Implicit-Solvent Coarse-Grained Simulations of Linear–Dendritic Block Copolymer Micelles
Abstract
1. Introduction
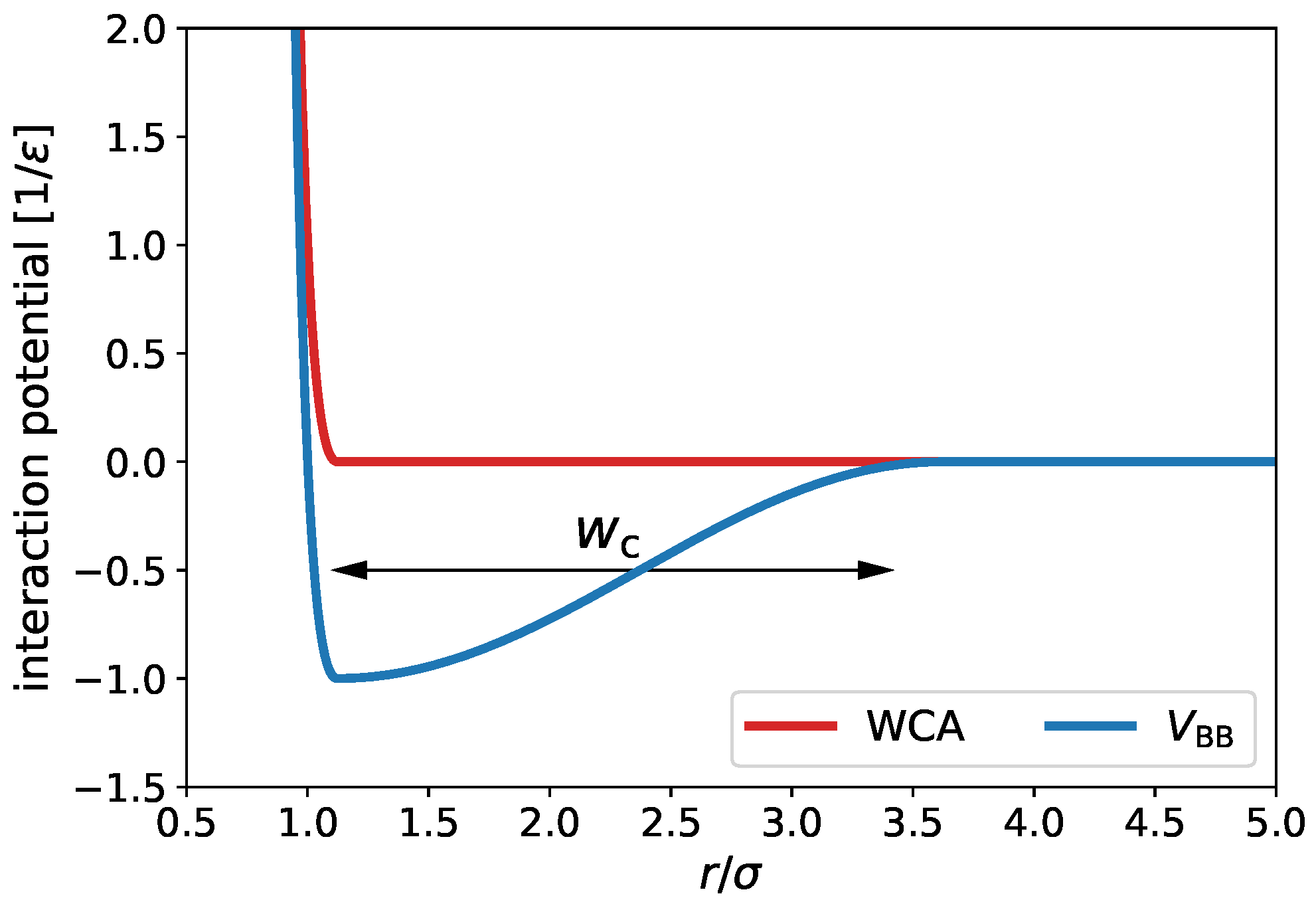
2. Simulation Model
3. Theory
4. Results
4.1. Model Parameters
4.2. Linear Block Copolymers,
4.3. Linear–Dendritic Block Copolymers
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lazzari, M.; Liu, G.; Lecommandoux, S. Block Copolymers in Nanoscience; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2006. [Google Scholar]
- Mane, S.R.; Sathyan, A.; Shunmugam, R. Biomedical Applications of pH-Responsive Amphiphilic Polymer Nanoassemblies. ACS Appl. Nano Mater. 2020, 3, 2104–2117. [Google Scholar] [CrossRef]
- Schacher, F.H.; Rupar, P.A.; Manners, I. Functional block copolymers: Nanostructured materials with emerging applications. Angew. Chem. Int. Ed. 2012, 51, 7898. [Google Scholar] [CrossRef]
- Tritschler, U.; Pearce, S.; Gwyther, J.; Whittell, G.R.; Manners, I. 50th Anniversary Perspective: Functional Nanoparticles from the Solution Self-Assembly of Block Copolymers. Macromolecules 2017, 50, 3439. [Google Scholar] [CrossRef]
- Rösler, A.; Vandermeulen, G.W.; Klok, H.A. Advanced drug delivery devices via self-assembly of amphiphilic block copolymers. Adv. Drug Deliv. Rev. 2012, 64, 270–279. [Google Scholar] [CrossRef]
- Elsabahy, M.; Wooley, K.L. Design of polymeric nanoparticles for biomedical delivery applications. Chem. Soc. Rev. 2012, 41, 2545–2561. [Google Scholar] [CrossRef]
- Lyu, Z.; Ding, L.; Tintaru, A.; Peng, L. Self-Assembling Supramolecular Dendrimers for Biomedical Applications: Lessons Learned from Poly(amidoamine) Dendrimers. Accounts Chem. Res. 2020, 53, 2936–2949. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, D.; Liu, X.; Peng, L. Amphiphilic Dendrimer Vectors for RNA Delivery: State-of-the-Art and Future Perspective. Accounts Mater. Res. 2022, 3, 484–497. [Google Scholar] [CrossRef]
- Israelachvili, J.N. Intermolecular and Surface Forces; Academic Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Israelachvili, J.N.; Mitchell, D.J.; Ninham, B.W. Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J. Chem. Soc. Faraday Trans. 2 1976, 72, 1525–1568. [Google Scholar] [CrossRef]
- Liggins, R.; Burt, H. Polyether–polyester diblock copolymers for the preparation of paclitaxel loaded polymeric micelle formulations. Adv. Drug Deliv. Rev. 2002, 54, 191–202. [Google Scholar] [CrossRef]
- Volkmar Weissig, T.E. Pharmaceutical Nanotechnology; Humana: New York, NY, USA, 2019. [Google Scholar]
