Emerging RUNX2-Mediated Gene Regulatory Mechanisms Consisting of Multi-Layered Regulatory Networks in Skeletal Development
Abstract
:1. Introduction
2. Overview of Gene Regulatory Networks
3. Cis-Regulatory Networks in Skeletal Cell Types
4. Essential Roles of RUNX2 in Skeletal Programs
5. Understanding of RUNX2 Association with DNA in Genome-Scale
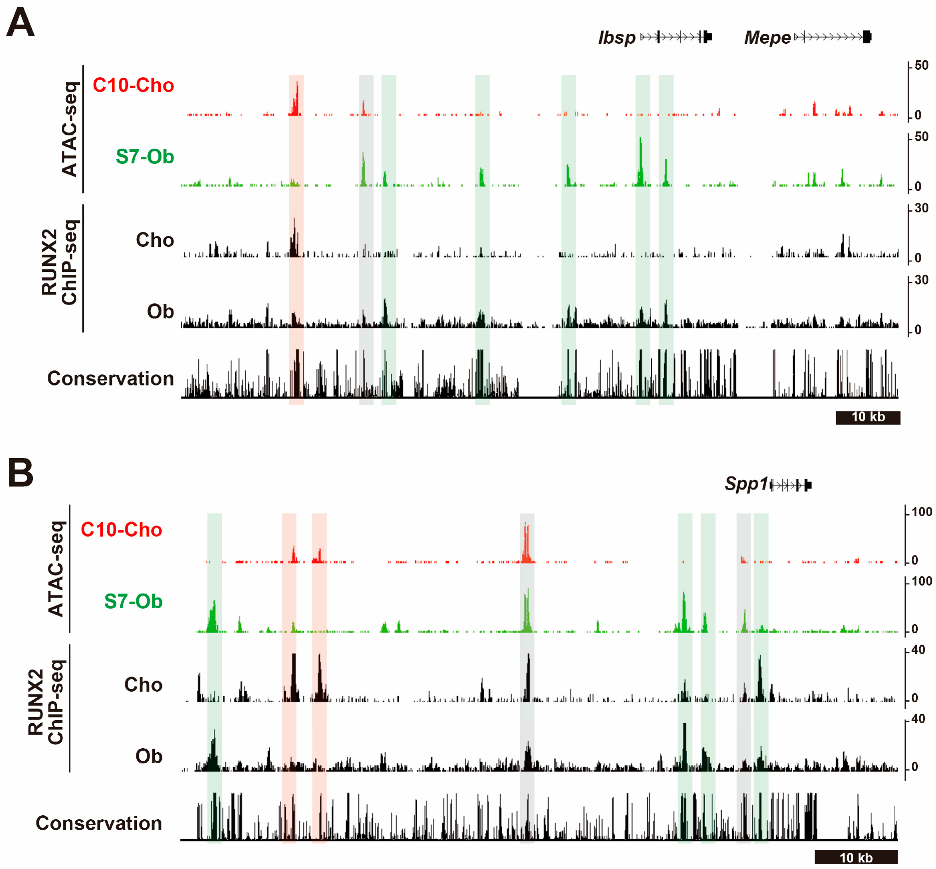
5.1. Runx2 Is Associated with CREs
5.2. The Mode of Action of RUNX2 (Figure 2)
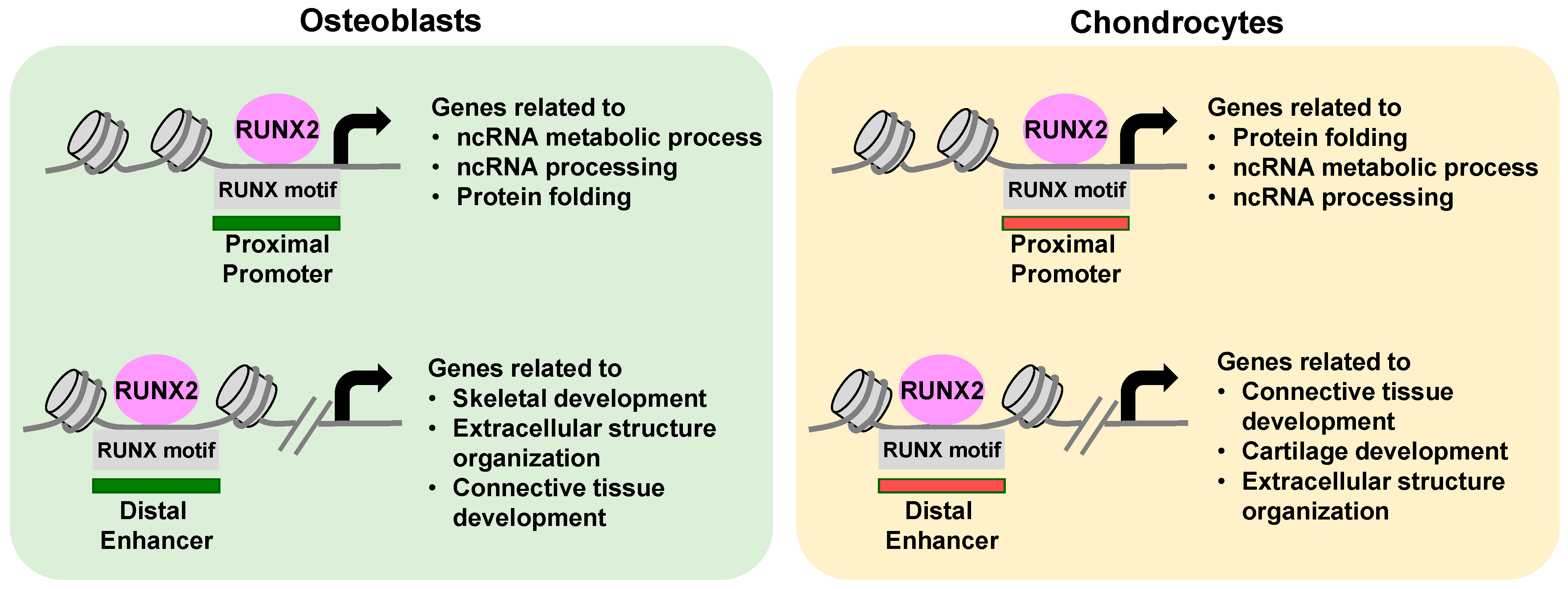
5.3. Molecular Mechanisms of the Cell Type-Distinct Roles of RUNX2
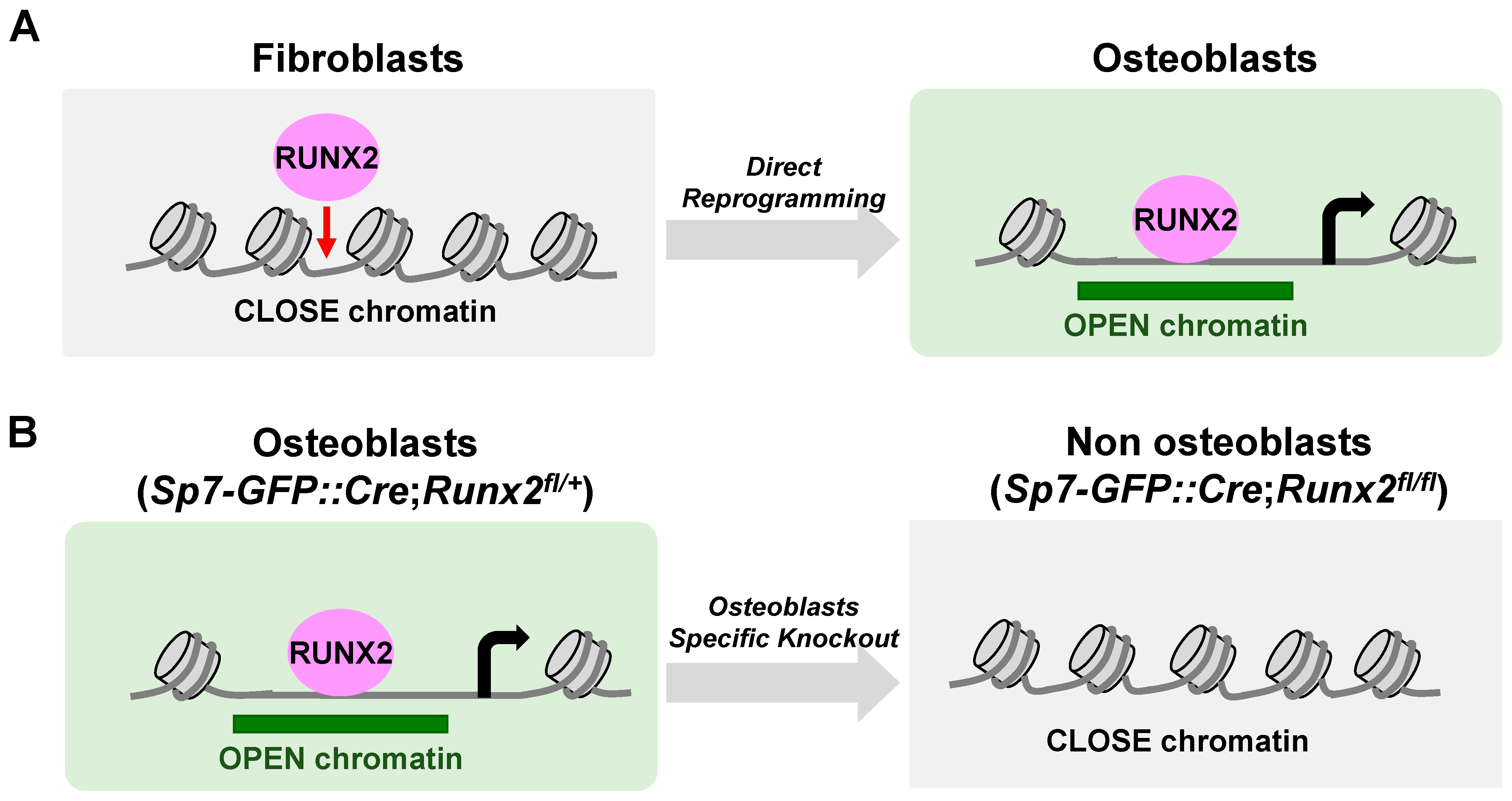
6. The Actions of RUNX2 in Chromatin Accessibility (Figure 4)
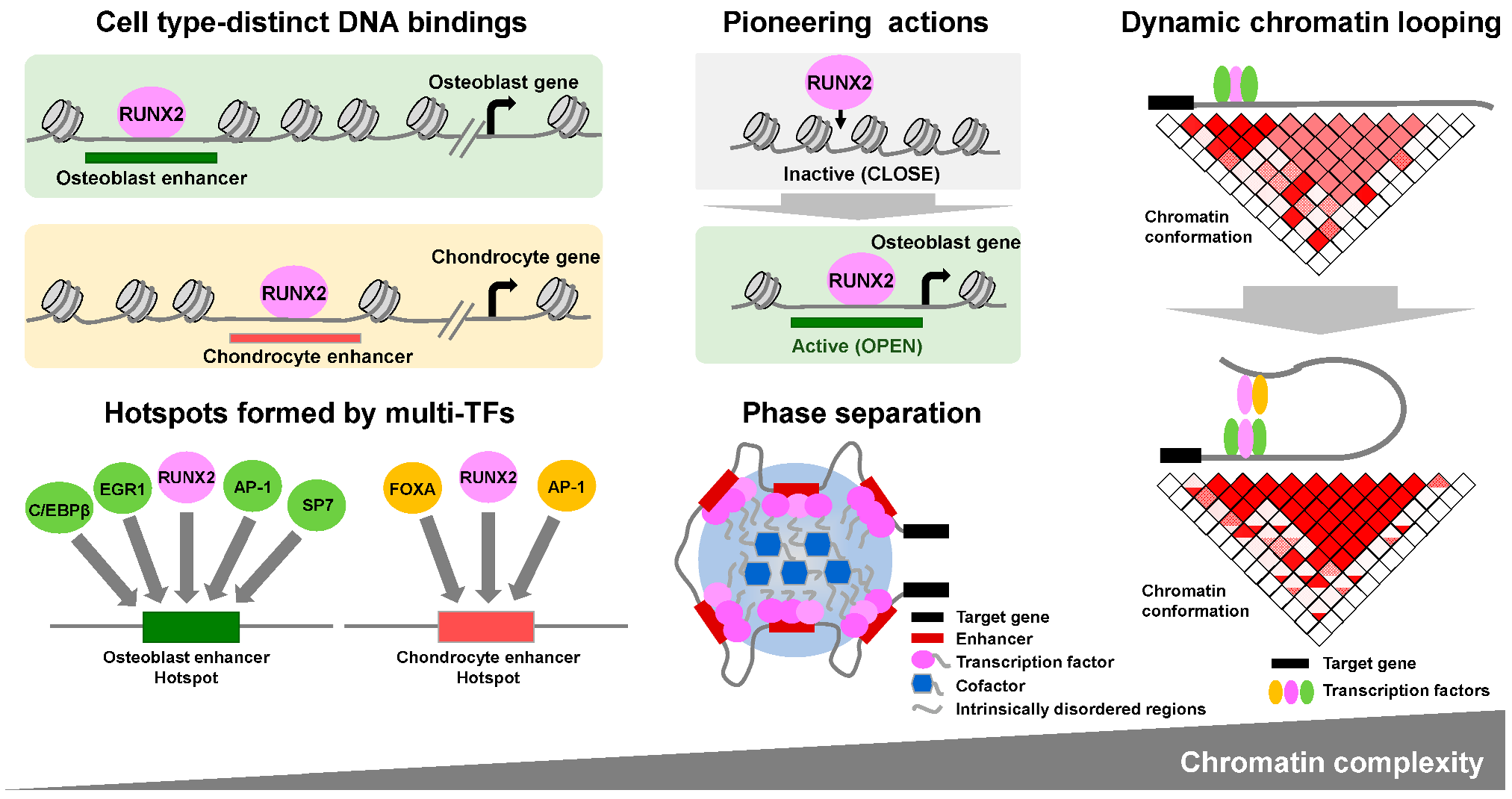
7. Higher Order Gene Regulation Mechanisms Where RUNX2 Is Involved
8. Concluding Remarks
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olsen, B.R. Vertebrate Skeletal Development, 1st ed.; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Kawasaki, K. The SCPP gene family and the complexity of hard tissues in vertebrates. Cells Tissues Organs 2011, 194, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Lettice, L.A.; Heaney, S.J.; Purdie, L.A.; Li, L.; de Beer, P.; Oostra, B.A.; Goode, D.; Elgar, G.; Hill, R.E.; de Graaff, E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003, 12, 1725–1735. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; Zhou, G.; Mukhopadhyay, K.; Smith, C.N.; Zhang, Z.; Eberspaecher, H.; Zhou, X.; Sinha, S.; Maity, S.N.; de Crombrugghe, B. An 18-base-pair sequence in the mouse proalpha1(II) collagen gene is sufficient for expression in cartilage and binds nuclear proteins that are selectively expressed in chondrocytes. Mol. Cell. Biol. 1996, 16, 4512–4523. [Google Scholar] [CrossRef]
- Zheng, Q.; Keller, B.; Zhou, G.; Napierala, D.; Chen, Y.; Zabel, B.; Parker, A.E.; Lee, B. Localization of the cis-enhancer element for mouse type X collagen expression in hypertrophic chondrocytes in vivo. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2009, 24, 1022–1032. [Google Scholar] [CrossRef]
- Bogdanovic, Z.; Bedalov, A.; Krebsbach, P.H.; Pavlin, D.; Woody, C.O.; Clark, S.H.; Thomas, H.F.; Rowe, D.W.; Kream, B.E.; Lichtler, A.C. Upstream regulatory elements necessary for expression of the rat COL1A1 promoter in transgenic mice. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 1994, 9, 285–292. [Google Scholar] [CrossRef]
- Ducy, P.; Karsenty, G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 1995, 15, 1858–1869. [Google Scholar] [CrossRef]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef]
- Ducy, P.; Zhang, R.; Geoffroy, V.; Ridall, A.L.; Karsenty, G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 1997, 89, 747–754. [Google Scholar] [CrossRef]
