; Del Lungo, S.; Ciolfi, M.; Olimpieri, I.; Tortolano, V.; Clark, J.; Hemery, G.E.; Mapelli, S.; et al. Rethinking the History of Common Walnut (Juglans regia L.) in Europe: Its Origins and Human Interactions. PLoS ONE 2017, 12, e0172541. [Google Scholar] [CrossRef]
- Guney, M.; Kafkas, S.; Keles, H.; Zarifikhosroshahi, M.; Gundesli, M.A.; Ercisli, S.; Necas, T.; Bujdoso, G. Genetic Diversity among Some Walnut (Juglans regia L.) Genotypes by SSR Markers. Sustainability 2021, 13, 6830. [Google Scholar] [CrossRef]
- Karimi, H.R.; Mirdehghan, S.H. Correlation between the Morphological Characters of Pomegranate (Punica granatum) Traits and Their Implications for Breeding. Turk. J. Bot. 2013, 37, 355–362. [Google Scholar] [CrossRef]
- Waits, L.P.; Luikart, G.; Taberlet, P. Estimating the Probability of Identity among Genotypes in Natural Populations: Cautions and Guidelines. Mol. Ecol. 2001, 10, 249–256. [Google Scholar] [CrossRef]
- Bernard, A.; Barreneche, T.; Lheureux, F.; Dirlewanger, E. Analysis of Genetic Diversity and Structure in a Worldwide Walnut (Juglans regia L.) Germplasm Using SSR Markers. PLoS ONE 2018, 13, e0208021. [Google Scholar] [CrossRef]
- Fatahi, R.; Ebrahimi, A.; Zamani, Z. Characterization of some Iranians and foreign walnut genotypes using morphological traits and RAPD markers. Hortic Environ. Biotechnol. 2010, 51, 51–60. [Google Scholar]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising How the Computer Program cervus Accommodates Genotyping Error Increases Success in Paternity Assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; A Series of Books in Biology; Freeman: San Francisco, CA, USA, 1973; ISBN 978-0-7167-0697-7. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
| Primer | Na | Ne | Ho | He | I | PIC | PI |
|---|---|---|---|---|---|---|---|
| WGA009 | 9.50 | 7.78 | 0.54 | 0.84 | 2.03 | 0.94 | 0.0073 |
| WGA321 | 9.50 | 8.07 | 0.62 | 0.85 | 2.06 | 0.95 | 0.0059 |
| WGA349 | 9.17 | 7.50 | 0.57 | 0.84 | 2.04 | 0.94 | 0.0071 |
| WGA069 | 9.83 | 8.46 | 0.60 | 0.87 | 2.17 | 0.95 | 0.0057 |
| WGA118 | 6.50 | 5.94 | 0.03 | 0.76 | 1.70 | 0.93 | 0.0102 |
| WGA202 | 6.67 | 6.20 | 0.00 | 0.78 | 1.73 | 0.94 | 0.0059 |
| WGA276 | 9.17 | 7.33 | 0.54 | 0.83 | 2.00 | 0.94 | 0.0073 |
| WGA001 | 12.83 | 11.27 | 0.84 | 0.88 | 2.37 | 0.97 | 0.0019 |
| WGA376 | 10.67 | 9.23 | 0.53 | 0.87 | 2.20 | 0.96 | 0.0035 |
| Mean | 9.3 | 7.97 | 0.47 | 0.84 | 2.03 | 0.94 | 0.0061 |
| No. | Local Walnut Selection | Latitude | Longitude | Altitude |
|---|---|---|---|---|
