Impact of ABCG2 and ABCB1 Polymorphisms on Imatinib Plasmatic Exposure: An Original Work and Meta-Analysis
Abstract
:1. Introduction
2. Results
2.1. Characteristics of the Patients
2.2. CRO-Aviano Study
ABC Genotypes and Imatinib Trough Concentrations
2.3. Meta-Analysis
2.3.1. Search Results and Study Characteristics
2.3.2. Quality Assessment
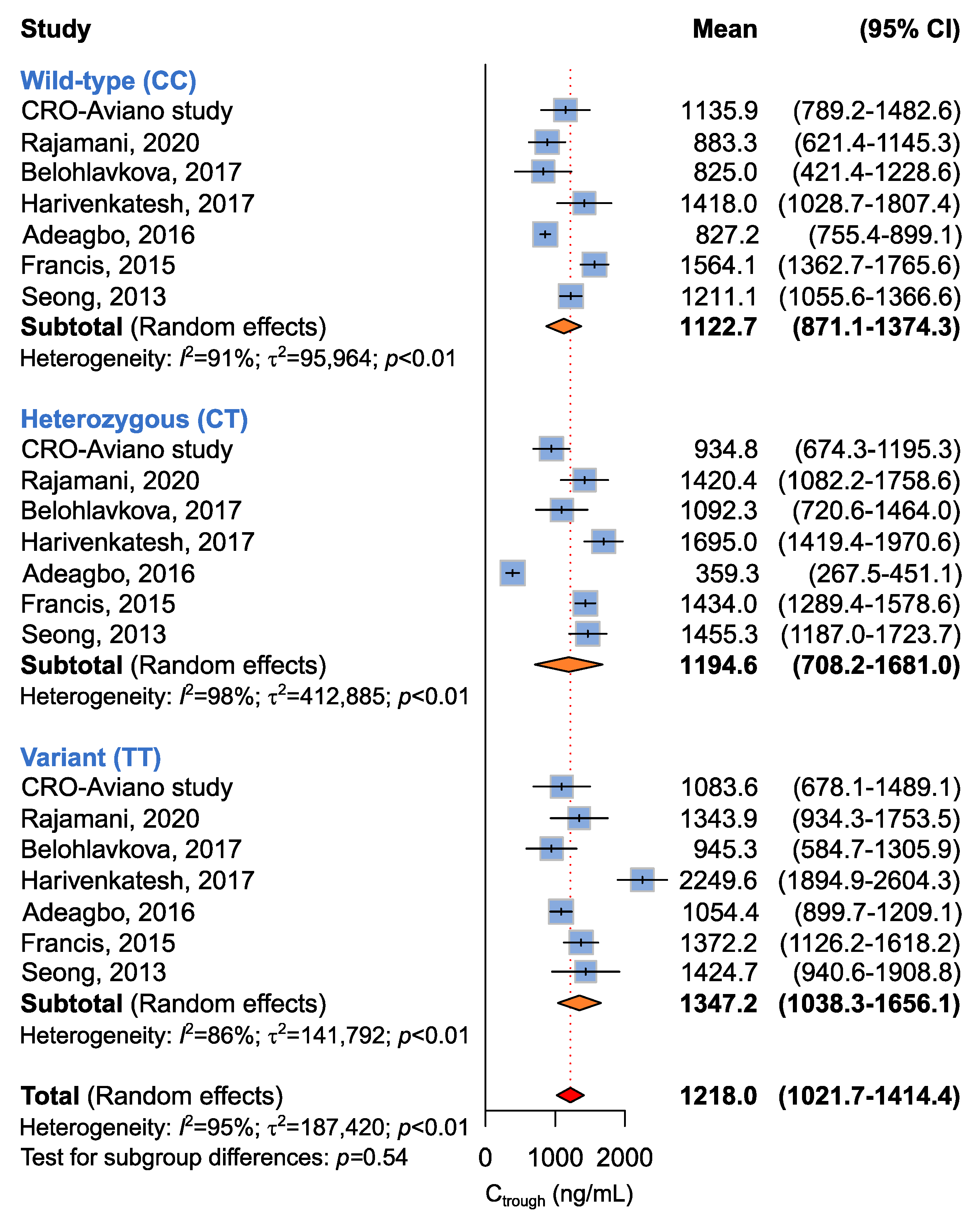
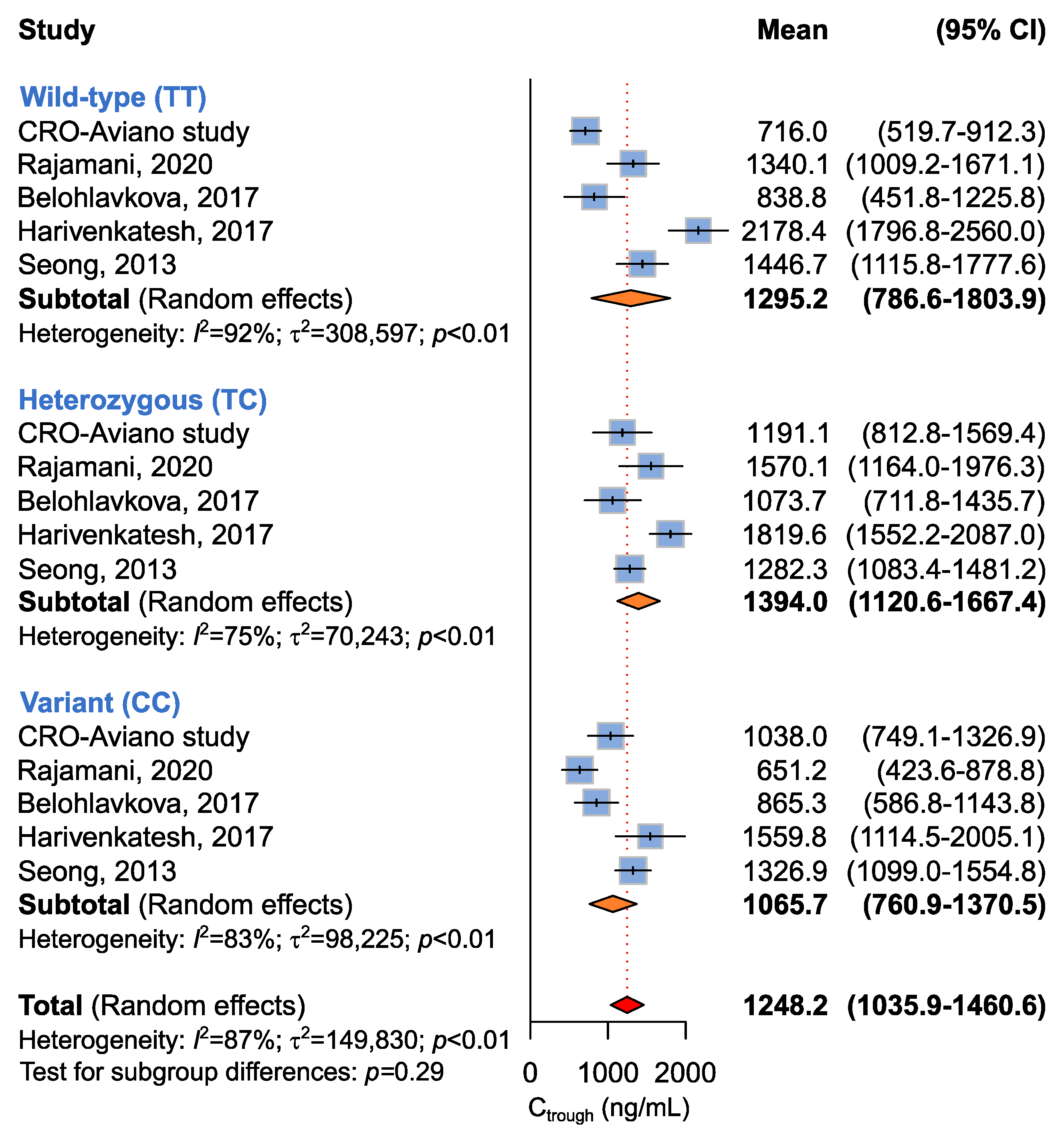
2.3.3. Association between ABCB1 c.3435C>T, c.2677G>T, and c.1236C>T and Imatinib Ctrough Levels
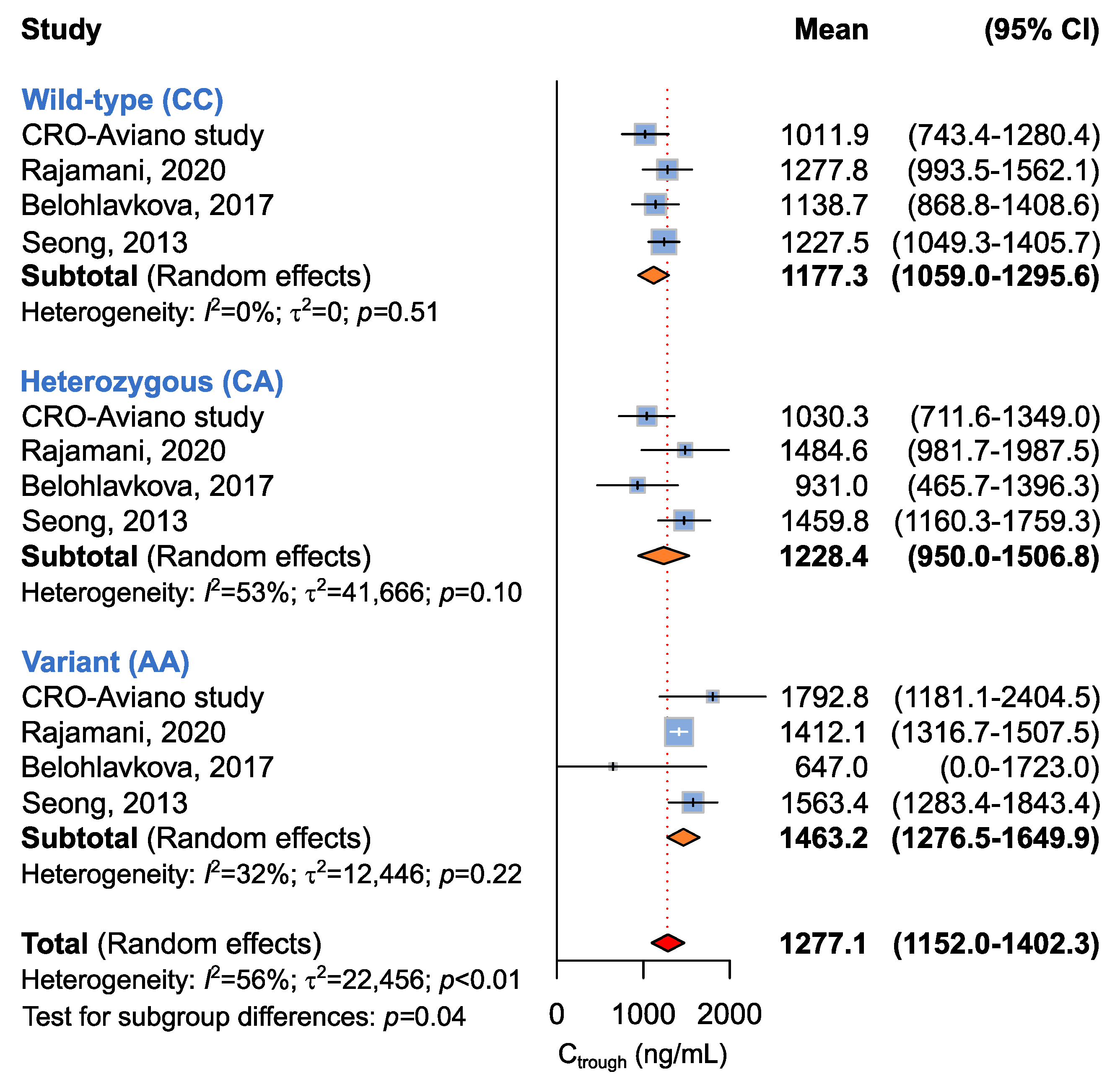
2.3.4. Association between ABCG2 c.412C>A and Imatinib Ctrough Levels
3. Discussion
4. Materials and Methods
4.1. Ethics Statements
4.2. Patient Selection
4.3. Blood Collection and Genotyping
4.4. Imatinib Quantification
4.5. Meta-Analysis
4.5.1. Search Strategy
4.5.2. Selection Criteria
4.5.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Demetri, G.D.; Benjamin, R.S.; Blanke, C.D.; Blay, J.