- Li, F.; Lu, Y.; Ding, M.; Napierala, D.; Abbassi, S.; Chen, Y.; Duan, X.; Wang, S.; Lee, B.; Zheng, Q. Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis-enhancer. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2011, 26, 2899–2910. [Google Scholar] [CrossRef]
- Akiyama, H.; Chaboissier, M.C.; Martin, J.F.; Schedl, A.; de Crombrugghe, B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002, 16, 2813–2828. [Google Scholar] [CrossRef]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef]
- Mundlos, S.; Otto, F.; Mundlos, C.; Mulliken, J.B.; Aylsworth, A.S.; Albright, S.; Lindhout, D.; Cole, W.G.; Henn, W.; Knoll, J.H.; et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 1997, 89, 773–779. [Google Scholar] [CrossRef]
- Otto, F.; Thornell, A.P.; Crompton, T.; Denzel, A.; Gilmour, K.C.; Rosewell, I.R.; Stamp, G.W.; Beddington, R.S.; Mundlos, S.; Olsen, B.R.; et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 1997, 89, 765–771. [Google Scholar] [CrossRef]
- Yoshida, C.A.; Yamamoto, H.; Fujita, T.; Furuichi, T.; Ito, K.; Inoue, K.; Yamana, K.; Zanma, A.; Takada, K.; Ito, Y.; et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004, 18, 952–963. [Google Scholar] [CrossRef]
- Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; Kaul, R.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef]
- Ohba, S.; He, X.; Hojo, H.; McMahon, A.P. Distinct Transcriptional Programs Underlie Sox9 Regulation of the Mammalian Chondrocyte. Cell Rep. 2015, 12, 229–243. [Google Scholar] [CrossRef]
- Liu, C.F.; Lefebvre, V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015, 43, 8183–8203. [Google Scholar] [CrossRef]
- Cheung, K.; Barter, M.J.; Falk, J.; Proctor, C.J.; Reynard, L.N.; Young, D.A. Histone ChIP-Seq identifies differential enhancer usage during chondrogenesis as critical for defining cell-type specificity. Faseb. J. 2020, 34, 5317–5331. [Google Scholar] [CrossRef]
- Meyer, M.B.; Benkusky, N.A.; Pike, J.W. The RUNX2 cistrome in osteoblasts: Characterization, down-regulation following differentiation, and relationship to gene expression. J. Biol. Chem. 2014, 289, 16016–16031. [Google Scholar] [CrossRef] [Green Version]
- Håkelien, A.M.; Bryne, J.C.; Harstad, K.G.; Lorenz, S.; Paulsen, J.; Sun, J.; Mikkelsen, T.S.; Myklebost, O.; Meza-Zepeda, L.A. The regulatory landscape of osteogenic differentiation. Stem Cells 2014, 32, 2780–2793. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Gordon, J.A.; Whitfield, T.W.; Tai, P.W.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Chromatin dynamics regulate mesenchymal stem cell lineage specification and differentiation to osteogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Whitfield, T.W.; Gordon, J.A.; Dobson, J.R.; Tai, P.W.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 2014, 15, R52. [Google Scholar] [CrossRef] [PubMed]
