Characterisation of the Paternal Influence on Intergenerational Offspring Cardiac and Brain Lipid Homeostasis in Mice
Abstract
1. Introduction
2. Results
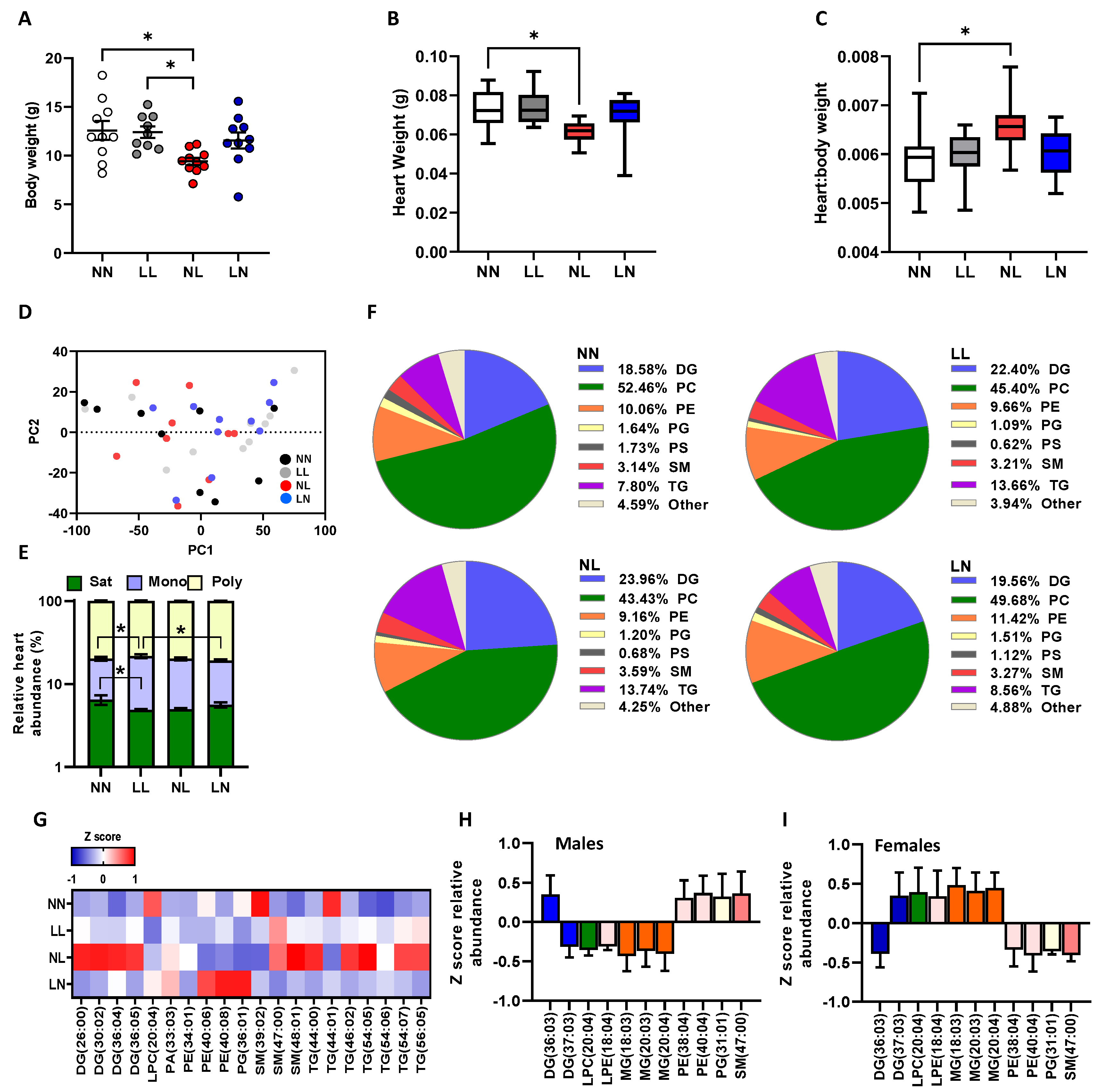
2.1. F1 Neonatal Lipids
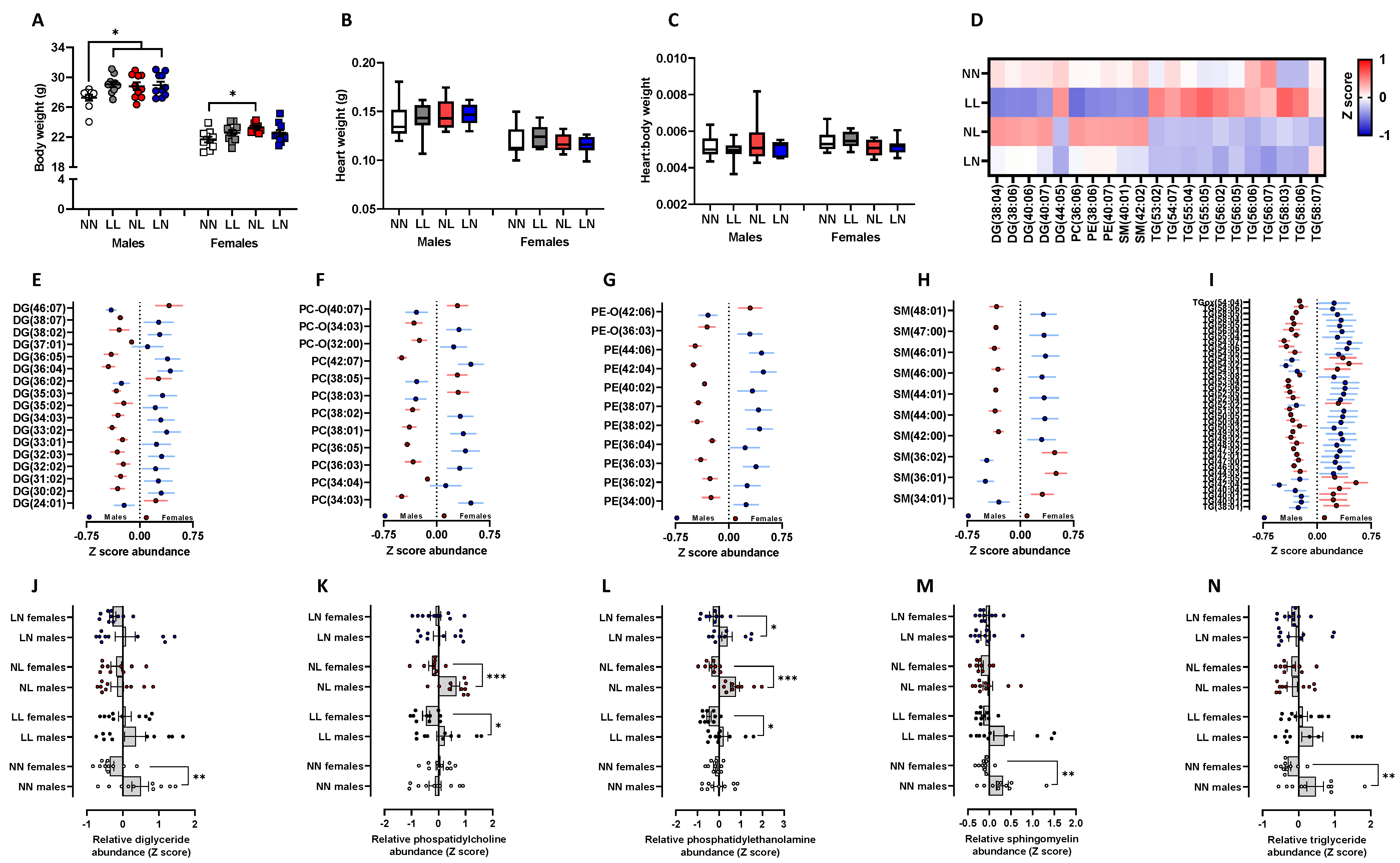
2.2. F1 Adult Offspring Lipids
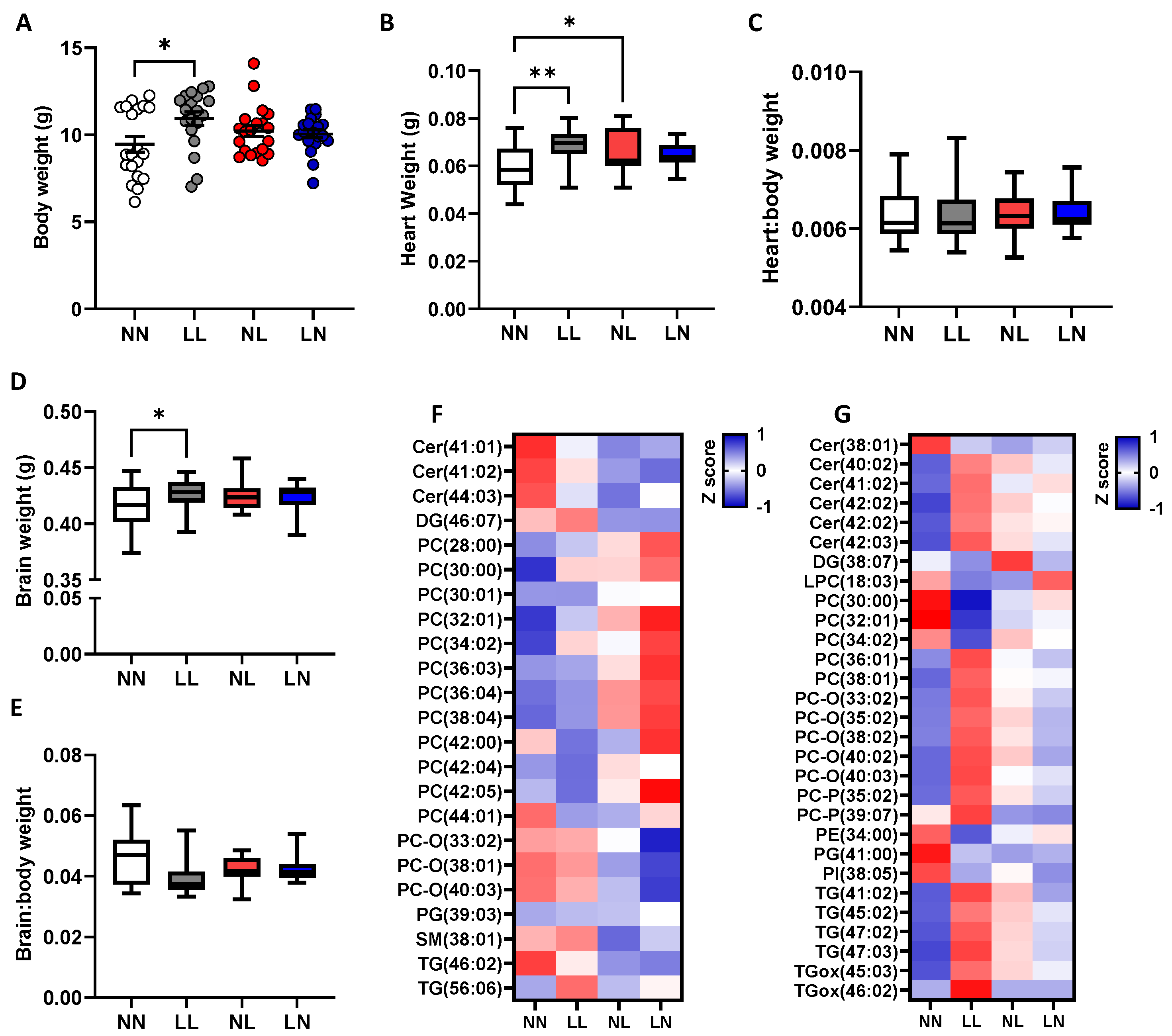
2.3. F2 Neonatal Offspring Lipids
3. Discussion
4. Materials and Methods
4.1. Animal Housing
4.2. Offspring Generation
4.3. Offspring Tissue Collection and Lipid Isolation
4.4. Offspring Tissue Lipidome Profiling
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Denisenko, Y.K.; Kytikova, O.Y.; Novgorodtseva, T.P.; Antonyuk, M.V.; Gvozdenko, T.A.; Kantur, T.A. Lipid-Induced Mechanisms of Metabolic Syndrome. J. Obes. 2020, 2020, 5762395. [Google Scholar] [CrossRef]
- Collab. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef]
- Hsu, C.N.; Hou, C.Y.; Hsu, W.H.; Tain, Y.L. Early-Life Origins of Metabolic Syndrome: Mechanisms and Preventive Aspects. Int. J. Mol. Sci. 2021, 22, 11872. [Google Scholar] [CrossRef] [PubMed]
- Crump, C.; Sundquist, J.; Sundquist, K. Association of preterm birth with lipid disorders in early adulthood: A Swedish cohort study. PLoS Med. 2019, 16, e1002947. [Google Scholar] [CrossRef]
- Rotteveel, J.; van Weissenbruch, M.M.; Twisk, J.W.; Delemarre-Van de Waal, H.A. Abnormal lipid profile and hyperinsulinaemia after a mixed meal: Additional cardiovascular risk factors in young adults born preterm. Diabetologia 2008, 51, 1269–1275. [Google Scholar] [CrossRef]
- Leunissen, R.W.; Kerkhof, G.F.; Stijnen, T.; Hokken-Koelega, A. Timing and tempo of first-year rapid growth in relation to cardiovascular and metabolic risk profile in early adulthood. JAMA 2009, 301, 2234–2242. [Google Scholar] [CrossRef]
- Yu, Z.B.; Han, S.P.; Zhu, G.Z.; Zhu, C.; Wang, X.J.; Cao, X.G.; Guo, X.R. Birth weight and subsequent risk of obesity: A systematic review and meta-analysis. Obes. Rev. 2011, 12, 525–542. [Google Scholar] [CrossRef]
- Dickinson, H.; Moss, T.J.; Gatford, K.L.; Moritz, K.M.; Akison, L.; Fullston, T.; Hryciw, D.H.; Maloney, C.A.; Morris, M.J.; Wooldridge, A.L.; et al. A review of fundamental principles for animal models of DOHaD research: An Australian perspective. J. Dev. Orig. Health Dis. 2016, 7, 449–472. [Google Scholar] [CrossRef]
- Furse, S.; Fernandez-Twinn, D.S.; Chiarugi, D.; Koulman, A.; Ozanne, S.E. Lipid Metabolism Is Dysregulated before, during and after Pregnancy in a Mouse Model of Gestational Diabetes. Int. J. Mol. Sci. 2021, 22, 7452. [Google Scholar] [CrossRef]
