The Dynamic and Crucial Role of the Arginine Methylproteome in Myoblast Cell Differentiation
Abstract
1. Introduction
2. Results
2.1. Quantitative Changes in the Cellular Proteome during Myoblast Differentiation
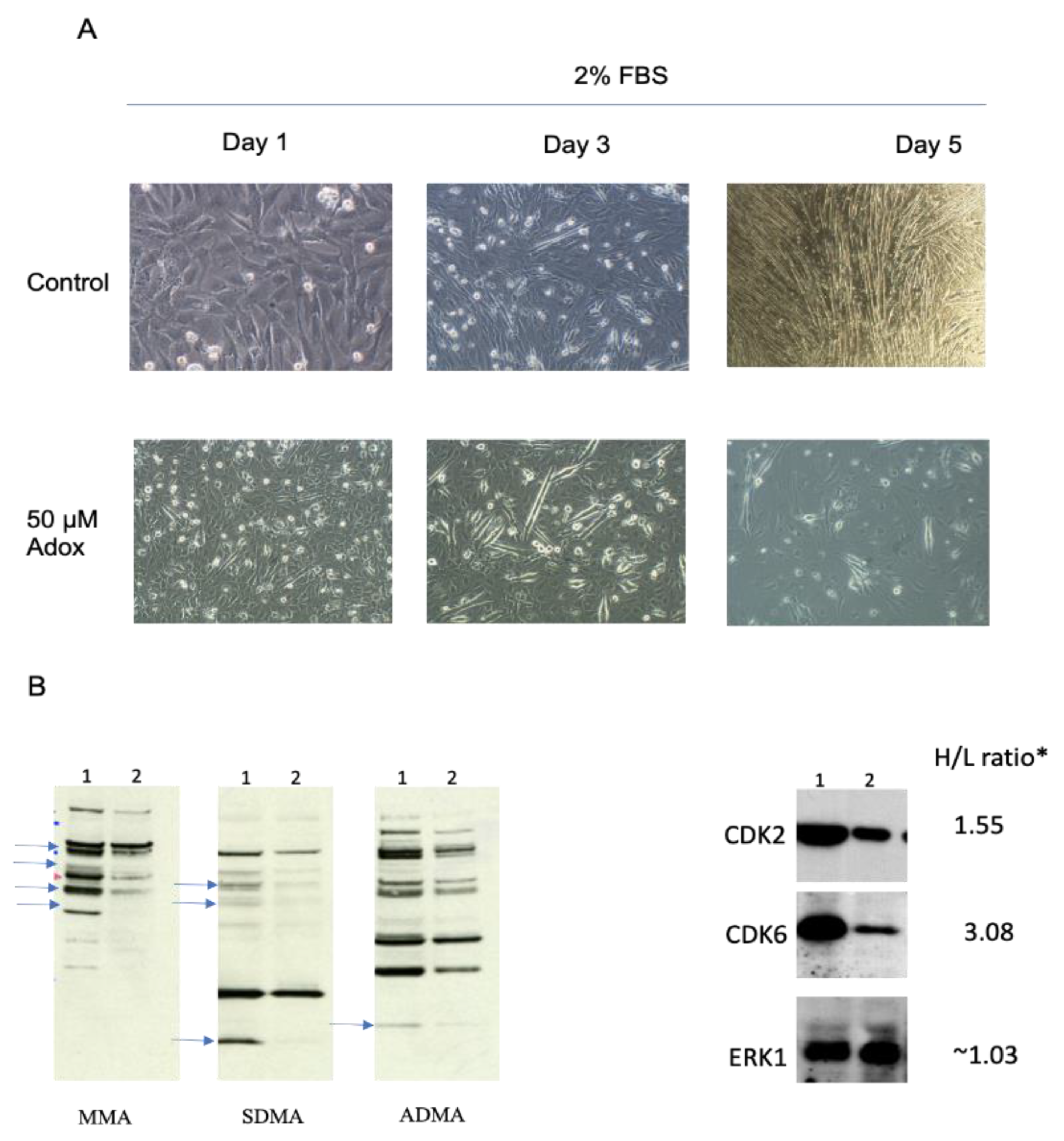
2.2. Arginine Methylation Is Important for Myoblast Differentiation
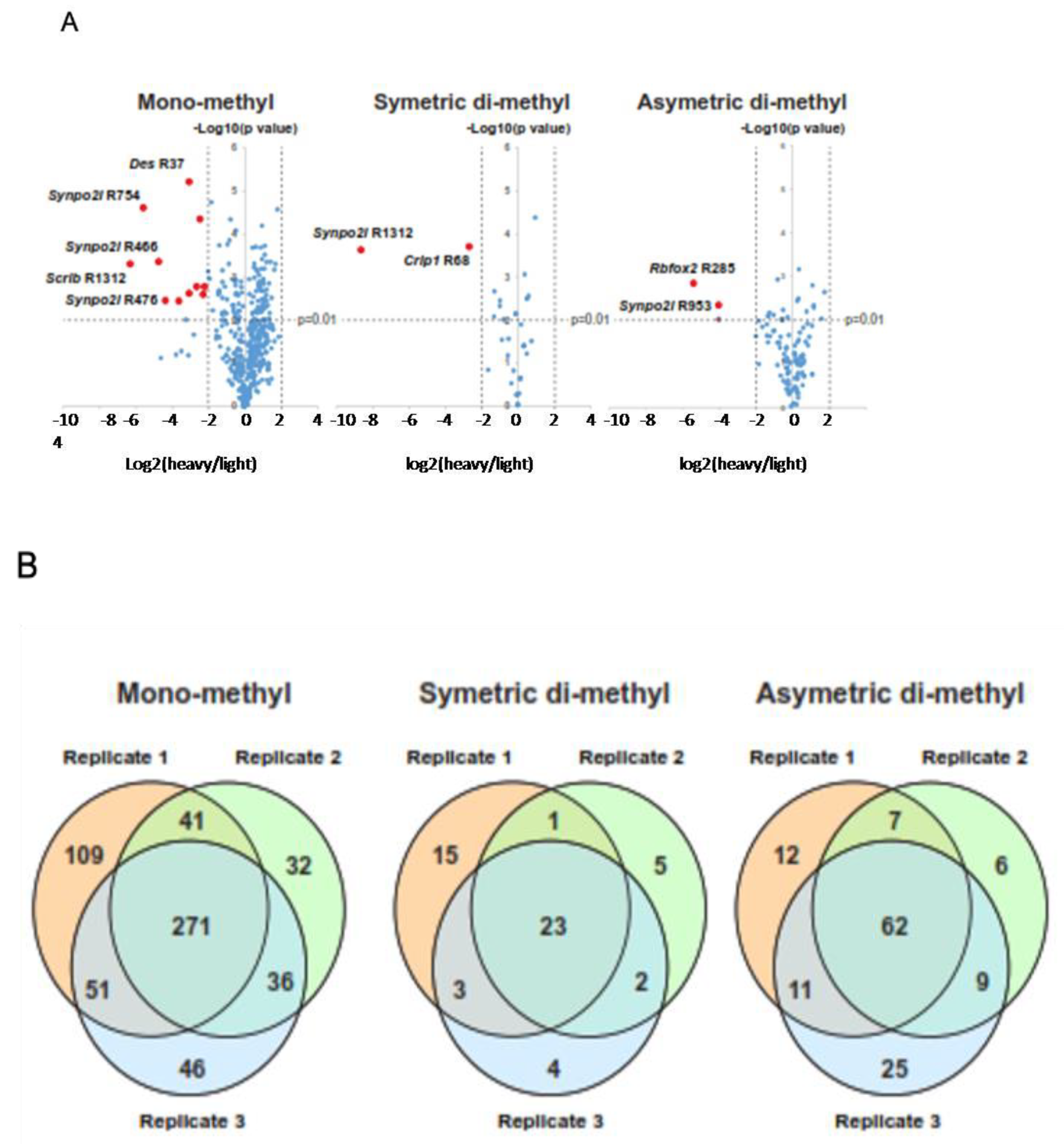
2.3. Alterations in the Arginine Methylproteome Revealed by Quantitative Proteomics
2.4. Identification of Differentially Methylated Proteins on Arginine
2.5. Transcription Factors and Histones Enriched in MMA and in DiMA Samples
2.6. Pathways Enriched in Down- or Upregulated Proteins in Whole Proteome with SILAC Proteomics
2.7. Pathways Down-or Upregulated for Arginine Monomethylation (MMA) in Differentiated Cells
2.8. Pathways Down- or Upregulated for Arginine Demethylation (DiMA) in Differentiated Cells
2.9. SLiMS and Protein Domains Enriched in MMA or DiMA Proteins
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Treatments and Immunoblot Analysis of Whole Cell Extracts for Arginine- or Lysine-Methylated Proteins
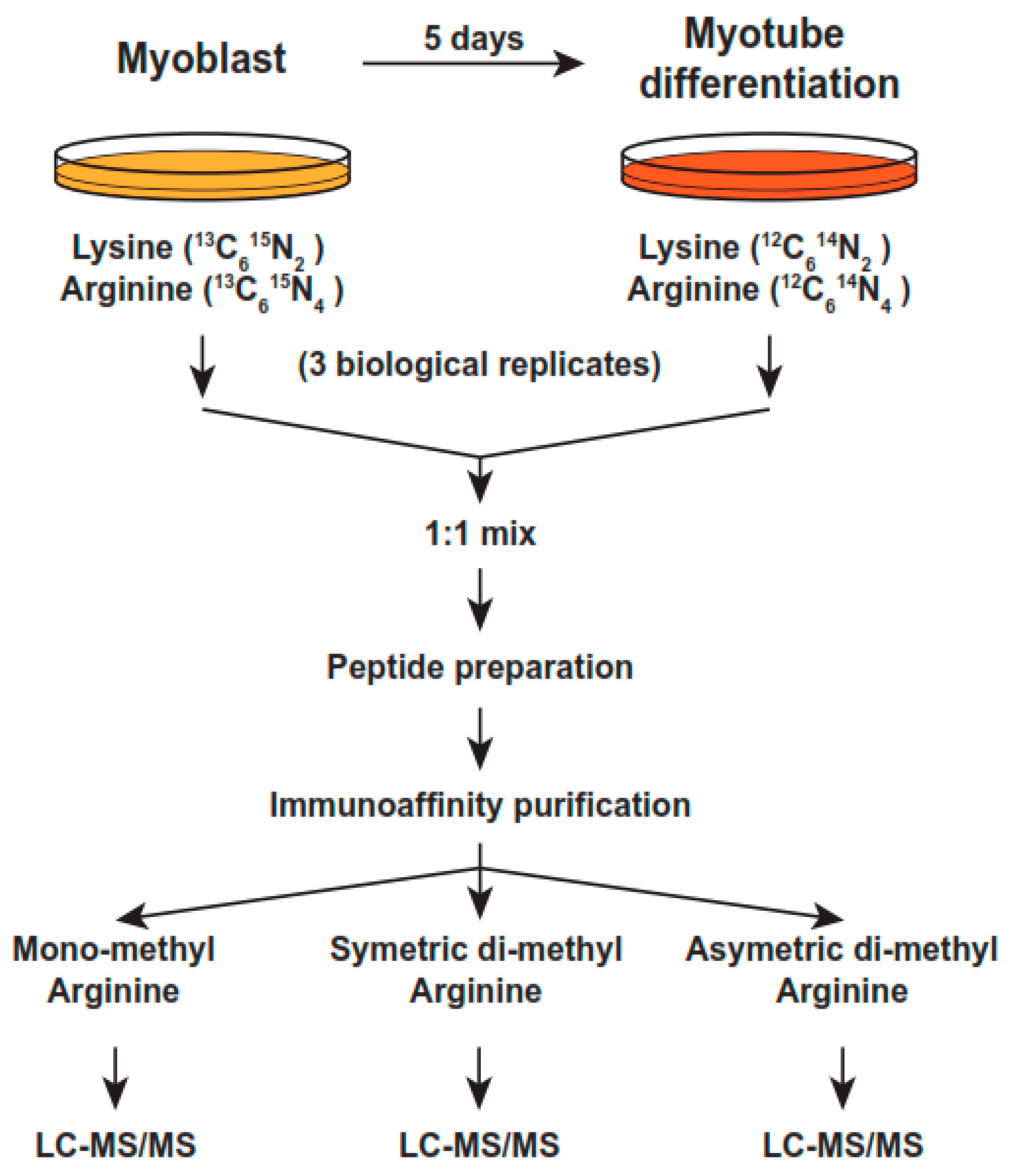
4.3. SILAC Labeling
4.4. Peptide Sample Preparation for Whole Cell Proteomic Analysis
4.5. Peptide Sample Preparation for Anti-Arginine Immunopreciptation
4.6. Immunoaffinity Purification of Methylated Peptides
4.7. Liquid Chromatography Tandem Mass Spectrometry
4.8. Mass Spectrometry Data analysis
4.9. Identification of Enriched Domains and Short Linear Motifs (SLiMs) in MMA or DiMA Proteins
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Kimura, M.; Takagi, S.; Toramoto, I.; Ishihama, Y. Identification of Mitosis-Specific Phosphorylation in Mitotic Chromosome-Associated Proteins. J. Proteome Res. 2016, 15, 3331–3341. [Google Scholar] [CrossRef] [PubMed]
- Al-Hamashi, A.A.; Diaz, K.; Huang, R. Non-Histone Arginine Methylation by Protein Arginine Methyltransferases. Curr. Protein Pept. Sci. 2020, 21, 699–712. [Google Scholar] [CrossRef]
- Alam, H.; Gu, B.; Lee, M.G. Histone methylation modifiers in cellular signaling pathways. Cell. Mol. Life Sci. 2015, 72, 4577–4592. [Google Scholar] [CrossRef] [PubMed]
- Aletta, J.M.; Hu, J.C. Protein arginine methylation in health and disease. Biotechnol. Annu. Rev. 2008, 14, 203–224. [Google Scholar]
- Auclair, Y.; Richard, S. The role of arginine methylation in the DNA damage response. DNA Repair 2013, 12, 459–465. [Google Scholar] [CrossRef]
- Carr, S.M.; Poppy Roworth, A.; Chan, C.; La Thangue, N.B. Post-translational control of transcription factors: Methylation ranks highly. FEBS J. 2015, 282, 4450–4465. [Google Scholar] [CrossRef]