- Zheng, X.; Xie, J.; Zhang, X.; Sun, W.; Zhao, H.; Li, Y.; Wang, C. An overview of polymeric nanomicelles in clinical trials and on the market. Chin. Chem. Lett. 2021, 32, 243–257. [Google Scholar] [CrossRef]
- Lee, C.C.; MacKay, J.A.; Fréchet, J.M.; Szoka, F.C. Designing dendrimers for biological applications. Nat. Biotechnol. 2005, 23, 1517. [Google Scholar] [CrossRef] [PubMed]
- Mane, S.R.; Sarkar, S.; Rao, V.N.; Sathyan, A.; Shunmugam, R. An Efficient Method to Prepare a New Class of Regioregular Graft Copolymer via a Click Chemistry Approach. RSC Adv. 2015, 5, 74159. [Google Scholar] [CrossRef]
- Vinciguerra, D.; Degrassi, A.; Mancini, L.; Mura, S.; Mougin, J.; Couvreur, P.; Nicolas, J. Drug-Initiated Synthesis of Heterotelechelic Polymer Prodrug Nanoparticles for in Vivo Imaging and Cancer Targeting. Biomacromolecules 2019, 20, 2464. [Google Scholar] [CrossRef] [PubMed]
- Matyjaszewski, K.; Spanswick, J. Controlled/living radical polymerization. Mater. Today 2005, 8, 26–33. [Google Scholar] [CrossRef]
- Hawker, C.J.; Wooley, K.L. The Convergence of Synthetic Organic and Polymer Chemistries. Science 2005, 309, 1200–1205. [Google Scholar] [CrossRef]
- Laurini, E.; Aulic, S.; Marson, D.; Fermeglia, M.; Pricl, S. Cationic Dendrimers for siRNA Delivery: An Overview of Methods for In Vitro/In Vivo Characterization. In Design and Delivery of SiRNA Therapeutics; Ditzel, H.J., Tuttolomondo, M., Kauppinen, S., Eds.; Springer: New York, NY, USA, 2021; pp. 209–244. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. A New Class of Polymers: Starburst-Dendritic Macromolecules. Polym. J. 1985, 17, 117–132. [Google Scholar] [CrossRef]
- Aharoni, S.; Murthy, N. SPHERICAL NON-DRAINING BOC-POLY (alpha, epsilon-L-LYSINE) MACROMOLECULES: SAXS AND VISCOSITY STUDIES. Polymer 1983, 24, 132–136. [Google Scholar]
- Neelov, I.; Falkovich, S.; Markelov, D.; Paci, E.; Darinskii, A.; Tenhu, H. Molecular Dynamics of Lysine Dendrimers. Computer Simulation and NMR. In Dendrimers in Biomedical Applications; The Royal Society of Chemistry: London, UK, 2013; pp. 99–114. [Google Scholar] [CrossRef]
- Moorefield, C.N.; Newkome, G.R. Unimolecular micelles: Supramolecular use of dendritic constructs to create versatile molecular containers. C. R. Chim. 2003, 6, 715–724. [Google Scholar] [CrossRef]
- Cao, W.; Zhu, L. Synthesis and Unimolecular Micelles of Amphiphilic Dendrimer-like Star Polymer with Various Functional Surface Groups. Macromolecules 2011, 44, 1500–1512. [Google Scholar] [CrossRef]