- Gould, J.M.; Smith, P.J.; Airey, C.J.; Mort, E.J.; Airey, L.E.; Warricker, F.D.M.; Pearson-Farr, J.E.; Weston, E.C.; Gould, P.J.W.; Semmence, O.G.; et al. Mouse maternal protein restriction during preimplantation alone permanently alters brain neuron proportion and adult short-term memory. Proc. Natl. Acad. Sci. USA 2018, 115, E7398–E7407. [Google Scholar] [CrossRef]
- Veena, S.R.; Gale, C.R.; Krishnaveni, G.V.; Kehoe, S.H.; Srinivasan, K.; Fall, C.H. Association between maternal nutritional status in pregnancy and offspring cognitive function during childhood and adolescence; a systematic review. BMC Pregnancy Childbirth 2016, 16, 220. [Google Scholar] [CrossRef]
- Watkins, A.J.; Ursell, E.; Panton, R.; Papenbrock, T.; Hollis, L.; Cunningham, C.; Wilkins, A.; Perry, V.H.; Sheth, B.; Kwong, W.Y.; et al. Adaptive responses by mouse early embryos to maternal diet protect fetal growth but predispose to adult onset disease. Biol. Reprod. 2008, 78, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Watkins, A.J.; Rubini, E.; Hosier, E.D.; Morgan, H.L. Paternal programming of offspring health. Early Hum. Dev. 2020, 150, 105185. [Google Scholar] [CrossRef]
- Derraik, J.G.B.; Pasupathy, D.; McCowan, L.M.E.; Poston, L.; Taylor, R.S.; Simpson, N.A.B.; Dekker, G.A.; Myers, J.; Vieira, M.C.; Cutfield, W.S.; et al. Paternal contributions to large-for-gestational-age term babies: Findings from a multicenter prospective cohort study. J. Dev. Orig. Health Dis. 2019, 10, 529–535. [Google Scholar] [CrossRef]
- McCowan, L.M.; North, R.A.; Kho, E.M.; Black, M.A.; Chan, E.H.; Dekker, G.A.; Poston, L.; Taylor, R.S.; Roberts, C.T. Paternal contribution to small for gestational age babies: A multicenter prospective study. Obesity 2011, 19, 1035–1039. [Google Scholar] [CrossRef]
- Gaillard, R.; Steegers, E.A.; Duijts, L.; Felix, J.F.; Hofman, A.; Franco, O.H.; Jaddoe, V.W. Childhood cardiometabolic outcomes of maternal obesity during pregnancy: The Generation R Study. Hypertension 2014, 63, 683–691. [Google Scholar] [CrossRef]
- McCarthy, K.; Ye, Y.L.; Yuan, S.; He, Q.Q. Parental weight status and offspring cardiovascular disease risks: A cross-sectional study of Chinese children. Prev. Chronic Dis. 2015, 12, E01. [Google Scholar] [CrossRef]
- Silva, D.R.; Werneck, A.O.; Collings, P.J.; Fernandes, R.A.; Barbosa, D.S.; Ronque, E.R.V.; Sardinha, L.B.; Cyrino, E.S. Family history of cardiovascular disease and parental lifestyle behaviors are associated with offspring cardiovascular disease risk markers in childhood. Am. J. Hum. Biol. 2017, 29, e22995. [Google Scholar] [CrossRef]