- Migliori, V.; Phalke, S.; Bezzi, M.; Guccione, E. Arginine/lysine-methyl/methyl switches: Biochemical role of histone arginine methylation in transcriptional regulation. Epigenomics 2010, 2, 119–137. [Google Scholar] [CrossRef]
- Xu, J.; Richard, S. Cellular pathways influenced by protein arginine methylation: Implications for cancer. Mol. Cell 2021, 81, 4357–4368. [Google Scholar] [CrossRef]
- Wu, J.; Chen, X.; Sehgal, P.; Zhang, T.; Jackson-Weaver, O.; Gou, Y.; Bautch, V.; Frenkel, B.; Sun, H.; Xu, J. Arginine methylation of R81 in Smad6 confines BMP-induced Smad1 signaling. J. Biol. Chem. 2021, 296, 100496. [Google Scholar] [CrossRef]
- Wei, H.-M.; Hu, H.-H.; Chang, G.-Y.; Lee, Y.-J.; Li, Y.-C.; Chang, H.-H.; Li, C. Arginine methylation of the cellular nucleic acid binding protein does not affect its subcellular localization but impedes RNA binding. FEBS Lett. 2014, 588, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- Poulard, C.; Corbo, L.; Le Romancer, M. Protein arginine methylation/demethylation and cancer. Oncotarget 2016, 7, 67532–67550. [Google Scholar] [CrossRef] [PubMed]
- Kowenz-Leutz, E.; Pless, O.; Dittmar, G.; Knoblich, M.; Leutz, A. Crosstalk between C/EBPbeta phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 2010, 29, 1105–1115. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, X.-L.; Cheng, M.-B.; Zhang, Y. PRMT1 activates myogenin transcription via MyoD arginine methylation at R121. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194442. [Google Scholar] [CrossRef] [PubMed]
- Blanc, R.S.; Vogel, G.; Li, X.; Yu, Z.; Li, S.; Richard, S. Arginine Methylation by PRMT1 Regulates Muscle Stem Cell Fate. Mol. Cell. Biol. 2017, 37, e00457-16. [Google Scholar] [CrossRef]
- Li, W.-J.; He, Y.-H.; Yang, J.-J.; Hu, G.-S.; Lin, Y.-A.; Ran, T.; Peng, B.-L.; Xie, B.-L.; Huang, M.-F.; Gao, X.; et al. Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth. Nat. Commun. 2021, 12, 1946. [Google Scholar] [CrossRef]
- Fong, J.Y.; Pignata, L.; Goy, P.-A.; Kawabata, K.C.; Lee, S.C.-W.; Koh, C.M.; Musiani, D.; Massignani, E.; Kotini, A.G.; Penson, A.; et al. Therapeutic Targeting of RNA Splicing Catalysis through Inhibition of Protein Arginine Methylation. Cancer Cell 2019, 36, 194–209.e9. [Google Scholar] [CrossRef]
- Urulangodi, M.; Mohanty, A. DNA damage response and repair pathway modulation by non-histone protein methylation: Implications in neurodegeneration. J. Cell Commun. Signal. 2020, 14, 31–45. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Z.; Han, L.; Guo, Z.; Yan, B.; Guo, L.; Zhao, H.; Wei, M.; Hou, N.; Ye, J.; et al. PRMT5 regulates RNA m6A demethylation for doxorubicin sensitivity in breast cancer. Mol. Ther. 2022, 30, 2603–2617. [Google Scholar] [CrossRef]