- Boris, D.; Rubinstein, M. A Self-Consistent Mean Field Model of a Starburst Dendrimer: Dense Core vs Dense Shell. Macromolecules 1996, 29, 7251–7260. [Google Scholar] [CrossRef]
- Murat, M.; Grest, G.S. Molecular Dynamics Study of Dendrimer Molecules in Solvents of Varying Quality. Macromolecules 1996, 29, 1278–1285. [Google Scholar] [CrossRef]
- Kłos, J.S.; Sommer, J.U. Properties of Dendrimers with Flexible Spacer-Chains: A Monte Carlo Study. Macromolecules 2009, 42, 4878–4886. [Google Scholar] [CrossRef]
- Okrugin, B.; Neelov, I.; Leermakers, F.; Borisov, O. Structure of asymmetrical peptide dendrimers: Insights given by self-consistent field theory. Polymer 2017, 125, 292–302. [Google Scholar] [CrossRef]
- Shavykin, O.; Mikhailov, I.; Darinskii, A.; Neelov, I.; Leermakers, F. Effect of an asymmetry of branching on structural characteristics of dendrimers revealed by Brownian dynamics simulations. Polymer 2018, 146, 256–266. [Google Scholar] [CrossRef]
- Gorzkiewicz, M.; Konopka, M.; Janaszewska, A.; Tarasenko, I.I.; Sheveleva, N.N.; Gajek, A.; Neelov, I.M.; Klajnert-Maculewicz, B. Application of new lysine-based peptide dendrimers D3K2 and D3G2 for gene delivery: Specific cytotoxicity to cancer cells and transfection in vitro. Bioorganic Chem. 2020, 95, 103504. [Google Scholar] [CrossRef]
- Gorzkiewicz, M.; Kopeć, O.; Janaszewska, A.; Konopka, M.; Pȩdziwiatr-Werbicka, E.; Tarasenko, I.I.; Bezrodnyi, V.V.; Neelov, I.M.; Klajnert-Maculewicz, B. Poly(lysine) Dendrimers Form Complexes with siRNA and Provide Its Efficient Uptake by Myeloid Cells: Model Studies for Therapeutic Nucleic Acid Delivery. Int. J. Mol. Sci. 2020, 21, 3138. [Google Scholar] [CrossRef]
- Shi, X.; Lesniak, W.; Islam, M.T.; MuÑiz, M.C.; Balogh, L.P.; Baker, J.R. Comprehensive characterization of surface-functionalized poly(amidoamine) dendrimers with acetamide, hydroxyl, and carboxyl groups. Colloids Surfaces A Physicochem. Eng. Asp. 2006, 272, 139–150. [Google Scholar] [CrossRef]
- Trinchi, A.; Muster, T.H. A Review of Surface Functionalized Amine Terminated Dendrimers for Application in Biological and Molecular Sensing. Supramol. Chem. 2007, 19, 431–445. [Google Scholar] [CrossRef]
- Caminade, A.M.; Turrin, C.O. Dendrimers for drug delivery. J. Mater. Chem. B 2014, 2, 4055–4066. [Google Scholar] [CrossRef]
- Sheveleva, N.N.; Markelov, D.A.; Vovk, M.A.; Mikhailova, M.E.; Tarasenko, I.I.; Neelov, I.M.; Lähderanta, E. NMR studies of excluded volume interactions in peptide dendrimers. Sci. Rep. 2018, 8, 8916. [Google Scholar] [CrossRef]