- Hojo, H.; Saito, T.; He, X.; Guo, Q.; Onodera, S.; Azuma, T.; Koebis, M.; Nakao, K.; Aiba, A.; Seki, M.; et al. Runx2 regulates chromatin accessibility to direct the osteoblast program at neonatal stages. Cell Rep. 2022, 40, 111315. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Chokalingam, K.; Juncosa-Melvin, N.; Hunter, S.A.; Gooch, C.; Frede, C.; Florert, J.; Bradica, G.; Wenstrup, R.; Butler, D.L. Tensile stimulation of murine stem cell-collagen sponge constructs increases collagen type I gene expression and linear stiffness. Tissue Eng. Part A 2009, 15, 2561–2570. [Google Scholar] [CrossRef]
- Maye, P.; Fu, Y.; Butler, D.L.; Chokalingam, K.; Liu, Y.; Floret, J.; Stover, M.L.; Wenstrup, R.; Jiang, X.; Gooch, C.; et al. Generation and characterization of Col10a1-mcherry reporter mice. Genesis 2011, 49, 410–418. [Google Scholar] [CrossRef]
- Rodda, S.J.; McMahon, A.P. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 2006, 133, 3231–3244. [Google Scholar] [CrossRef]
- Hojo, H.; Ohba, S.; He, X.; Lai, L.P.; McMahon, A.P. Sp7/Osterix Is Restricted to Bone-Forming Vertebrates where It Acts as a Dlx Co-factor in Osteoblast Specification. Dev. Cell 2016, 37, 238–253. [Google Scholar] [CrossRef] [Green Version]
- Nagata, K.; Hojo, H.; Chang, S.H.; Okada, H.; Yano, F.; Chijimatsu, R.; Omata, Y.; Mori, D.; Makii, Y.; Kawata, M.; et al. Runx2 and Runx3 differentially regulate articular chondrocytes during surgically induced osteoarthritis development. Nat. Commun. 2022, 13, 6187. [Google Scholar] [CrossRef]
- Komori, T. Whole Aspect of Runx2 Functions in Skeletal Development. Int. J. Mol. Sci. 2022, 23, 5776. [Google Scholar] [CrossRef]
- Otto, F.; Kanegane, H.; Mundlos, S. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum. Mutat. 2002, 19, 209–216. [Google Scholar] [CrossRef]
- Stricker, S.; Fundele, R.; Vortkamp, A.; Mundlos, S. Role of Runx genes in chondrocyte differentiation. Dev. Biol. 2002, 245, 95–108. [Google Scholar] [CrossRef]
- Takeda, S.; Bonnamy, J.P.; Owen, M.J.; Ducy, P.; Karsenty, G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001, 15, 467–481. [Google Scholar] [CrossRef]
- Ueta, C.; Iwamoto, M.; Kanatani, N.; Yoshida, C.; Liu, Y.; Enomoto-Iwamoto, M.; Ohmori, T.; Enomoto, H.; Nakata, K.; Takada, K.; et al. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J. Cell Biol. 2001, 153, 87–100. [Google Scholar] [CrossRef]
