Gut Microbiota Deficiency Exacerbates Liver Injury in Bile Duct Ligated Mice via Inflammation and Lipid Metabolism
Abstract
:1. Introduction
2. Results
2.1. AIMD Aggravates Liver Injury after BDL
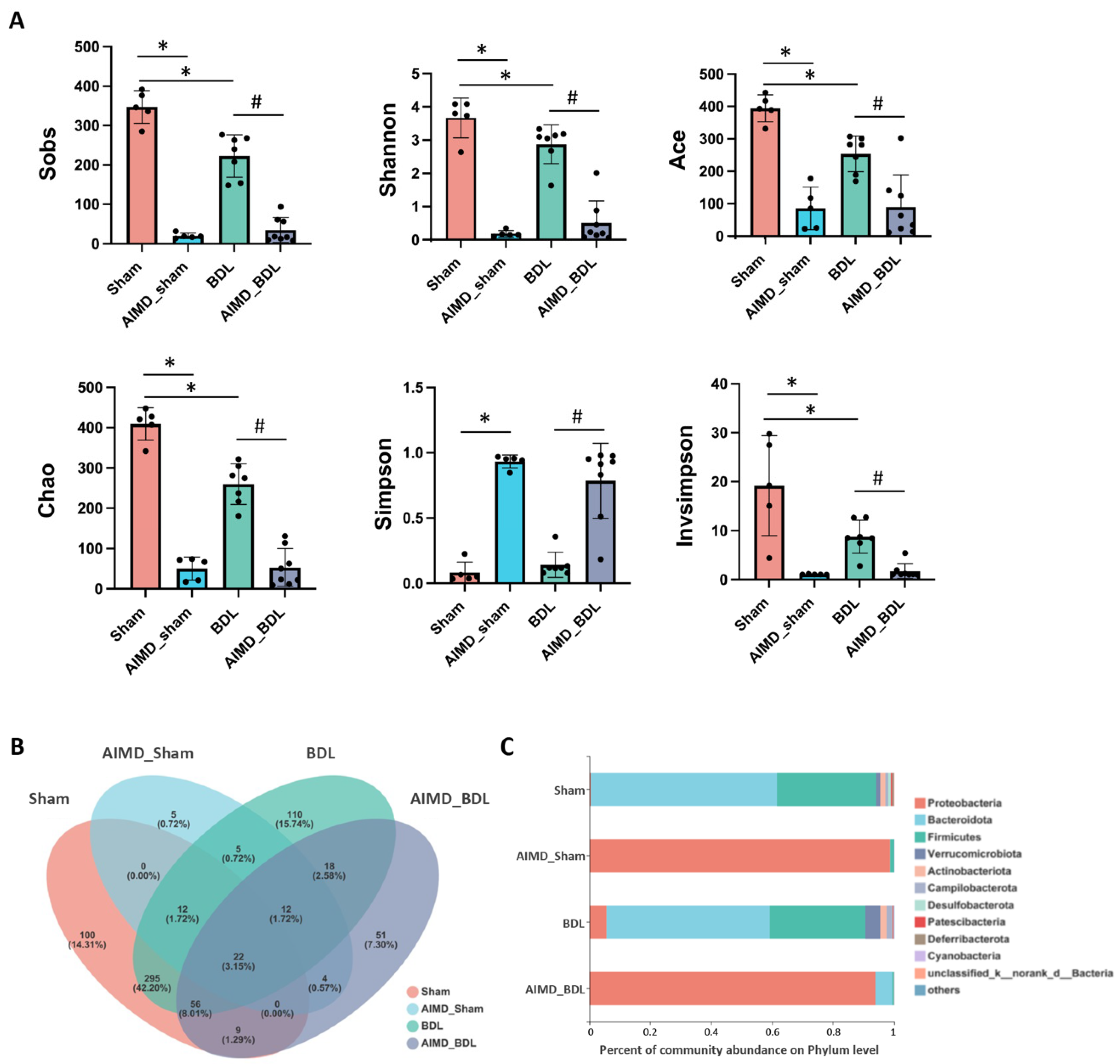
2.2. AIMD Alters Gut Microbiota Alpha Diversity in Mice
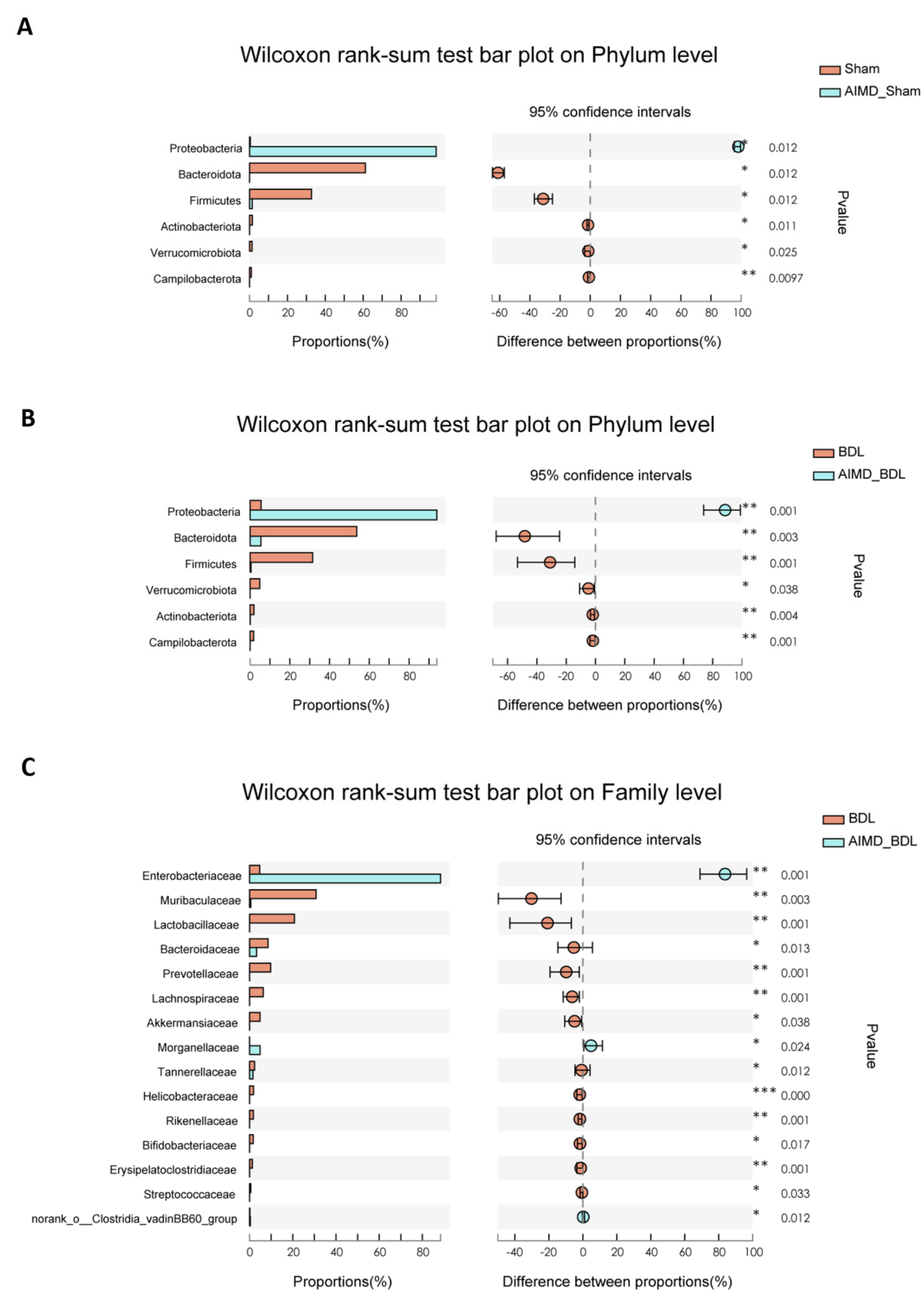
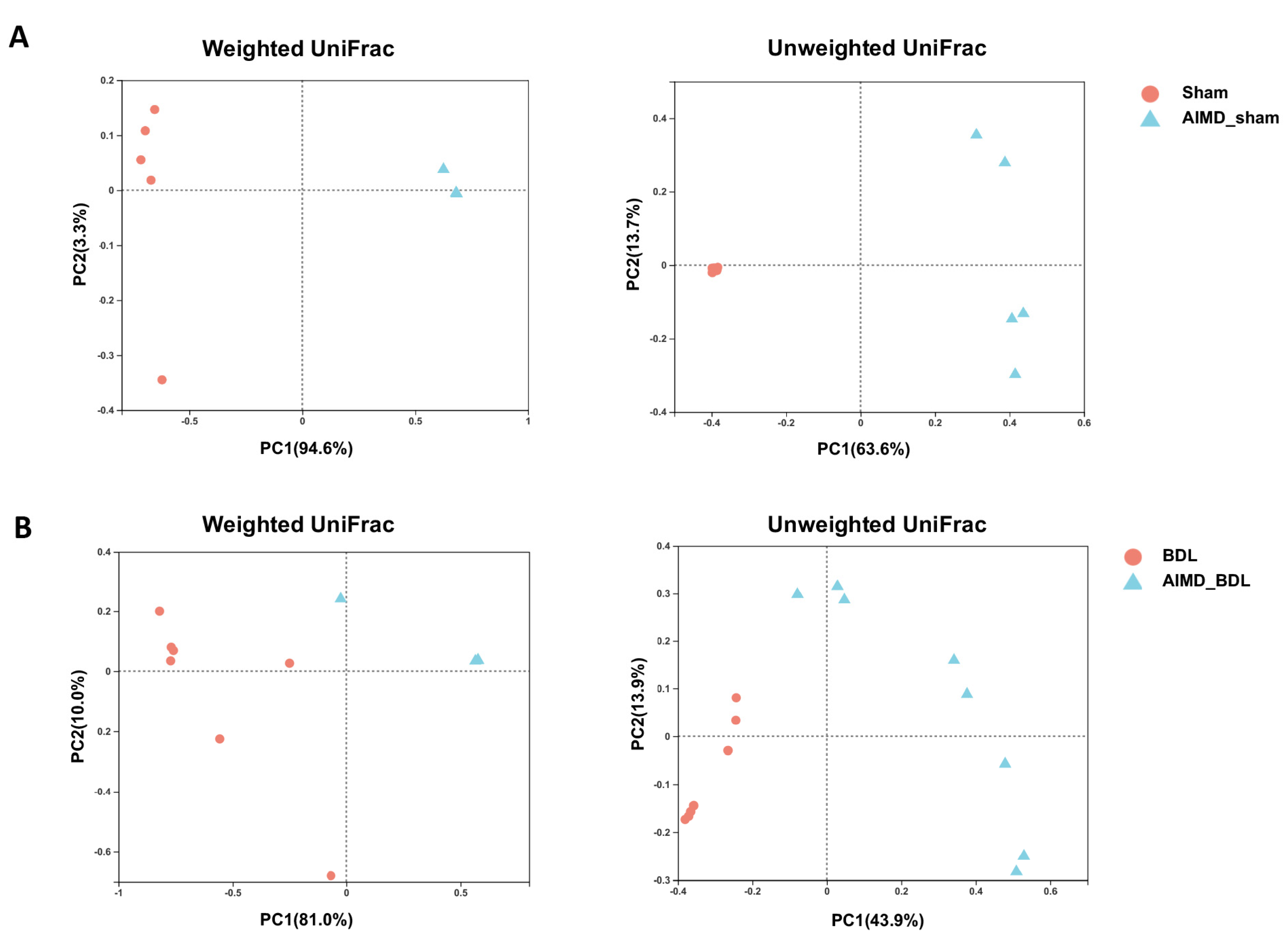
2.3. AIMD_BDL Mice Harbor Different OTUs of Gut Microbiota versus BDL Mice
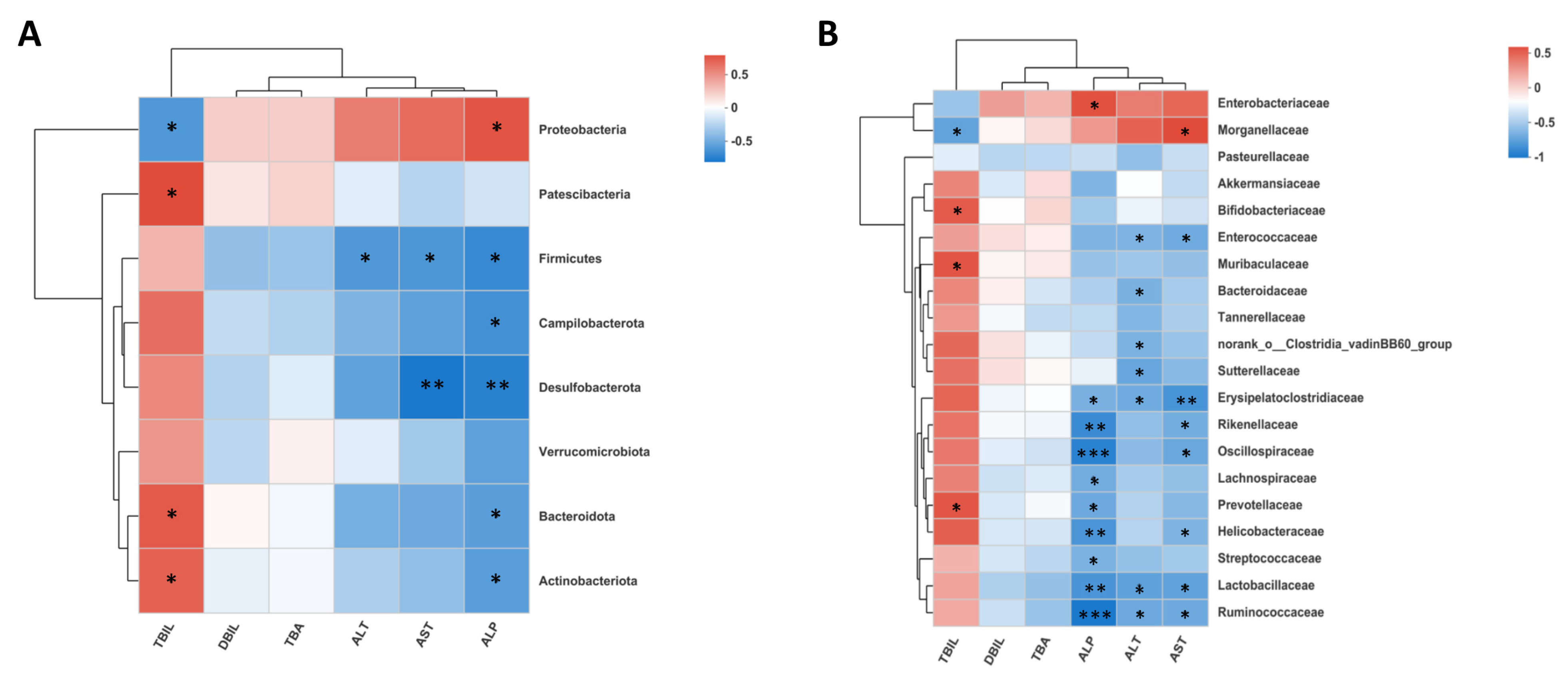
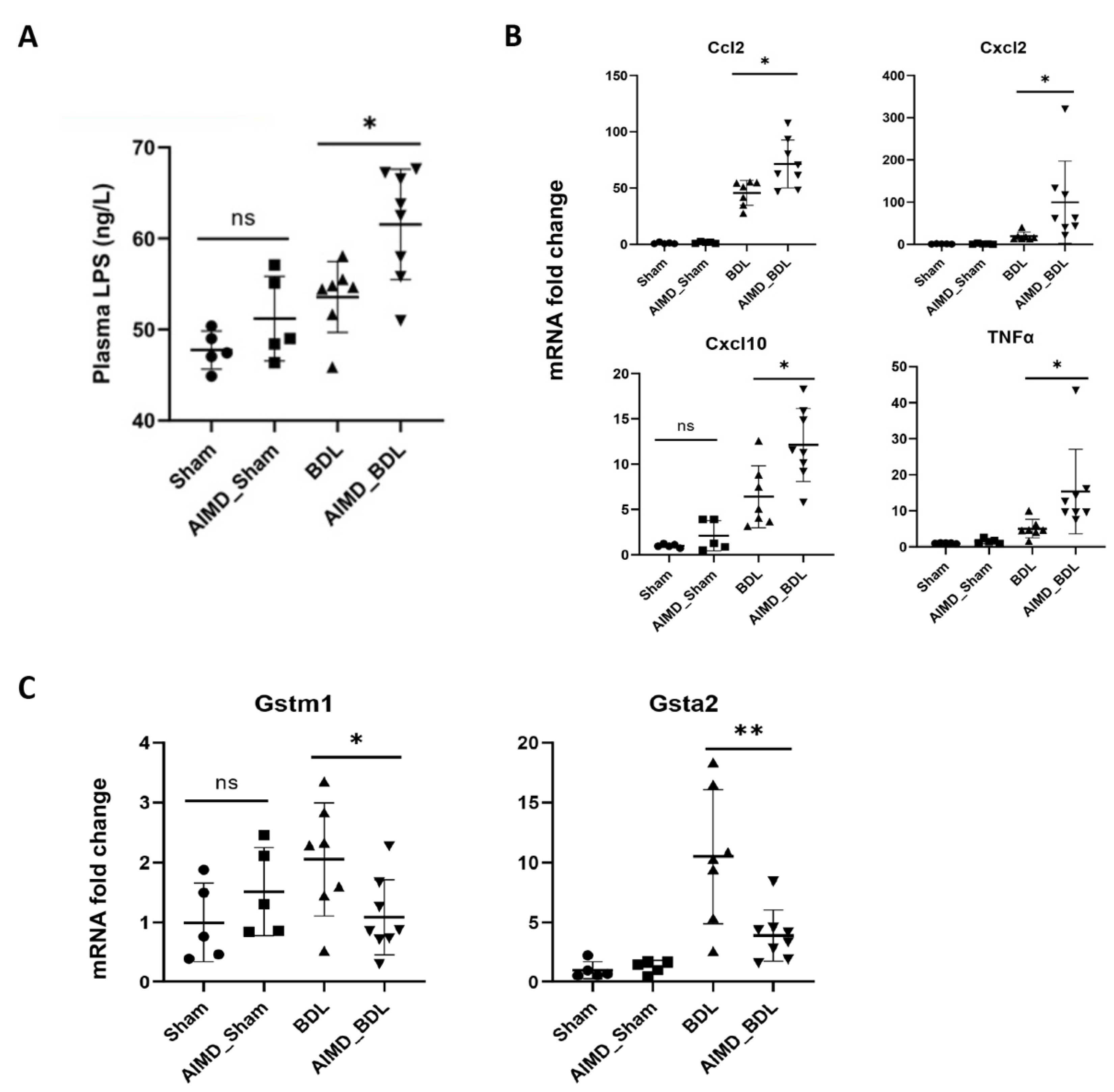
2.4. Gut Microbiota Deficiency Is Associated with Cholestatic Liver Injury
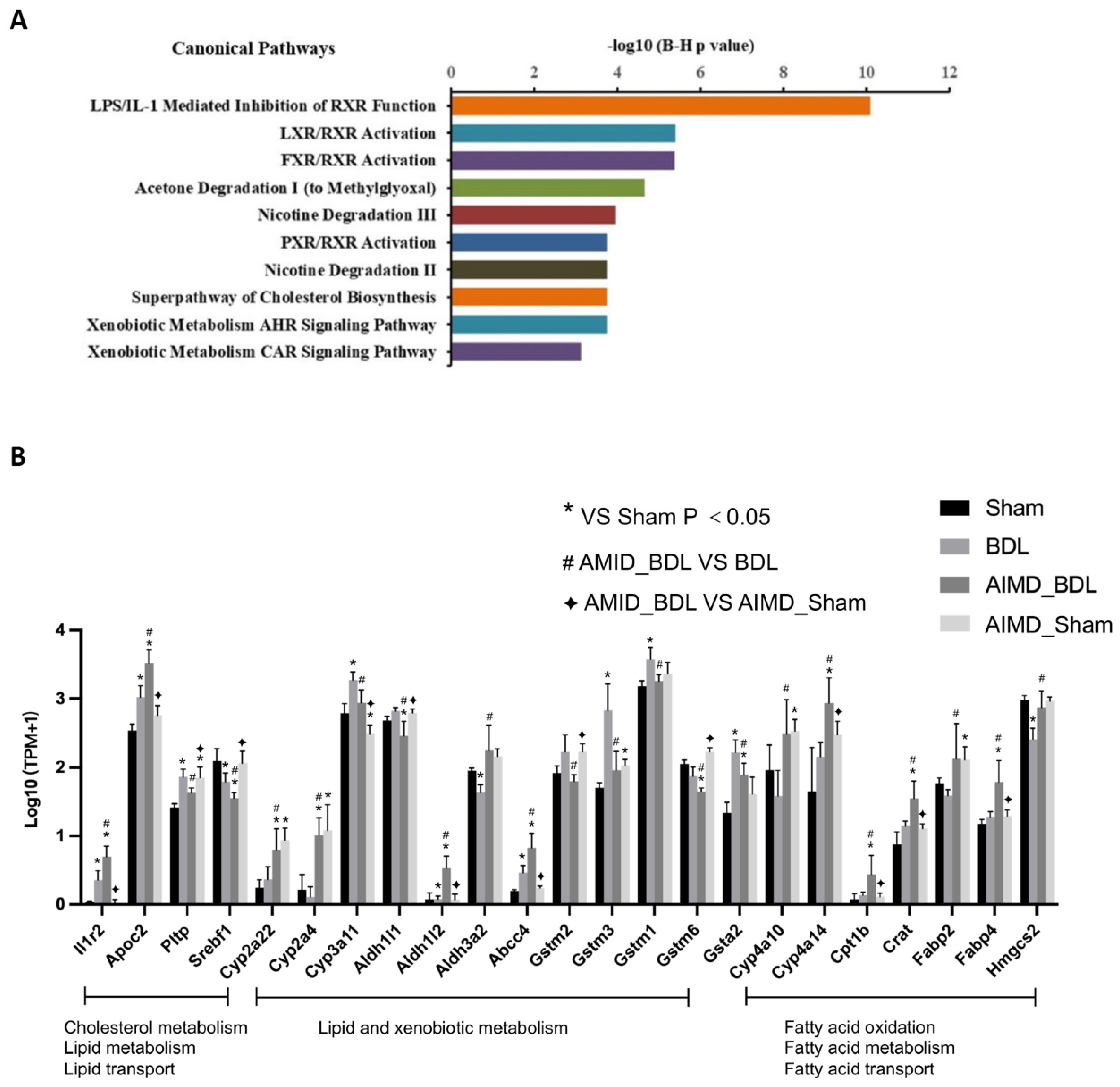
2.5. Gut Microbiota Deficiency Altered Gene Expression in the Livers of BDL Mice
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Antibiotic Pretreatment and Procedure of BDL in Mice
4.3. Hematoxylin-Eosin (HE) Staining and Liver Histology
4.4. Measurement of Lipopolysaccharide (LPS) Level in Plasma
4.5. 16S rRNA Gene Sequence Analysis
4.6. RNA Sequencing
4.7. Real-Time Quantitative Polymerase Chain Reaction (RT-PCR)