- Sheveleva, N.N.; Markelov, D.A.; Vovk, M.A.; Tarasenko, I.I.; Mikhailova, M.E.; Ilyash, M.Y.; Neelov, I.M.; Lahderanta, E. Stable Deuterium Labeling of Histidine-Rich Lysine-Based Dendrimers. Molecules 2019, 24, 2481. [Google Scholar] [CrossRef] [PubMed]
- Sheveleva, N.N.; Markelov, D.A.; Vovk, M.A.; Mikhailova, M.E.; Tarasenko, I.I.; Tolstoy, P.M.; Neelov, I.M.; Lähderanta, E. Lysine-based dendrimer with double arginine residues. RSC Adv. 2019, 9, 18018–18026. [Google Scholar] [CrossRef]
- Yang, H.; Lopina, S.T. Penicillin V-conjugated PEG-PAMAM star polymers. J. Biomater. Sci. Polym. Ed. 2003, 14, 1043–1056. [Google Scholar] [CrossRef]
- Luong, D.; Kesharwani, P.; Deshmukh, R.; Mohd Amin, M.C.I.; Gupta, U.; Greish, K.; Iyer, A.K. PEGylated PAMAM dendrimers: Enhancing efficacy and mitigating toxicity for effective anticancer drug and gene delivery. Acta Biomater. 2016, 43, 14–29. [Google Scholar] [CrossRef]
- Liu, X.; Liu, C.; Zhou, J.; Chen, C.; Qu, F.; Rossi, J.J.; Rocchi, P.; Peng, L. Promoting siRNA delivery via enhanced cellular uptake using an arginine-decorated amphiphilic dendrimer. Nanoscale 2015, 7, 3867–3875. [Google Scholar] [CrossRef]
- Xiong, Y.; Ke, R.; Zhang, Q.; Lan, W.; Yuan, W.; Chan, K.N.I.; Roussel, T.; Jiang, Y.; Wu, J.; Liu, S.; et al. Small Activating RNA Modulation of the G Protein-Coupled Receptor for Cancer Treatment. Adv. Sci. 2022, 9, 2200562. [Google Scholar] [CrossRef]
- Borisov, O.V.; Zhulina, E.B.; Leermakers, F.A.; Müller, A.H. Self-assembled structures of amphiphilic ionic block copolymers: Theory, self-consistent field modeling and experiment. In Self Organized Nanostructures of Amphiphilic Block Copolymers I; Muller, A.H., Borisov, O.V., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 57–129. [Google Scholar] [CrossRef]
- Zhang, Q.; Lin, J.; Wang, L.; Xu, Z. Theoretical modeling and simulations of self-assembly of copolymers in solution. Prog. Polym. Sci. 2017, 75, 1–30. [Google Scholar] [CrossRef]
- Diaz, J.; Pinna, M.; Zvelindovsky, A.V.; Pagonabarraga, I. Hybrid Time-Dependent Ginzburg-Landau Simulations of Block Copolymer Nanocomposites: Nanoparticle Anisotropy. Polymers 2022, 14, 1910. [Google Scholar] [CrossRef]
- Tan, H.; Wang, W.; Yu, C.; Zhou, Y.; Lu, Z.; Yan, D. Dissipative particle dynamics simulation study on self-assembly of amphiphilic hyperbranched multiarm copolymers with different degrees of branching. Soft Matter 2015, 11, 8460–8470. [Google Scholar] [CrossRef]
- Tan, H.; Yu, C.; Lu, Z.; Zhou, Y.; Yan, D. A dissipative particle dynamics simulation study on phase diagrams for the self-assembly of amphiphilic hyperbranched multiarm copolymers in various solvents. Soft Matter 2017, 13, 6178–6188. [Google Scholar] [CrossRef]