- Wang, C.; Yatsuya, H.; Tamakoshi, K.; Toyoshima, H.; Wada, K.; Li, Y.; Hilawe, E.H.; Uemura, M.; Chiang, C.; Zhang, Y.; et al. Association between parental history of diabetes and the incidence of type 2 diabetes mellitus differs according to the sex of the parent and offspring’s body weight: A finding from a Japanese worksite-based cohort study. Prev. Med. 2015, 81, 49–53. [Google Scholar] [CrossRef]
- Sorensen, H.J.; Pedersen, C.B.; Nordentoft, M.; Mortensen, P.B.; Ehrenstein, V.; Petersen, L. Effects of paternal age and offspring cognitive ability in early adulthood on the risk of schizophrenia and related disorders. Schizophr. Res. 2014, 160, 131–135. [Google Scholar] [CrossRef]
- Ng, S.F.; Lin, R.C.; Laybutt, D.R.; Barres, R.; Owens, J.A.; Morris, M.J. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 2010, 467, 963–966. [Google Scholar] [CrossRef] [PubMed]
- Carone, B.R.; Fauquier, L.; Habib, N.; Shea, J.M.; Hart, C.E.; Li, R.; Bock, C.; Li, C.; Gu, H.; Zamore, P.D.; et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 2010, 143, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Watkins, A.J.; Sinclair, K.D. Paternal low protein diet affects adult offspring cardiovascular and metabolic function in mice. American journal of physiology. Heart Circ. Physiol. 2014, 306, H1444–H1452. [Google Scholar] [CrossRef]
- Lambrot, R.; Xu, C.; Saint-Phar, S.; Chountalos, G.; Cohen, T.; Paquet, M.; Suderman, M.; Hallett, M.; Kimmins, S. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat. Commun. 2013, 4, 2889. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.L.; Watkins, A.J. The influence of seminal plasma on offspring development and health. Semin. Cell Dev. Biol. 2020, 97, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Donkin, I.; Barres, R. Sperm epigenetics and influence of environmental factors. Mol. Metab. 2018, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Schjenken, J.E.; Robertson, S.A. The Female Response to Seminal Fluid. Physiol. Rev. 2020, 100, 1077–1117. [Google Scholar] [CrossRef]
- Furse, S.; Watkins, A.J.; Hojat, N.; Smith, J.; Williams, H.E.L.; Chiarugi, D.; Koulman, A. Lipid Traffic Analysis reveals the impact of high paternal carbohydrate intake on offsprings lipid metabolism. Commun. Biol. 2021, 4, 163. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.L.; Furse, S.; Dias, I.H.K.; Shabir, K.; Castellanos, M.; Khan, I.; May, S.T.; Holmes, N.; Carlile, M.; Sang, F.; et al. Paternal low protein diet perturbs inter-generational metabolic homeostasis in a tissue-specific manner in mice. Commun. Biol. 2022, 5, 929. [Google Scholar] [CrossRef]