- Gong, F.; Miller, K.M. Histone methylation and the DNA damage response. Mutat. Res. Rev. Mutat. Res. 2019, 780, 37–47. [Google Scholar] [CrossRef]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell. Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef]
- Katsuno, Y.; Qin, J.; Oses-Prieto, J.; Wang, H.; Jackson-Weaver, O.; Zhang, T.; Lamouille, S.; Wu, J.; Burlingame, A.; Xu, J.; et al. Arginine methylation of SMAD7 by PRMT1 in TGF-β-induced epithelial-mesenchymal transition and epithelial stem-cell generation. J. Biol. Chem. 2018, 293, 13059–13072. [Google Scholar] [CrossRef] [PubMed]
- Blanc, R.S.; Richard, S. Regenerating muscle with arginine methylation. Transcription 2017, 8, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, H.; Zhao, X.; Luo, Q.; Wang, Q.; Tan, K.; Wang, Z.; Jiang, J.; Cui, J.; Du, E.; et al. Arginine methylation of METTL14 promotes RNA N6-methyladenosine modification and endoderm differentiation of mouse embryonic stem cells. Nat. Commun. 2021, 12, 3780. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, E.; Ceman, S. Arginine methylation of RNA-binding proteins regulates cell function and differentiation. Mol. Reprod. Dev. 2012, 79, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-H.; Lee, R.; Kim, E.; Choi, Y.E.; Choi, E.-J. PRMT1 negatively regulates activation-induced cell death in macrophages by arginine methylation of GAPDH. Exp. Cell Res. 2018, 368, 50–58. [Google Scholar] [CrossRef]
- Fay, M.M.; Clegg, J.M.; Uchida, K.A.; Powers, M.A.; Ullman, K.S. Enhanced arginine methylation of programmed cell death 4 protein during nutrient deprivation promotes tumor cell viability. J. Biol. Chem. 2014, 289, 17541–17552. [Google Scholar] [CrossRef] [PubMed]
- Cakouros, D.; Mills, K.; Denton, D.; Paterson, A.; Daish, T.; Kumar, S. dLKR/SDH regulates hormone-mediated histone arginine methylation and transcription of cell death genes. J. Cell Biol. 2008, 182, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Calnan, B.J.; Tidor, B.; Biancalana, S.; Hudson, D.; Frankel, A.D. Arginine-mediated RNA recognition: The arginine fork. Science 1991, 252, 1167–1171. [Google Scholar] [CrossRef]
- Churcher, M.J.; Lamont, C.; Hamy, F.; Dingwall, C.; Green, S.M.; Lowe, A.D.; Butler, J.G.; Gait, M.J.; Karn, J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J. Mol. Biol. 1993, 230, 90–110. [Google Scholar] [CrossRef]