- Aghajanian, P.; Mohan, S. The art of building bone: Emerging role of chondrocyte-to-osteoblast transdifferentiation in endochondral ossification. Bone Res. 2018, 6, 19. [Google Scholar] [CrossRef]
- Qin, X.; Jiang, Q.; Nagano, K.; Moriishi, T.; Miyazaki, T.; Komori, H.; Ito, K.; Mark, K.V.; Sakane, C.; Kaneko, H.; et al. Runx2 is essential for the transdifferentiation of chondrocytes into osteoblasts. PLoS Genet. 2020, 16, e1009169. [Google Scholar] [CrossRef]
- Ionescu, A.; Kozhemyakina, E.; Nicolae, C.; Kaestner, K.H.; Olsen, B.R.; Lassar, A.B. FoxA family members are crucial regulators of the hypertrophic chondrocyte differentiation program. Dev. Cell 2012, 22, 927–939. [Google Scholar] [CrossRef]
- Tan, Z.; Niu, B.; Tsang, K.Y.; Melhado, I.G.; Ohba, S.; He, X.; Huang, Y.; Wang, C.; McMahon, A.P.; Jauch, R.; et al. Synergistic co-regulation and competition by a SOX9-GLI-FOXA phasic transcriptional network coordinate chondrocyte differentiation transitions. PLoS Genet. 2018, 14, e1007346. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, M.; Hata, K.; Takashima, R.; Ono, K.; Nakamura, E.; Takahata, Y.; Murakami, T.; Iseki, S.; Takano-Yamamoto, T.; Nishimura, R.; et al. The transcription factor Foxc1 is necessary for Ihh-Gli2-regulated endochondral ossification. Nat. Commun. 2015, 6, 6653. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Yu, H.V.; Tseng, K.C.; Flath, M.; Fabian, P.; Segil, N.; Crump, J.G. Foxc1 establishes enhancer accessibility for craniofacial cartilage differentiation. Elife 2021, 10, e63595. [Google Scholar] [CrossRef] [PubMed]
- Robledo, R.F.; Rajan, L.; Li, X.; Lufkin, T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002, 16, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Bozec, A.; Bakiri, L.; Jimenez, M.; Schinke, T.; Amling, M.; Wagner, E.F. Fra-2/AP-1 controls bone formation by regulating osteoblast differentiation and collagen production. J. Cell Biol. 2010, 190, 1093–1106. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Ohba, S.; Hojo, H.; McMahon, A.P. AP-1 family members act with Sox9 to promote chondrocyte hypertrophy. Development 2016, 143, 3012–3023. [Google Scholar] [CrossRef]
- Karreth, F.; Hoebertz, A.; Scheuch, H.; Eferl, R.; Wagner, E.F. The AP1 transcription factor Fra2 is required for efficient cartilage development. Development 2004, 131, 5717–5725. [Google Scholar] [CrossRef]
- Wang, W.; Lian, N.; Li, L.; Moss, H.E.; Wang, W.; Perrien, D.S.; Elefteriou, F.; Yang, X. Atf4 regulates chondrocyte proliferation and differentiation during endochondral ossification by activating Ihh transcription. Development 2009, 136, 4143–4153. [Google Scholar] [CrossRef]