-Y.; Casali, P.; Choi, H.; Corless, C.L.; Debiec-Rychter, M.; DeMatteo, R.P.; Ettinger, D.S.; et al. NCCN Task Force Report: Management of Patients with Gastrointestinal Stromal Tumor (GIST)--Update of the NCCN Clinical Practice Guidelines. J. Natl. Compr. Canc. Netw. 2007, 5 (Suppl. S2), S1–S29; quiz S30. [Google Scholar] [CrossRef]
- Hochhaus, A.; Larson, R.A.; Guilhot, F.; Radich, J.P.; Branford, S.; Hughes, T.P.; Baccarani, M.; Deininger, M.W.; Cervantes, F.; Fujihara, S.; et al. Long-Term Outcomes of Imatinib Treatment for Chronic Myeloid Leukemia. N. Engl. J. Med. 2017, 376, 917–927. [Google Scholar] [CrossRef]
- O’Brien, S.G.; Guilhot, F.; Larson, R.A.; Gathmann, I.; Baccarani, M.; Cervantes, F.; Cornelissen, J.J.; Fischer, T.; Hochhaus, A.; Hughes, T.; et al. Imatinib Compared with Interferon and Low-Dose Cytarabine for Newly Diagnosed Chronic-Phase Chronic Myeloid Leukemia. N. Engl. J. Med. 2003, 348, 994–1004. [Google Scholar] [CrossRef]
- Gramza, A.W.; Corless, C.L.; Heinrich, M.C. Resistance to Tyrosine Kinase Inhibitors in Gastrointestinal Stromal Tumors. Clin. Cancer Res. 2009, 15, 7510–7518. [Google Scholar] [CrossRef]