- Liu, Q.; Dreyfuss, G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 1995, 15, 2800–2808. [Google Scholar] [CrossRef] [PubMed]
- Luscombe, N.M.; Laskowski, R.A.; Thornton, J.M. Amino acid-base interactions: A three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001, 29, 2860–2874. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb. Perspect. Biol. 2014, 6, a018747. [Google Scholar] [CrossRef]
- Sun, L.; Groover, O.A.; Canfield, J.M.; Warncke, K. Critical role of arginine 160 of the EutB protein subunit for active site structure and radical catalysis in coenzyme B12-dependent ethanolamine ammonia-lyase. Biochemistry 2008, 47, 5523–5535. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.M.; Waters, M.L. Arginine methylation in a beta-hairpin peptide: Implications for Arg-pi interactions, DeltaCp(o), and the cold denatured state. J. Am. Chem. Soc. 2006, 128, 12735–12742. [Google Scholar] [CrossRef] [PubMed]
- Sprangers, R.; Groves, M.R.; Sinning, I.; Sattler, M. High-resolution X-ray and NMR structures of the SMN Tudor domain: Conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 2003, 327, 507–520. [Google Scholar] [CrossRef]
- Black, B.L.; Olson, E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998, 14, 167–196. [Google Scholar] [CrossRef]
- Parker, M.H.; Seale, P.; Rudnicki, M.A. Looking back to the embryo: Defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 2003, 4, 497–507. [Google Scholar] [CrossRef]
- Watt, K.I.; Judson, R.; Medlow, P.; Reid, K.; Kurth, T.B.; Burniston, J.G.; Ratkevicius, A.; De Bari, C.; Wackerhage, H. Yap is a novel regulator of C2C12 myogenesis. Biochem. Biophys. Res. Commun. 2010, 393, 619–624. [Google Scholar] [CrossRef]
- Haldar, M.; Karan, G.; Tvrdik, P.; Capecchi, M.R. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev. Cell 2008, 14, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, B.; Mafi, R.; Mafi, P.; Khan, W.S. The Regulation of Differentiation of Mesenchymal Stem-cells into Skeletal Muscle: A Look at Signalling Molecules Involved in Myogenesis. Curr. Stem Cell Res. Ther. 2018, 13, 384–407. [Google Scholar] [CrossRef] [PubMed]
- Pajalunga, D.; Crescenzi, M. Restoring the Cell Cycle and Proliferation Competence in Terminally Differentiated Skeletal Muscle Myotubes. Cells 2021, 10, 2753. [Google Scholar] [CrossRef]
- Langlois, S.; Cowan, K.N. Regulation of Skeletal Muscle Myoblast Differentiation and Proliferation by Pannexins. Adv. Exp. Med. Biol. 2017, 925, 57–73. [Google Scholar]