- Morgan, H.L.; Paganopoulou, P.; Akhtar, S.; Urquhart, N.; Philomin, R.; Dickinson, Y.; Watkins, A.J. Paternal diet impairs F1 and F2 offspring vascular function through sperm and seminal plasma specific mechanisms in mice. J. Physiol. 2020, 598, 699–715. [Google Scholar] [CrossRef]
- Watkins, A.J.; Dias, I.; Tsuro, H.; Allen, D.; Emes, R.D.; Moreton, J.; Wilson, R.; Ingram, R.J.M.; Sinclair, K.D. Paternal diet programs offspring health through sperm- and seminal plasma-specific pathways in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 10064–10069. [Google Scholar] [CrossRef] [PubMed]
- Bruce, K.D.; Zsombok, A.; Eckel, R.H. Lipid Processing in the Brain: A Key Regulator of Systemic Metabolism. Front. Endocrinol. 2017, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Soppert, J.; Lehrke, M.; Marx, N.; Jankowski, J.; Noels, H. Lipoproteins and lipids in cardiovascular disease: From mechanistic insights to therapeutic targeting. Adv. Drug Deliv. Rev. 2020, 159, 4–33. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Yang, S.; Zhuang, Q.; Sun, J.; Wei, P.; Zhao, X.; Chen, Y.; Chen, X.; Li, M.; Wei, L.; et al. The Associations of Lipid Profiles With Cardiovascular Diseases and Death in a 10-Year Prospective Cohort Study. Front. Cardiovasc. Med. 2021, 8, 745539. [Google Scholar] [CrossRef]
- Watkins, A.J.; Sirovica, S.; Stokes, B.; Isaacs, M.; Addison, O.; Martin, R.A. Paternal low protein diet programs preimplantation embryo gene expression, fetal growth and skeletal development in mice. Biochim. Biophys. Acta 2017, 1863, 1371–1381. [Google Scholar] [CrossRef]
- Bromfield, J.J.; Schjenken, J.E.; Chin, P.Y.; Care, A.S.; Jasper, M.J.; Robertson, S.A. Maternal tract factors contribute to paternal seminal fluid impact on metabolic phenotype in offspring. Proc. Natl. Acad. Sci. USA 2014, 111, 2200–2205. [Google Scholar] [CrossRef]
- Martinez, C.A.; Cambra, J.M.; Gil, M.A.; Parrilla, I.; Alvarez-Rodriguez, M.; Rodriguez-Martinez, H.; Cuello, C.; Martinez, E.A. Seminal Plasma Induces Overexpression of Genes Associated with Embryo Development and Implantation in Day-6 Porcine Blastocysts. Int. J. Mol. Sci. 2020, 21, 3362. [Google Scholar] [CrossRef]
- Mateo-Otero, Y.; Sanchez, J.M.; Recuero, S.; Bages-Arnal, S.; McDonald, M.; Kenny, D.A.; Yeste, M.; Lonergan, P.; Fernandez-Fuertes, B. Effect of Exposure to Seminal Plasma Through Natural Mating in Cattle on Conceptus Length and Gene Expression. Front. Cell Dev. Biol. 2020, 8, 341. [Google Scholar] [CrossRef]
- Abel, E.D.; O’Shea, K.M.; Ramasamy, R. Insulin resistance: Metabolic mechanisms and consequences in the heart. Arter. Thromb. Vasc. Biol. 2012, 32, 2068–2076. [Google Scholar] [CrossRef]