- Yang, X.; Matsuda, K.; Bialek, P.; Jacquot, S.; Masuoka, H.C.; Schinke, T.; Li, L.; Brancorsini, S.; Sassone-Corsi, P.; Townes, T.M.; et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 2004, 117, 387–398. [Google Scholar] [CrossRef]
- Zhang, Q.; Zuo, H.; Yu, S.; Lin, Y.; Chen, S.; Liu, H.; Chen, Z. RUNX2 co-operates with EGR1 to regulate osteogenic differentiation through Htra1 enhancers. J. Cell. Physiol. 2020, 235, 8601–8612. [Google Scholar] [CrossRef]
- Long, H.K.; Prescott, S.L.; Wysocka, J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 2016, 167, 1170–1187. [Google Scholar] [CrossRef] [Green Version]
- Rickels, R.; Shilatifard, A. Enhancer Logic and Mechanics in Development and Disease. Trends Cell Biol. 2018, 28, 608–630. [Google Scholar] [CrossRef]
- Siersbæk, R.; Rabiee, A.; Nielsen, R.; Sidoli, S.; Traynor, S.; Loft, A.; Poulsen, L.C.; Rogowska-Wrzesinska, A.; Jensen, O.N.; Mandrup, S. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014, 7, 1443–1455. [Google Scholar] [CrossRef]
- Meyer, M.B.; Benkusky, N.A.; Sen, B.; Rubin, J.; Pike, J.W. Epigenetic Plasticity Drives Adipogenic and Osteogenic Differentiation of Marrow-derived Mesenchymal Stem Cells. J. Biol. Chem. 2016, 291, 17829–17847. [Google Scholar] [CrossRef]
- Rauch, A.; Haakonsson, A.K.; Madsen, J.G.S.; Larsen, M.; Forss, I.; Madsen, M.R.; Van Hauwaert, E.L.; Wiwie, C.; Jespersen, N.Z.; Tencerova, M.; et al. Osteogenesis depends on commissioning of a network of stem cell transcription factors that act as repressors of adipogenesis. Nat. Genet. 2019, 51, 716–727. [Google Scholar] [CrossRef]
- Kundu, M.; Javed, A.; Jeon, J.P.; Horner, A.; Shum, L.; Eckhaus, M.; Muenke, M.; Lian, J.B.; Yang, Y.; Nuckolls, G.H.; et al. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat. Genet. 2002, 32, 639–644. [Google Scholar] [CrossRef]
- Yoshida, C.A.; Furuichi, T.; Fujita, T.; Fukuyama, R.; Kanatani, N.; Kobayashi, S.; Satake, M.; Takada, K.; Komori, T. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat. Genet. 2002, 32, 633–638. [Google Scholar] [CrossRef]
- Bialek, P.; Kern, B.; Yang, X.; Schrock, M.; Sosic, D.; Hong, N.; Wu, H.; Yu, K.; Ornitz, D.M.; Olson, E.N.; et al. A twist code determines the onset of osteoblast differentiation. Dev. Cell 2004, 6, 423–435. [Google Scholar] [CrossRef]
- Kim, S.; Koga, T.; Isobe, M.; Kern, B.E.; Yokochi, T.; Chin, Y.E.; Karsenty, G.; Taniguchi, T.; Takayanagi, H. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003, 17, 1979–1991. [Google Scholar] [CrossRef]
- Jones, D.C.; Wein, M.N.; Oukka, M.; Hofstaetter, J.G.; Glimcher, M.J.; Glimcher, L.H. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science 2006, 312, 1223–1227. [Google Scholar] [CrossRef]