- Ciemerych, M.A.; Archacka, K.; Grabowska, I.; Przewoźniak, M. Cell cycle regulation during proliferation and differentiation of mammalian muscle precursor cells. Results Probl. Cell Differ. 2011, 53, 473–527. [Google Scholar]
- Shawber, C.; Nofziger, D.; Hsieh, J.J.; Lindsell, C.; Bögler, O.; Hayward, D.; Weinmaster, G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development 1996, 122, 3765–3773. [Google Scholar] [CrossRef]
- Puri, P.L.; Sartorelli, V.; Yang, X.J.; Hamamori, Y.; Ogryzko, V.V.; Howard, B.H.; Kedes, L.; Wang, J.Y.; Graessmann, A.; Nakatani, Y.; et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1997, 1, 35–45. [Google Scholar] [CrossRef]
- Lechner, C.; Zahalka, M.A.; Giot, J.F.; Møller, N.P.; Ullrich, A. ERK6, a mitogen-activated protein kinase involved in C2C12 myoblast differentiation. Proc. Natl. Acad. Sci. USA 1996, 93, 4355–4359. [Google Scholar] [CrossRef]
- Ornatsky, O.I.; Cox, D.M.; Tangirala, P.; Andreucci, J.J.; Quinn, Z.A.; Wrana, J.L.; Prywes, R.; Yu, Y.T.; McDermott, J.C. Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res. 1999, 27, 2646–2654. [Google Scholar] [CrossRef]
- Tao, Y.; Neppl, R.L.; Huang, Z.-P.; Chen, J.; Tang, R.-H.; Cao, R.; Zhang, Y.; Jin, S.-W.; Wang, D.-Z. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J. Cell Biol. 2011, 194, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Dacwag, C.S.; Ohkawa, Y.; Pal, S.; Sif, S.; Imbalzano, A.N. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol. Cell Biol. 2007, 27, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.-S.; Sim, Y.-J.; Jeong, H.-S.; Kim, S.-J.; Yun, Y.-J.; Song, J.-H.; Jeon, S.-H.; Choe, C.; Park, K.-T.; Kim, C.-H.; et al. Jmjd2C increases MyoD transcriptional activity through inhibiting G9a-dependent MyoD degradation. Biochim. Biophys Acta 2015, 1849, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Ling BM, T.; Bharathy, N.; Chung, T.-K.; Kok, W.K.; Li, S.; Tan, Y.H.; Rao, V.K.; Gopinadhan, S.; Sartorelli, V.; Walsh, M.J.; et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc. Natl. Acad. Sci. USA 2012, 109, 841–846. [Google Scholar] [CrossRef]
- Bharathy, N.; Taneja, R. Methylation muscles into transcription factor silencing. Transcription 2012, 3, 215–220. [Google Scholar] [CrossRef]
- Ong, S.-E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell.Proteom. 2002, 1, 376–386. [Google Scholar] [CrossRef]
- Pimienta, G.; Chaerkady, R.; Pandey, A. SILAC for global phosphoproteomic analysis. Methods Mol. Biol. 2009, 527, 107–116. [Google Scholar]
- Amanchy, R.; Kalume, D.E.; Pandey, A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci. STKE 2005, 2005, pl2. [Google Scholar] [CrossRef]