- Yu, C.; Chen, Y.; Cline, G.W.; Zhang, D.; Zong, H.; Wang, Y.; Bergeron, R.; Kim, J.K.; Cushman, S.W.; Cooney, G.J.; et al. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J. Biol. Chem. 2002, 277, 50230–50236. [Google Scholar] [CrossRef]
- Hayashi, K.; Okumura, K.; Matsui, H.; Murase, K.; Kamiya, H.; Saburi, Y.; Numaguchi, Y.; Toki, Y.; Hayakawa, T. Involvement of 1,2-diacylglycerol in improvement of heart function by etomoxir in diabetic rats. Life Sci. 2001, 68, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Baxter, G.F.; Mocanu, M.M.; Yellon, D.M. Attenuation of myocardial ischaemic injury 24 h after diacylglycerol treatment in vivo. J. Mol. Cell. Cardiol. 1997, 29, 1967–1975. [Google Scholar] [CrossRef] [PubMed]
- Luchtman, D.W.; Song, C. Cognitive enhancement by omega-3 fatty acids from child-hood to old age: Findings from animal and clinical studies. Neuropharmacology 2013, 64, 550–565. [Google Scholar] [CrossRef] [PubMed]
- Cermenati, G.; Mitro, N.; Audano, M.; Melcangi, R.C.; Crestani, M.; De Fabiani, E.; Caruso, D. Lipids in the nervous system: From biochemistry and molecular biology to patho-physiology. Biochim. Biophys. Acta 2015, 1851, 51–60. [Google Scholar] [CrossRef]
- Aureli, M.; Grassi, S.; Prioni, S.; Sonnino, S.; Prinetti, A. Lipid membrane domains in the brain. Biochim. Biophys. Acta 2015, 1851, 1006–1016. [Google Scholar] [CrossRef]
- Tabatadze, N.; Savonenko, A.; Song, H.; Bandaru, V.V.; Chu, M.; Haughey, N.J. Inhibition of neutral sphingomyelinase-2 perturbs brain sphingolipid balance and spatial memory in mice. J. Neurosci. Res. 2010, 88, 2940–2951. [Google Scholar] [CrossRef]
- Darios, F.; Wasser, C.; Shakirzyanova, A.; Giniatullin, A.; Goodman, K.; Munoz-Bravo, J.L.; Raingo, J.; Jorgacevski, J.; Kreft, M.; Zorec, R.; et al. Sphingosine facilitates SNARE complex assembly and activates synaptic vesicle exocytosis. Neuron 2009, 62, 683–694. [Google Scholar] [CrossRef]
- Chumak, T.; Lecuyer, M.J.; Nilsson, A.K.; Faustino, J.; Ardalan, M.; Svedin, P.; Sjobom, U.; Ek, J.; Obenaus, A.; Vexler, Z.S.; et al. Maternal n-3 Polyunsaturated Fatty Acid Enriched Diet Commands Fatty Acid Composition in Postnatal Brain and Protects from Neonatal Arterial Focal Stroke. Transl. Stroke Res. 2022, 13, 449–461. [Google Scholar] [CrossRef]
- Liu, Y.; Neumann, D.; Glatz, J.F.C.; Luiken, J. Molecular mechanism of lipid-induced cardiac insulin resistance and contractile dysfunction. Prostaglandins Leukot. Essent. Fat. Acids 2018, 136, 131–141. [Google Scholar] [CrossRef]