- Eadie, L.N.; Hughes, T.P.; White, D.L. Interaction of the Efflux Transporters ABCB1 and ABCG2 With Imatinib, Nilotinib, and Dasatinib. Clin. Pharmacol. Ther. 2014, 95, 294–306. [Google Scholar] [CrossRef]
- Hoffmeyer, S.; Burk, O.; von Richter, O.; Arnold, H.P.; Brockmöller, J.; Johne, A.; Cascorbi, I.; Gerloff, T.; Roots, I.; Eichelbaum, M.; et al. Functional Polymorphisms of the Human Multidrug-Resistance Gene: Multiple Sequence Variations and Correlation of One Allele with P-Glycoprotein Expression and Activity in Vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 3473–3478. [Google Scholar] [CrossRef]
- Vivona, D.; Lima, L.T.; Rodrigues, A.C.; Bueno, C.T.; Alcantara, G.K.S.; Barros, L.S.R.; DE Moraes Hungria, V.T.; Chiattone, C.S.; DE Lourdes Lopes Ferrari Chauffaille, M.; Guerra-Shinohara, E.M. ABCB1 Haplotypes Are Associated with P-Gp Activity and Affect a Major Molecular Response in Chronic Myeloid Leukemia Patients Treated with a Standard Dose of Imatinib. Oncol. Lett. 2014, 7, 1313–1319. [Google Scholar] [CrossRef]