- Lebedeva, I.O.; Zhulina, E.B.; Borisov, O.V. Theory of linear–dendritic block copolymer micelles. ACS Macro Lett. 2018, 7, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, I.O.; Zhulina, E.B.; Borisov, O.V. Self-assembly of linear-dendritic and double dendritic block copolymers: From dendromicelles to dendrimersomes. Macromolecules 2019, 52, 3655–3667. [Google Scholar] [CrossRef]
- Suek, N.W.; Lamm, M.H. Computer Simulation of Architectural and Molecular Weight Effects on the Assembly of Amphiphilic Linear-Dendritic Block Copolymers in Solution. Langmuir 2008, 24, 3030–3036. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Chang, H.Y.; Sheng, Y.J.; Tsao, H.K. Photoresponsive Polymersomes Formed by Amphiphilic Linear–Dendritic Block Copolymers: Generation-Dependent Aggregation Behavior. Macromolecules 2012, 45, 7143–7156. [Google Scholar] [CrossRef]
- Lin, C.M.; Li, C.S.; Sheng, Y.J.; Wu, D.T.; Tsao, H.K. Size-Dependent Properties of Small Unilamellar Vesicles Formed by Model Lipids. Langmuir 2012, 28, 689–700. [Google Scholar] [CrossRef]
- Márquez-Miranda, V.; Araya-Durán, I.; Camarada, M.B.; Comer, J.; Valencia-Gallegos, J.A.; González-Nilo, F.D. Self-Assembly of Amphiphilic Dendrimers: The Role of Generation and Alkyl Chain Length in siRNA Interaction. Sci. Rep. 2016, 6, 29436. [Google Scholar] [CrossRef]
- Milchev, A.; Bhattacharya, A.; Binder, K. Formation of Block Copolymer Micelles in Solution: A Monte Carlo Study of Chain Length Dependence. Macromolecules 2001, 34, 1881–1893. [Google Scholar] [CrossRef]
- Cooke, I.R.; Deserno, M. Solvent-free model for self-assembling fluid bilayer membranes: Stabilization of the fluid phase based on broad attractive tail potentials. J. Chem. Phys. 2005, 123, 224710. [Google Scholar] [CrossRef]
- Cooke, I.R.; Kremer, K.; Deserno, M. Tunable generic model for fluid bilayer membranes. Phys. Rev. E 2005, 72, 011506. [Google Scholar] [CrossRef]
- Kremer, K.; Grest, G.S. Dynamics of entangled linear polymer melts: A molecular-dynamics simulation. J. Chem. Phys. 1990, 92, 5057–5086. [Google Scholar] [CrossRef]
- Weik, F.; Weeber, R.; Szuttor, K.; Breitsprecher, K.; de Graaf, J.; Kuron, M.; Landsgesell, J.; Menke, H.; Sean, D.; Holm, C. ESPResSo 4.0—An extensible software package for simulating soft matter systems. Eur. Phys. J. Spec. Top. 2019, 227, 1789–1816. [Google Scholar] [CrossRef]
- Available online: www.espressomd.org (accessed on 29 January 2023).