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, H.X.; Keane, R.; Sheng, L.; Wan, Y.J. Implications of microbiota and bile acid in liver injury and regeneration. J. Hepatol. 2015, 63, 1502–1510. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyotylainen, T.; Hamalainen, A.M.; Peet, A.; Tillmann, V.; Poho, P.; Mattila, I.; et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef]
- Zheng, P.; Zeng, B.; Zhou, C.; Liu, M.; Fang, Z.; Xu, X.; Zeng, L.; Chen, J.; Fan, S.; Du, X.; et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 2016, 21, 786–796. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Lindheim, L.; Bashir, M.; Munzker, J.; Trummer, C.; Zachhuber, V.; Leber, B.; Horvath, A.; Pieber, T.R.; Gorkiewicz, G.; Stadlbauer, V.; et al. Alterations in Gut Microbiome Composition and Barrier Function Are Associated with Reproductive and Metabolic Defects in Women with Polycystic Ovary Syndrome (PCOS): A Pilot Study. PLoS ONE 2017, 12, e0168390. [Google Scholar] [CrossRef] [PubMed]
- Kwong, E.K.; Puri, P. Gut microbiome changes in Nonalcoholic fatty liver disease & alcoholic liver disease. Transl. Gastroenterol. Hepatol. 2021, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Gawlik-Kotelnicka, O.; Strzelecki, D. Adiposity in Depression or Depression in Adiposity? The Role of Immune-Inflammatory-Microbial Overlap. Life 2021, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.C.H.; Sung, J.J.; Yu, J. Gut microbiota: Impacts on gastrointestinal cancer immunotherapy. Gut Microbes 2021, 13, 1–21. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, E.; Narbad, A.; Rodriguez, J.M. Autism Spectrum Disorder Associated With Gut Microbiota at Immune, Metabolomic, and Neuroactive Level. Front. Neurosci. 2020, 14, 578666. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Backhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Wong, S.H.; Zhao, L.; Zhang, X.; Nakatsu, G.; Han, J.; Xu, W.; Xiao, X.; Kwong, T.N.Y.; Tsoi, H.; Wu, W.K.K.; et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017, 153, 1621–1633.e1626. [Google Scholar] [CrossRef] [PubMed]
- Zarrinpar, A.; Chaix, A.; Xu, Z.Z.; Chang, M.W.; Marotz, C.A.; Saghatelian, A.; Knight, R.; Panda, S. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 2018, 9, 2872. [Google Scholar] [CrossRef]
- Wichmann, A.; Allahyar, A.; Greiner, T.U.; Plovier, H.; Lunden, G.O.; Larsson, T.; Drucker, D.J.; Delzenne, N.M.; Cani, P.D.; Backhed, F. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe 2013, 14, 582–590. [Google Scholar] [CrossRef]
- Kusaczuk, M. Tauroursodeoxycholate-Bile Acid with Chaperoning Activity: Molecular and Cellular Effects and Therapeutic Perspectives. Cells 2019, 8, 1471. [Google Scholar] [CrossRef] [PubMed]
- Benakis, C.; Martin-Gallausiaux, C.; Trezzi, J.P.; Melton, P.; Liesz, A.; Wilmes, P. The microbiome-gut-brain axis in acute and chronic brain diseases. Curr. Opin. Neurobiol. 2020, 61, 1–9. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Patterson, A.M.; Cotter, P.D.; Beraza, N. Cholestasis induced by bile duct ligation promotes changes in the intestinal microbiome in mice. Sci. Rep. 2019, 9, 12324. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017, 25, 1054–1062.e1055. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; White, M.B.; Monteith, P.; Noble, N.A.; Unser, A.B.; Daita, K.; Fisher, A.R.; et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 2014, 60, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; You, H.J.; Bajaj, J.S.; Joo, S.K.; Yu, J.; Park, S.; Kang, H.; Park, J.H.; Kim, J.H.; Lee, D.H.; et al. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 2020, 11, 4982. [Google Scholar] [CrossRef]