- Freedman, D.S.; Otvos, J.D.; Jeyarajah, E.J.; Shalaurova, I.; Cupples, L.A.; Parise, H.; D’Agostino, R.B.; Wilson, P.W.; Schaefer, E.J. Sex and age differences in lipoprotein subclasses measured by nuclear magnetic resonance spectroscopy: The Framingham Study. Clin. Chem. 2004, 50, 1189–1200. [Google Scholar] [CrossRef]
- Stevenson, J.C.; Crook, D.; Godsland, I.F. Influence of age and menopause on serum lipids and lipoproteins in healthy women. Atherosclerosis 1993, 98, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Steegenga, W.T.; Mischke, M.; Lute, C.; Boekschoten, M.V.; Pruis, M.G.; Lendvai, A.; Verkade, H.J.; Boekhorst, J.; Timmerman, H.M.; Plosch, T.; et al. Sexually dimorphic characteristics of the small intestine and colon of prepubescent C57BL/6 mice. Biol. Sex Differ. 2014, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Mischke, M.; Pruis, M.G.; Boekschoten, M.V.; Groen, A.K.; Fitri, A.R.; van de Heijning, B.J.; Verkade, H.J.; Muller, M.; Plosch, T.; Steegenga, W.T. Maternal Western-style high fat diet induces sex-specific physiological and molecular changes in two-week-old mouse offspring. PLoS ONE 2013, 8, e78623. [Google Scholar] [CrossRef]
- Isensee, J.; Witt, H.; Pregla, R.; Hetzer, R.; Regitz-Zagrosek, V.; Noppinger, P.R. Sexually dimorphic gene expression in the heart of mice and men. J. Mol. Med. 2008, 86, 61–74. [Google Scholar] [CrossRef]
- Welle, S.; Tawil, R.; Thornton, C.A. Sex-related differences in gene expression in human skeletal muscle. PLoS ONE 2008, 3, e1385. [Google Scholar] [CrossRef]
- Yang, X.; Schadt, E.E.; Wang, S.; Wang, H.; Arnold, A.P.; Ingram-Drake, L.; Drake, T.A.; Lusis, A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006, 16, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, M.E.; Karas, R.H. Molecular and cellular basis of cardiovascular gender differences. Science 2005, 308, 1583–1587. [Google Scholar] [CrossRef]
- Ahluwalia, A.; Clodfelter, K.H.; Waxman, D.J. Sexual dimorphism of rat liver gene expression: Regulatory role of growth hormone revealed by deoxyribonucleic Acid microarray analysis. Mol. Endocrinol. 2004, 18, 747–760. [Google Scholar] [CrossRef]
- MacCannell, A.D.V.; Futers, T.S.; Whitehead, A.; Moran, A.; Witte, K.K.; Roberts, L.D. Sexual dimorphism in adipose tissue mitochondrial function and metabolic flexibility in obesity. Int. J. Obes. 2021, 45, 1773–1781. [Google Scholar] [CrossRef]
- Brunner, F.J.; Waldeyer, C.; Ojeda, F.; Salomaa, V.; Kee, F.; Sans, S.; Thorand, B.; Giampaoli, S.; Brambilla, P.; Tunstall-Pedoe, H.; et al. Application of non-HDL cholesterol for population-based cardiovascular risk stratification: Results from the Multinational Cardiovascular Risk Consortium. Lancet 2019, 394, 2173–2183. [Google Scholar] [CrossRef]