- Dobreva, G.; Chahrour, M.; Dautzenberg, M.; Chirivella, L.; Kanzler, B.; Farinas, I.; Karsenty, G.; Grosschedl, R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 2006, 125, 971–986. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.H.; Hwang, E.S.; McManus, M.T.; Amsterdam, A.; Tian, Y.; Kalmukova, R.; Mueller, E.; Benjamin, T.; Spiegelman, B.M.; Sharp, P.A.; et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 2005, 309, 1074–1078. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yao, X.; Yan, G.; Xu, Y.; Yan, J.; Zou, W.; Wang, G. Mediator MED23 cooperates with RUNX2 to drive osteoblast differentiation and bone development. Nat. Commun. 2016, 7, 11149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ting, K.; Bessette, C.M.; Culiat, C.T.; Sung, S.J.; Lee, H.; Chen, F.; Shen, J.; Wang, J.J.; Kuroda, S.; et al. Nell-1, a key functional mediator of Runx2, partially rescues calvarial defects in Runx2(+/−) mice. J. Bone Min. Res. 2011, 26, 777–791. [Google Scholar] [CrossRef] [PubMed]
- Hesse, E.; Saito, H.; Kiviranta, R.; Correa, D.; Yamana, K.; Neff, L.; Toben, D.; Duda, G.; Atfi, A.; Geoffroy, V.; et al. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J. Cell Biol. 2010, 191, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Mohanapriya, R.; Akshaya, R.L.; Selvamurugan, N. A regulatory role of circRNA-miRNA-mRNA network in osteoblast differentiation. Biochimie 2022, 193, 137–147. [Google Scholar] [CrossRef]
- Yoon, D.S.; Kim, E.J.; Cho, S.; Jung, S.; Lee, K.M.; Park, K.H.; Lee, J.W.; Kim, S.H. RUNX2 stabilization by long non-coding RNAs contributes to hypertrophic changes in human chondrocytes. Int. J. Biol. Sci. 2023, 19, 13–33. [Google Scholar] [CrossRef]
- Iwafuchi-Doi, M.; Zaret, K.S. Cell fate control by pioneer transcription factors. Development 2016, 143, 1833–1837. [Google Scholar] [CrossRef]
- Donaghey, J.; Thakurela, S.; Charlton, J.; Chen, J.S.; Smith, Z.D.; Gu, H.; Pop, R.; Clement, K.; Stamenova, E.K.; Karnik, R.; et al. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 2018, 50, 250–258. [Google Scholar] [CrossRef]
- Korinfskaya, S.; Parameswaran, S.; Weirauch, M.T.; Barski, A. Runx Transcription Factors in T Cells-What Is Beyond Thymic Development? Front. Immunol. 2021, 12, 701924. [Google Scholar] [CrossRef]
- Mevel, R.; Draper, J.E.; Lie, A.L.M.; Kouskoff, V.; Lacaud, G. RUNX transcription factors: Orchestrators of development. Development 2019, 146, dev148296. [Google Scholar] [CrossRef] [Green Version]
- Lichtinger, M.; Ingram, R.; Hannah, R.; Müller, D.; Clarke, D.; Assi, S.A.; Lie, A.L.M.; Noailles, L.; Vijayabaskar, M.S.; Wu, M.; et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. Embo. J. 2012, 31, 4318–4333. [Google Scholar] [CrossRef]