| 1 | GR-PARNON-01 | 37°15′13.8″ N | 22°33′14.2″ E | 1031 |
| 2 | GR-PARNON-02 | 37°15′13.3″ N | 22°33′14.2″ E | 1011 |
| 3 | GR-PARNON-03 | 37°14′27.1″ N | 22°33′26.5″ E | 1079 |
| 4 | GR-PARNON-04 | 37°14′18.1″ N | 22°33′25.0″ E | 1045 |
| 5 | GR-PARNON-05 | 37°14′36.3″ N | 22°30′22.4″ E | 720 |
| 6 | GR-PARNON-06 | 37°14′42.4″ N | 22°30′15.5″ E | 755 |
| 7 | GR-PARNON-07 | 37°14′39.6″ N | 22°30′35.4″ E | 730 |
| 8 | GR-PARNON-08 | 37°14′54.1″ N | 22°34′05.3″ E | 1062 |
| 9 | GR-PARNON-09 | 37°14′55.8″ N | 22°34′34.6″ E | 1224 |
| 10 | GR-PARNON-10 | 37°14′55.7″ N | 22°34′37.3″ E | 1247 |
| 11 | GR-PARNON-11 | 37°14′28.5″ N | 22°34′31.6″ E | 1297 |
| 12 | GR-PARNON-12 | 37°14′27.3″ N | 22°34′42.7″ E | 1280 |
| 13 | GR-PARNON-13 | 37°14′22.9″ N | 22°34′36.6″ E | 1278 |
| 14 | GR-PARNON-14 | 37°14′35.0″ N | 22°34′20.3″ E | 1212 |
| 15 | GR-PARNON-15 | 37°16′22.1″ N | 22°31′32.1″ E | 1020 |
| 16 | GR-PARNON-16 | 37°15′49.1″ N | 22°31′58.9″ E | 950 |
| 17 | GR-PARNON-17 | 37°15′47.4″ N | 22°32′02.3″ E | 980 |
| 18 | GR-PARNON-18 | 37°15′44.2″ N | 22°31′57.6″ E | 940 |
| 19 | GR-PARNON-19 | 37°15′55.4″ N | 22°32′50.1″ E | 1135 |
| 20 | GR-PARNON-20 | 37°15′55.0″ N | 22°32′51.0″ E | 1105 |
| 21 | GR-PARNON-21 | 37°15′58.4″ N | 22°33′10.5″ E | 1100 |
| 22 | GR-PARNON-22 | 37°14′47.2″ N | 22°32′30.1″ E | 865 |
| 23 | GR-PARNON-23 | 37°15′21.7″ N | 22°31′33.2″ E | 910 |
| 24 | GR-PARNON-24 | 37°15′19.5″ N | 22°31′32.7″ E | 878 |
| 25 | GR-PARNON-25 | 37°14′57.0″ N | 22°32′25.2″ E | 840 |
| 26 | GR-PARNON-26 | 37°15′57.6″ N | 22°32′58.4″ E | 1127 |
| 27 | GR-PARNON-27 | 37°16′08.6″ N | 22°31′53.3″ E | 1110 |
| 28 | GR-PARNON-28 | 37°14′20.3″ N | 22°33′04.1″ E | 921 |
| 29 | GR-PARNON-29 | 37°16′24.3″ N | 22°32′42.9″ E | 1279 |
| 30 | GR-PARNON-30 | 37°14′37.5″ N | 22°32′40.8″ E | 884 |
| 31 | GR-PARNON-31 | 37°16′09.3″ N | 22°32′19.1″ E | 1204 |
| 32 | GR-PARNON-32 | 37°16′15.1″ N | 22°32′06.1″ E | 1217 |
| 33 | GR-PARNON-33 | 37°16′17.8″ N | 22°32′08.3″ E | 1232 |
| 34 | GR-PARNON-34 | 37°14′40.6″ N | 22°33′09.9″ E | 985 |
| 35 | GR-PARNON-35 | 37°14′38.7″ N | 22°33′09.5″ E | 1001 |
| 36 | GR-PARNON-36 | 37°14′38.6″ N | 22°33′05.4″ E | 950 |
| 37 | GR-PARNON-37 | 37°14′45.9″ N | 22°34′07.8″ E | 1078 |
| 38 | GR-PARNON-38 | 37°14′46.6″ N | 22°34′51.7″ E | 1290 |
| 39 | GR-PARNON-39 | 37°14′54.1″ N | 22°34′05.4″ E | 1061 |
| 40 | GR-PARNON-40 | 37°14′39.7″ N | 22°33′09.5″ E | 803 |
| 41 | GR-PARNON-41 | 37°15′01.2″ N | 22°34′34.9″ E | 1287 |
| 42 | GR-PARNON-42 | 37°17′18.0″ N | 22°29′31.0″ E | 905 |