- Munukka, E.; Rintala, A.; Toivonen, R.; Nylund, M.; Yang, B.; Takanen, A.; Hanninen, A.; Vuopio, J.; Huovinen, P.; Jalkanen, S.; et al. Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high-fat fed mice. ISME J. 2017, 11, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.R.; Haileselassie, Y.; Nguyen, L.P.; Tropini, C.; Wang, M.; Becker, L.S.; Sim, D.; Jarr, K.; Spear, E.T.; Singh, G.; et al. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe 2020, 27, 659–670.e655. [Google Scholar] [CrossRef] [PubMed]
- Kakiyama, G.; Pandak, W.M.; Gillevet, P.M.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; Takei, H.; Muto, A.; Nittono, H.; Ridlon, J.M.; et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 2013, 58, 949–955. [Google Scholar] [CrossRef]
- Maharshak, N.; Packey, C.D.; Ellermann, M.; Manick, S.; Siddle, J.P.; Huh, E.Y.; Plevy, S.; Sartor, R.B.; Carroll, I.M. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes 2013, 4, 316–324. [Google Scholar] [CrossRef]
- Selvanantham, T.; Lin, Q.; Guo, C.X.; Surendra, A.; Fieve, S.; Escalante, N.K.; Guttman, D.S.; Streutker, C.J.; Robertson, S.J.; Philpott, D.J.; et al. NKT Cell-Deficient Mice Harbor an Altered Microbiota That Fuels Intestinal Inflammation during Chemically Induced Colitis. J. Immunol. 2016, 197, 4464–4472. [Google Scholar] [CrossRef]