- Richardson, T.G.; Sanderson, E.; Palmer, T.M.; Ala-Korpela, M.; Ference, B.A.; Davey Smith, G.; Holmes, M.V. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 2020, 17, e1003062. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Navas, C.; Morselli, E.; Clegg, D.J. Sexually dimorphic brain fatty acid composition in low and high fat diet-fed mice. Mol. Metab. 2016, 5, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Lundsgaard, A.M.; Kiens, B. Gender differences in skeletal muscle substrate metabolism—Molecular mechanisms and insulin sensitivity. Front. Endocrinol. 2014, 5, 195. [Google Scholar] [CrossRef]
- Marks, K.A.; Kitson, A.P.; Stark, K.D. Hepatic and plasma sex differences in saturated and monounsaturated fatty acids are associated with differences in expression of elongase 6, but not stearoyl-CoA desaturase in Sprague-Dawley rats. Genes Nutr. 2013, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- De Castro Barbosa, T.; Ingerslev, L.R.; Alm, P.S.; Versteyhe, S.; Massart, J.; Rasmussen, M.; Donkin, I.; Sjogren, R.; Mudry, J.M.; Vetterli, L.; et al. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol. Metab. 2016, 5, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Cropley, J.E.; Eaton, S.A.; Aiken, A.; Young, P.E.; Giannoulatou, E.; Ho, J.W.K.; Buckland, M.E.; Keam, S.P.; Hutvagner, G.; Humphreys, D.T.; et al. Male-lineage transmission of an acquired metabolic phenotype induced by grand-paternal obesity. Mol. Metab. 2016, 5, 699–708. [Google Scholar] [CrossRef]
- Fullston, T.; Palmer, N.O.; Owens, J.A.; Mitchell, M.; Bakos, H.W.; Lane, M. Diet-induced paternal obesity in the absence of diabetes diminishes the reproductive health of two subsequent generations of mice. Hum. Reprod. 2012, 27, 1391–1400. [Google Scholar] [CrossRef]
- Billah, M.M.; Khatiwada, S.; Morris, M.J.; Maloney, C.A. Effects of paternal overnutrition and interventions on future generations. Int. J. Obes. 2022, 46, 901–917. [Google Scholar] [CrossRef]
- Furse, S.; Fernandez-Twinn, D.; Jenkins, B.; Meek, C.L.; Williams, H.E.; Smith, G.C.S.; Charnock-Jones, D.S.; Ozanne, S.E.; Koulman, A. A high throughput platform for detailed lipidomic analysis of a range of mouse and human tissues. Anal. Bioanal. Chem. 2020, 412, 2851–2862. [Google Scholar] [CrossRef]
- Harshfield, E.L.; Koulman, A.; Ziemek, D.; Marney, L.; Fauman, E.B.; Paul, D.S.; Stacey, D.; Rasheed, A.; Lee, J.-J.; Shah, N.; et al. An Unbiased Lipid Phenotyping Approach To Study the Genetic Determinants of Lipids and Their Association with Coronary Heart Disease Risk Factors. J. Proteome Res. 2019, 18, 2397–2410. [Google Scholar] [CrossRef]