| 43 | GR-PARNON-43 | 37°15′30.4″ N | 22°32′37.7″ E | 1041 |
| 44 | GR-PARNON-44 | 37°16′37.9″ N | 22°31′04.2″ E | 1053 |
| 45 | GR-PARNON-45 | 37°17′21.8″ N | 22°30′17.7″ E | 880 |
| 46 | GR-PARNON-46 | 37°16′30.7″ N | 22°31′37.3″ E | 1052 |
| 47 | GR-PARNON-47 | 37°16′06.8″ N | 22°31′48.9″ E | 1036 |
| Primer | Primer Sequence (5′–3′) | Repeat Motif | Annealing Temperature (°C) | Product Size Range (bp) |
|---|---|---|---|---|
| WGA009 | F CATCAAAGCAAGCAATGGG | (GA)16 | 51 | 231–245 |
| R CCATTGCTCTGTGATTGGG | ||||
| WGA321 | F TCCAATCGAAACTCCAAAGG | (GA)14 | 51 | 223–245 |
| R TGTCCAAAGACGATGATGGA | ||||
| WGA349 | F GTGGCGAAAGTTTATTTTTTGC | (CT)14 | 52 | 262–274 |
| R ACAAATGCACAGCAGCAAAC | ||||
| WGA069 | F TTAGTTAGCAAACCCACCCG | (GA)4ATATAA(GA)1 | 54 | 160–182 |
| R AGATGCACAGACCAACCCTC | ||||
| WGA118 | F TGTGCTCTGATCTGCCTCC | (GA)18(GT)11 | 54 | 186–200 |
| R GGGTGGGTGAAAAGTAGCAA | ||||
| WGA202 | F CCCATCTACCGTTGCACTTT | (GA)11 | 54 | 259–295 |
| R GCTGGTGGTTCTATCATGGG | ||||
| WGA276 | F CTCACTTTCTCGGCTCTTCC | (GA)14 | 55 | 168–194 |
| R GGTCTTATGTGGGCAGTCGT | ||||
| WGA001 | F ATTGGAAGGGAAGGGAAATG | (GA)5GCA(GA)3GCA(GA)3 | 52 | 180–192 |
| R CGCGCACATACGTAAATCAC | ||||
| WGA376 | F GCCCTCAAAGTGATGAACGT | (AG)2AA(AG)6 | 54 | 230–265 |
| R TCATCCATATTTACCCCTTTCG |
| Groups of Samples | Altitude Range |
|---|---|
| 1 | 720–755 m |
| 2 | 803–884 m |
| 3 | 905–985 m |
| 4 | 1001–1079 m |
| 5 | 1100–1135 m |
| 6 | 1204–1297 m |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manthos, I.; Sotiropoulos, T.; Karapetsi, L.; Ganopoulos, I.; Pratsinakis, E.D.; Maloupa, E.; Madesis, P. Molecular Characterization of Local Walnut (Juglans regia) Genotypes in the North-East Parnon Mountain Region of Greece. Int. J. Mol. Sci. 2023, 24, 17230. https://doi.org/10.3390/ijms242417230
Manthos I, Sotiropoulos T, Karapetsi L, Ganopoulos I, Pratsinakis ED, Maloupa E, Madesis P. Molecular Characterization of Local Walnut (Juglans regia) Genotypes in the North-East Parnon Mountain Region of Greece. International Journal of Molecular Sciences. 2023; 24(24):17230. https://doi.org/10.3390/ijms242417230
Chicago/Turabian StyleManthos, Ioannis, Thomas Sotiropoulos, Lefkothea Karapetsi, Ioannis Ganopoulos, Emmanouil D. Pratsinakis, Eleni Maloupa, and Panagiotis Madesis. 2023. "Molecular Characterization of Local Walnut (Juglans regia) Genotypes in the North-East Parnon Mountain Region of Greece" International Journal of Molecular Sciences 24, no. 24: 17230. https://doi.org/10.3390/ijms242417230