- Zhulina, E.B.; Borisov, O.V. Theory of Block Polymer Micelles: Recent Advances and Current Challenges. Macromolecules 2012, 45, 4429–4440. [Google Scholar] [CrossRef]
- de Gennes, P.J. Solid State Physics; Academic Press: New York, NY, USA, 1978; p. 1. [Google Scholar]
- Zhulina, Y.B.; Birshtein, T.M. Conformations of block-copolymer molecules in selective solvents (micellar structures). Polym. Sci. USSR 1985, 27, 570. [Google Scholar] [CrossRef]
- Halperin, A. Polymeric micelles: A star model. Macromolecules 1987, 20, 2943. [Google Scholar] [CrossRef]
- Birshtein, T.M.; Zhulina, E.B. Scaling theory of supermolecular structures in block copolymer-solvent systems: 1. Model of micellar structures. Polymer 1989, 30, 170. [Google Scholar] [CrossRef]
- Evans, D.F.; Wennerström, H. The Colloidal Domain: Where Physics, Chemistry, Biology, and Technology Meet; Wiley: Hoboken, NJ, USA, 1999. [Google Scholar]
- Shavykin, O.V.; Leermakers, F.A.M.; Neelov, I.M.; Darinskii, A.A. Self-Assembly of Lysine-Based Dendritic Surfactants Modeled by the Self-Consistent Field Approach. Langmuir 2018, 34, 1613–1626. [Google Scholar] [CrossRef]
- LaRue, I.; Adam, M.; Zhulina, E.B.; Rubinstein, M.; Pitsikalis, M.; Hadjichristidis, N.; Ivanov, D.A.; Gearba, R.I.; Anokhin, D.V.; Sheiko, S.S. Effect of the Soluble Block Size on Spherical Diblock Copolymer Micelles. Macromolecules 2008, 41, 6555–6563. [Google Scholar] [CrossRef]
- Pickett, G.T. Classical Path Analysis of end-Grafted Dendrimers: Dendrimer Forest. Macromolecules 2001, 34, 8784. [Google Scholar] [CrossRef]
- Zook, T.C.; Pickett, G.T. Hollow-Core Dendrimers Revised. Phys. Rev. Lett. 2003, 90, 015502. [Google Scholar] [CrossRef]
- Zhulina, E.B.; Leermakers, F.A.M.; Borisov, O.V. Ideal mixing in multicomponent brushes of branched macromolecules. Macromolecules 2015, 48, 8025. [Google Scholar] [CrossRef]












| System 1 | System 2 | System 3 | System 4 | |
|---|---|---|---|---|
| Case 1 |  |  |  | |
| , | , | , , | , , | |
| Case 2 |  |  |  |  |
| , | , | , , | , , | , , |
| [1/e] | |||
|---|---|---|---|
| Case 1 | system 1 | ||
| system 2 | |||
| system 3 | |||
| Case 2 | system 1 | ||
| system 2 | |||
| system 3 | |||
| system 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brito, M.E.; Mikhtaniuk, S.E.; Neelov, I.M.; Borisov, O.V.; Holm, C. Implicit-Solvent Coarse-Grained Simulations of Linear–Dendritic Block Copolymer Micelles. Int. J. Mol. Sci. 2023, 24, 2763. https://doi.org/10.3390/ijms24032763
Brito ME, Mikhtaniuk SE, Neelov IM, Borisov OV, Holm C. Implicit-Solvent Coarse-Grained Simulations of Linear–Dendritic Block Copolymer Micelles. International Journal of Molecular Sciences. 2023; 24(3):2763. https://doi.org/10.3390/ijms24032763
Chicago/Turabian StyleBrito, Mariano E., Sofia E. Mikhtaniuk, Igor M. Neelov, Oleg V. Borisov, and Christian Holm. 2023. "Implicit-Solvent Coarse-Grained Simulations of Linear–Dendritic Block Copolymer Micelles" International Journal of Molecular Sciences 24, no. 3: 2763. https://doi.org/10.3390/ijms24032763
APA StyleBrito, M. E., Mikhtaniuk, S. E., Neelov, I. M., Borisov, O. V., & Holm, C. (2023). Implicit-Solvent Coarse-Grained Simulations of Linear–Dendritic Block Copolymer Micelles. International Journal of Molecular Sciences, 24(3), 2763. https://doi.org/10.3390/ijms24032763