- Liu, T.; Yu, H.; Zhang, Z.; Xie, Y.; Yang, L.; You, F. Intestinal ELF4 Deletion Exacerbates Alcoholic Liver Disease by Disrupting Gut Homeostasis. Int. J. Mol. Sci. 2022, 23, 4825. [Google Scholar] [CrossRef]
- Mao, J.; Zhan, H.; Meng, F.; Wang, G.; Huang, D.; Liao, Z.; Chen, M. Costunolide protects against alcohol-induced liver injury by regulating gut microbiota, oxidative stress and attenuating inflammation in vivo and in vitro. Phytotherapy Res. 2022, 36, 1268–1283. [Google Scholar] [CrossRef] [PubMed]
- Gupta, H.; Suk, K.T.; Kim, D.J. Gut Microbiota at the Intersection of Alcohol, Brain, and the Liver. J. Clin. Med. 2021, 10, 541. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Hakoupian, M.; Ferino, E.; Jickling, G.C.; Amini, H.; Stamova, B.; Ander, B.P.; Alomar, N.; Sharp, F.R.; Zhan, X. Bacterial lipopolysaccharide is associated with stroke. Sci. Rep. 2021, 11, 6570. [Google Scholar] [CrossRef]
- Choi, S.H.; Kim, S.G. Lipopolysaccharide inhibition of rat hepatic microsomal epoxide hydrolase and glutathione S-transferase gene expression irrespective of nuclear factor-kappaB activation. Biochem. Pharmacol. 1998, 56, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Nyagode, B.A.; Williams, I.R.; Morgan, E.T. Altered inflammatory responses to Citrobacter rodentium infection, but not bacterial lipopolysaccharide, in mice lacking the Cyp4a10 or Cyp4a14 genes. Inflammation 2014, 37, 893–907. [Google Scholar] [CrossRef]