- Furse, S.; Koulman, A. The Lipid and Glyceride Profiles of Infant Formula Differ by Manufacturer, Region and Date Sold. Nutrients 2019, 11, 1122. [Google Scholar] [CrossRef]
- Bosco, M.; Culeddu, N.; Toffanin, R.; Pollesello, P. Organic solvent systems for 31P nuclear magnetic resonance analysis of lecithin phospholipids: Applications to two-dimensional gradient-enhanced 1H-detected heteronuclear multiple quantum coherence experiments. Anal. Biochem. 1997, 245, 38–47. [Google Scholar] [CrossRef]
- Furse, S.; Liddell, S.; Ortori, C.A.; Williams, H.; Neylon, D.C.; Scott, D.J.; Barrett, D.A.; Gray, D.A. The lipidome and proteome of oil bodies from Helianthus annuus (common sunflower). J. Chem. Biol. 2013, 6, 63–76. [Google Scholar] [CrossRef]
- Furse, S.; Williams, H.E.L.; Watkins, A.J.; Virtue, S.; Vidal-Puig, A.; Amarsi, R.; Charalambous, M.; Koulman, A. A pipeline for making (31)P NMR accessible for small- and large-scale lipidomics studies. Anal. Bioanal. Chem. 2021, 413, 4763–4773. [Google Scholar] [CrossRef] [PubMed]
- Murgia, S.; Mele, S.; Monduzzi, M. Quantitative characterization of phospholipids in milk fat via 31P NMR using a monophasic solvent mixture. Lipids 2003, 38, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Dearden, L.; Bouret, S.G.; Ozanne, S.E. Sex and gender differences in developmental programming of metabolism. Mol. Metab. 2018, 15, 8–19. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furse, S.; Morgan, H.L.; Koulman, A.; Watkins, A.J. Characterisation of the Paternal Influence on Intergenerational Offspring Cardiac and Brain Lipid Homeostasis in Mice. Int. J. Mol. Sci. 2023, 24, 1814. https://doi.org/10.3390/ijms24031814
Furse S, Morgan HL, Koulman A, Watkins AJ. Characterisation of the Paternal Influence on Intergenerational Offspring Cardiac and Brain Lipid Homeostasis in Mice. International Journal of Molecular Sciences. 2023; 24(3):1814. https://doi.org/10.3390/ijms24031814
Chicago/Turabian StyleFurse, Samuel, Hannah L. Morgan, Albert Koulman, and Adam J. Watkins. 2023. "Characterisation of the Paternal Influence on Intergenerational Offspring Cardiac and Brain Lipid Homeostasis in Mice" International Journal of Molecular Sciences 24, no. 3: 1814. https://doi.org/10.3390/ijms24031814
APA StyleFurse, S., Morgan, H. L., Koulman, A., & Watkins, A. J. (2023). Characterisation of the Paternal Influence on Intergenerational Offspring Cardiac and Brain Lipid Homeostasis in Mice. International Journal of Molecular Sciences, 24(3), 1814. https://doi.org/10.3390/ijms24031814