- Chen, S.L.; Loffler, K.A.; Chen, D.; Stallcup, M.R.; Muscat, G.E.O. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J. Biol. Chem. 2002, 277, 4324–4333. [Google Scholar] [CrossRef]
- Tsakona, D.; Galliou, P.A.; Papanikolaou, N.A. Identification with SILAC Proteomics of Novel Short Linear Motifs in Demethylase Enzymes Regulated During Myoblast Differentiation. Cell Dev. Biol. 2018, 7, 2. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.G. Protein methylation at the surface and buried deep: Thinking outside the histone box. Trends Biochem. Sci. 2013, 38, 243–252. [Google Scholar] [CrossRef] [PubMed]
- So, H.-K.; Kim, S.; Kang, J.-S.; Lee, S.-J. Role of Protein Arginine Methyltransferases and Inflammation in Muscle Pathophysiology. Front. Physiol. 2021, 12, 712389. [Google Scholar] [CrossRef]
- Wesche, J.; Kühn, S.; Kessler, B.M.; Salton, M.; Wolf, A. Protein arginine methylation: A prominent modification and its demethylation. Cell Mol. Life Sci. 2017, 74, 3305–3315. [Google Scholar] [CrossRef]
- Yang, J.; Chen, S.; Yang, Y.; Ma, X.; Shao, B.; Yang, S.; Wei, Y.; Wei, X. Jumonji domain-containing protein 6 protein and its role in cancer. Cell Prolif. 2020, 53, e12747. [Google Scholar] [CrossRef]
- Walport, L.J.; Hopkinson, R.J.; Chowdhury, R.; Schiller, R.; Ge, W.; Kawamura, A.; Schofield, C.J. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 2016, 7, 11974. [Google Scholar] [CrossRef]
- Song, W.; Zsindely, N.; Faragó, A.; Marsh, J.L.; Bodai, L. Systematic genetic interaction studies identify histone demethylase Utx as potential target for ameliorating Huntington’s disease. Hum. Mol. Genet. 2018, 27, 649–666. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Schapira, M.; Arrowsmith, C.H.; Barsyte-Lovejoy, D. Protein arginine methylation: From enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 2021, 20, 509–530. [Google Scholar] [CrossRef]
- Choi, J.; Jang, H.; Kim, H.; Lee, J.-H.; Kim, S.-T.; Cho, E.-J.; Youn, H.-D. Modulation of lysine methylation in myocyte enhancer factor 2 during skeletal muscle cell differentiation. Nucleic Acids Res. 2014, 42, 224–234. [Google Scholar] [CrossRef]
- Cloos PA, C.; Christensen, J.; Agger, K.; Helin, K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008, 22, 1115–1140. [Google Scholar] [CrossRef]
- Infantino, S.; Light, A.; O’Donnell, K.; Bryant, V.; Avery, D.T.; Elliott, M.; Tangye, S.G.; Belz, G.; Mackay, F.; Richard, S.; et al. Arginine methylation catalyzed by PRMT1 is required for B cell activation and differentiation. Nat. Commun. 2017, 8, 891. [Google Scholar] [CrossRef] [PubMed]