- Robertson, G.; Leclercq, I.; Farrell, G.C. Nonalcoholic steatosis and steatohepatitis. II. Cytochrome P-450 enzymes and oxidative stress. Am. J. Physiol. Liver Physiol. 2001, 281, G1135–G1139. [Google Scholar] [CrossRef]
- Kennedy, E.A.; King, K.Y.; Baldridge, M.T. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front. Physiol. 2018, 9, 1534. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Sham | AIMD_Sham | BDL | AIMD_BDL | |
|---|---|---|---|---|
| Subjects | n = 5 | n = 5 | n = 7 | n = 8 |
| ALT (IU/L) | 17.28 ± 3.87 | 20.44 ± 3.26 | 365.49 ± 162.389 * | 628.50 ± 256.92 *,# |
| AST (IU/L) | 99.76 ± 10.91 | 109.60 ± 14.63 | 675.95 ± 328.53 * | 1509.95 ± 565.15 *,# |
| ALP (IU/L) | 116.00 ± 12.13 | 124.00 ± 21.91 | 413.71 ± 136.99 * | 675.00 ± 242.84 *,# |
| TBA (μmol/L) | 1.50 ± 1.06 | 1.88 ± 1.20 | 368.02 ± 211.81 * | 449.65 ± 231.31 * |
| TBIL (μmol/L) | 7.28 ± 4.65 | 5.68 ± 1.87 | 181.49 ± 42.73 * | 139.46 ± 23.23 *,# |