- Hass, M.R.; Brissette, D.; Parameswaran, S.; Pujato, M.; Donmez, O.; Kottyan, L.C.; Weirauch, M.T.; Kopan, R. Runx1 shapes the chromatin landscape via a cascade of direct and indirect targets. PLoS Genet. 2021, 17, e1009574. [Google Scholar] [CrossRef]
- Lee, J.W.; Kim, D.M.; Jang, J.W.; Park, T.G.; Song, S.H.; Lee, Y.S.; Chi, X.Z.; Park, I.Y.; Hyun, J.W.; Ito, Y.; et al. RUNX3 regulates cell cycle-dependent chromatin dynamics by functioning as a pioneer factor of the restriction-point. Nat. Commun. 2019, 10, 1897. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef]
- Basu, S.; Mackowiak, S.D.; Niskanen, H.; Knezevic, D.; Asimi, V.; Grosswendt, S.; Geertsema, H.; Ali, S.; Jerković, I.; Ewers, H.; et al. Unblending of Transcriptional Condensates in Human Repeat Expansion Disease. Cell 2020, 181, 1062–1079.e1030. [Google Scholar] [CrossRef]
- Peng, L.; Li, E.M.; Xu, L.Y. From start to end: Phase separation and transcriptional regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194641. [Google Scholar] [CrossRef]
- Andrey, G.; Mundlos, S. The three-dimensional genome: Regulating gene expression during pluripotency and development. Development 2017, 144, 3646–3658. [Google Scholar] [CrossRef]
- Hao, R.H.; Guo, Y.; Wang, C.; Chen, F.; Di, C.X.; Dong, S.S.; Cao, Q.L.; Guo, J.; Rong, Y.; Yao, S.; et al. Lineage-specific rearrangement of chromatin loops and epigenomic features during adipocytes and osteoblasts commitment. Cell Death Differ. 2022, 29, 2503–2518. [Google Scholar] [CrossRef]
- Tosa, I.; Yamada, D.; Yasumatsu, M.; Hinoi, E.; Ono, M.; Oohashi, T.; Kuboki, T.; Takarada, T. Postnatal Runx2 deletion leads to low bone mass and adipocyte accumulation in mice bone tissues. Biochem. Biophys. Res. Commun. 2019, 516, 1229–1233. [Google Scholar] [CrossRef] [PubMed]
- Catheline, S.E.; Hoak, D.; Chang, M.; Ketz, J.P.; Hilton, M.J.; Zuscik, M.J.; Jonason, J.H. Chondrocyte-Specific RUNX2 Overexpression Accelerates Post-traumatic Osteoarthritis Progression in Adult Mice. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2019, 34, 1676–1689. [Google Scholar] [CrossRef] [PubMed]
- Wilflingseder, J.; Willi, M.; Lee, H.K.; Olauson, H.; Jankowski, J.; Ichimura, T.; Erben, R.; Valerius, M.T.; Hennighausen, L.; Bonventre, J.V. Enhancer and super-enhancer dynamics in repair after ischemic acute kidney injury. Nat. Commun. 2020, 11, 3383. [Google Scholar] [CrossRef]