- Daubner, G.M.; Cléry, A.; Allain, F.H.-T. RRM-RNA recognition: NMR or crystallography…and new findings. Curr. Opin. Struct. Biol. 2013, 23, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Korn, S.M.; Ulshöfer, C.J.; Schneider, T.; Schlundt, A. Structures and target RNA preferences of the RNA-binding protein family of IGF2BPs:An overview. Structure 2021, 29, 787–803. [Google Scholar] [CrossRef] [PubMed]
- Maris, C.; Dominguez, C.; Allain, F.H.-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- She, X.; Lin, Y.; Liang, R.; Liu, Z.; Gao, X.; Ye, J. RNA-Binding Motif Protein 38 as a Potential Biomarker and Therapeutic Target in Cancer. Onco. Targets Ther. 2020, 13, 13225–13236. [Google Scholar] [CrossRef]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef]
- Martinez-Contreras, R.; Cloutier, P.; Shkreta, L.; Fisette, J.-F.; Revil, T.; Chabot, B. hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 2007, 623, 123–147. [Google Scholar] [PubMed]
- Poulard, C.; Jacquemetton, J.; Pham, T.H.; Le Romancer, M. Using proximity ligation assay to detect protein arginine methylation. Methods 2020, 175, 66–71. [Google Scholar] [CrossRef]
- Gao, Q.; Zhou, R.; Meng, Y.; Duan, R.; Wu, L.; Li, R.; Deng, F.; Lin, C.; Zhao, L. Long noncoding RNA CMPK2 promotes colorectal cancer progression by activating the FUBP3-c-Myc axis. Oncogene 2020, 39, 3926–3938. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Guo, K.; Wu, S.; Guo, C.; Zhang, X.; Wang, Z. Identification of a key glioblastoma candidate gene, FUBP3, based on weighted gene co-expression network analysis. BMC Neurol. 2022, 22, 139. [Google Scholar] [CrossRef]
- Sharma, M.; Anandram, S.; Ross, C.; Srivastava, S. FUBP3 regulates chronic myeloid leukaemia progression through PRC2 complex regulated PAK1-ERK signalling. J. Cell. Mol. Med. 2022, 27, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Gau, B.-H.; Chen, T.-M.; Shih, Y.-H.J.; Sun, H.S. FUBP3 interacts with FGF9 3’ microsatellite and positively regulates FGF9 translation. Nucleic Acids Res. 2011, 39, 3582–3593. [Google Scholar] [CrossRef] [PubMed]
- Bonello, T.T.; Peifer, M. Scribble: A master scaffold in polarity, adhesion, synaptogenesis, and proliferation. J. Cell Biol. 2019, 218, 742–756. [Google Scholar] [CrossRef] [PubMed]
- Mantsou, A.; Koutsogiannouli, E.; Haitoglou, C.; Papavassiliou, A.G.; Papanikolaou, N.A. Regulation of expression of the p21CIP1 gene by the transcription factor ZNF217 and MDM2. Biochem. Cell Biol. 2016, 94, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Petritis, K.; Shen, Y.; Camp, D.G.; Moore, R.J.; Smith, R.D. Phosphopeptide elution times in reversed-phase liquid chromatography. J. Chromatogr. A 2007, 1172, 9–18. [Google Scholar] [CrossRef]