| DBIL (μmol/L) | 4.71 ± 3.28 | 3.91 ± 2.15 | 60.59 ± 29.62 * | 84.48 ± 38.38 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Zhang, X.; Zhao, N.; Zhang, L.; Qiu, W.; Song, C.; Chai, J.; Cai, S.; Chen, W. Gut Microbiota Deficiency Exacerbates Liver Injury in Bile Duct Ligated Mice via Inflammation and Lipid Metabolism. Int. J. Mol. Sci. 2023, 24, 3180. https://doi.org/10.3390/ijms24043180
Zhou X, Zhang X, Zhao N, Zhang L, Qiu W, Song C, Chai J, Cai S, Chen W. Gut Microbiota Deficiency Exacerbates Liver Injury in Bile Duct Ligated Mice via Inflammation and Lipid Metabolism. International Journal of Molecular Sciences. 2023; 24(4):3180. https://doi.org/10.3390/ijms24043180
Chicago/Turabian StyleZhou, Xueqian, Xiaoxun Zhang, Nan Zhao, Liangjun Zhang, Wen Qiu, Chunwei Song, Jin Chai, Shiying Cai, and Wensheng Chen. 2023. "Gut Microbiota Deficiency Exacerbates Liver Injury in Bile Duct Ligated Mice via Inflammation and Lipid Metabolism" International Journal of Molecular Sciences 24, no. 4: 3180. https://doi.org/10.3390/ijms24043180
APA StyleZhou, X., Zhang, X., Zhao, N., Zhang, L., Qiu, W., Song, C., Chai, J., Cai, S., & Chen, W. (2023). Gut Microbiota Deficiency Exacerbates Liver Injury in Bile Duct Ligated Mice via Inflammation and Lipid Metabolism. International Journal of Molecular Sciences, 24(4), 3180. https://doi.org/10.3390/ijms24043180