- Kang, J.; Hu, J.; Karra, R.; Dickson, A.L.; Tornini, V.A.; Nachtrab, G.; Gemberling, M.; Goldman, J.A.; Black, B.L.; Poss, K.D. Modulation of tissue repair by regeneration enhancer elements. Nature 2016, 532, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Cigliola, V.; Oonk, K.A.; Petrover, Z.; DeLuca, S.; Wolfson, D.W.; Vekstein, A.; Mendiola, M.A.; Devlin, G.; Bishawi, M.; et al. An enhancer-based gene-therapy strategy for spatiotemporal control of cargoes during tissue repair. Cell Stem Cell 2023, 30, 96–111.e6. [Google Scholar] [CrossRef]
- Jiang, S.; Fagman, J.B.; Chen, C.; Alberti, S.; Liu, B. Protein phase separation and its role in tumorigenesis. Elife 2020, 9, e60264. [Google Scholar] [CrossRef]
- Boer, C.G.; Hatzikotoulas, K.; Southam, L.; Stefánsdóttir, L.; Zhang, Y.; Coutinho de Almeida, R.; Wu, T.T.; Zheng, J.; Hartley, A.; Teder-Laving, M.; et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell 2021, 184, 4784–4818.e17. [Google Scholar] [CrossRef]
- Zhu, X.; Bai, W.; Zheng, H. Twelve years of GWAS discoveries for osteoporosis and related traits: Advances, challenges and applications. Bone Res. 2021, 9, 23. [Google Scholar] [CrossRef]
- Nasser, J.; Bergman, D.T.; Fulco, C.P.; Guckelberger, P.; Doughty, B.R.; Patwardhan, T.A.; Jones, T.R.; Nguyen, T.H.; Ulirsch, J.C.; Lekschas, F.; et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 2021, 593, 238–243. [Google Scholar] [CrossRef]
- Chesi, A.; Wagley, Y.; Johnson, M.E.; Manduchi, E.; Su, C.; Lu, S.; Leonard, M.E.; Hodge, K.M.; Pippin, J.A.; Hankenson, K.D.; et al. Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat. Commun. 2019, 10, 1260. [Google Scholar] [CrossRef] [Green Version]
- Morris, J.A.; Kemp, J.P.; Youlten, S.E.; Laurent, L.; Logan, J.G.; Chai, R.C.; Vulpescu, N.A.; Forgetta, V.; Kleinman, A.; Mohanty, S.T.; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019, 51, 258–266. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hojo, H. Emerging RUNX2-Mediated Gene Regulatory Mechanisms Consisting of Multi-Layered Regulatory Networks in Skeletal Development. Int. J. Mol. Sci. 2023, 24, 2979. https://doi.org/10.3390/ijms24032979
Hojo H. Emerging RUNX2-Mediated Gene Regulatory Mechanisms Consisting of Multi-Layered Regulatory Networks in Skeletal Development. International Journal of Molecular Sciences. 2023; 24(3):2979. https://doi.org/10.3390/ijms24032979
Chicago/Turabian StyleHojo, Hironori. 2023. "Emerging RUNX2-Mediated Gene Regulatory Mechanisms Consisting of Multi-Layered Regulatory Networks in Skeletal Development" International Journal of Molecular Sciences 24, no. 3: 2979. https://doi.org/10.3390/ijms24032979
APA StyleHojo, H. (2023). Emerging RUNX2-Mediated Gene Regulatory Mechanisms Consisting of Multi-Layered Regulatory Networks in Skeletal Development. International Journal of Molecular Sciences, 24(3), 2979. https://doi.org/10.3390/ijms24032979