- Shen, Y.; Tolić, N.; Piehowski, P.D.; Shukla, A.K.; Kim, S.; Zhao, R.; Qu, Y.; Robinson, E.; Smith, R.D.; Paša-Tolić, L. High-resolution ultrahigh-pressure long column reversed-phase liquid chromatography for top-down proteomics. J. Chromatogr. A 2017, 1498, 99–110. [Google Scholar] [CrossRef]
- Bailey, T.L.; Baker, M.E.; Elkan, C.P. An artificial intelligence approach to motif discovery in protein sequences: Application to steriod dehydrogenases. J. Steroid Biochem. Mol. Biol. 1997, 62, 29–44. [Google Scholar] [CrossRef]
- Bailey, T.L.; Gribskov, M. Methods and statistics for combining motif match scores. J. Comput. Biol. 1998, 5, 211–221. [Google Scholar] [CrossRef]



| MMA | Domain | Occurrences | |
|---|---|---|---|
| Downregulated | RRM_1 | 12 | |
| WD-40 | 1 | ||
| Upregulated | PDZ | 2 | |
| MORN | 1 | ||
| Mito-car | 1 | ||
| RRM_1 | 1 | ||
| DiMA | |||
| Downregulated | KH_1 | 12 | |
| RRM_1 | 4 | ||
| DUF1897 | 4 | ||
| Mito-car | 3 | ||
| WD-40 | 2 | ||
| Upregulated | RRM_1 | 10 | |
| PDZ | 4 | ||
| LRR_8 | 2 | ||
| MIF4G | 2 | ||
| PAM2 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papanikolaou, N.A.; Nikolaidis, M.; Amoutzias, G.D.; Fouza, A.; Papaioannou, M.; Pandey, A.; Papavassiliou, A.G. The Dynamic and Crucial Role of the Arginine Methylproteome in Myoblast Cell Differentiation. Int. J. Mol. Sci. 2023, 24, 2124. https://doi.org/10.3390/ijms24032124
Papanikolaou NA, Nikolaidis M, Amoutzias GD, Fouza A, Papaioannou M, Pandey A, Papavassiliou AG. The Dynamic and Crucial Role of the Arginine Methylproteome in Myoblast Cell Differentiation. International Journal of Molecular Sciences. 2023; 24(3):2124. https://doi.org/10.3390/ijms24032124
Chicago/Turabian StylePapanikolaou, Nikolaos A., Marios Nikolaidis, Grigorios D. Amoutzias, Ariadni Fouza, Maria Papaioannou, Akhilesh Pandey, and Athanasios G. Papavassiliou. 2023. "The Dynamic and Crucial Role of the Arginine Methylproteome in Myoblast Cell Differentiation" International Journal of Molecular Sciences 24, no. 3: 2124. https://doi.org/10.3390/ijms24032124
APA StylePapanikolaou, N. A., Nikolaidis, M., Amoutzias, G. D., Fouza, A., Papaioannou, M., Pandey, A., & Papavassiliou, A. G. (2023). The Dynamic and Crucial Role of the Arginine Methylproteome in Myoblast Cell Differentiation. International Journal of Molecular Sciences, 24(3), 2124. https://doi.org/10.3390/ijms24032124