- Imai, Y.; Nakane, M.; Kage, K.; Tsukahara, S.; Ishikawa, E.; Tsuruo, T.; Miki, Y.; Sugimoto, Y. C421A Polymorphism in the Human Breast Cancer Resistance Protein Gene Is Associated with Low Expression of Q141K Protein and Low-Level Drug Resistance. Mol. Cancer 2002, 1, 611–616. [Google Scholar]
- Prasad, B.; Lai, Y.; Lin, Y.; Unadkat, J.D. Interindividual Variability in the Hepatic Expression of the Human Breast Cancer Resistance Protein (BCRP/ABCG2): Effect of Age, Sex, and Genotype. J. Pharm. Sci. 2013, 102, 787–793. [Google Scholar] [CrossRef]
- Chen, S.; Sutiman, N.; Chowbay, B. Pharmacogenetics of Drug Transporters in Modulating Imatinib Disposition and Treatment Outcomes in Chronic Myeloid Leukemia & Gastrointestinal Stromal Tumor Patients. Pharmacogenomics 2016, 17, 1941–1955. [Google Scholar] [CrossRef]
- Hamada, A.; Miyano, H.; Watanabe, H.; Saito, H. Interaction of Imatinib Mesilate with Human P-Glycoprotein. J. Pharmacol. Exp. Ther. 2003, 307, 824–828. [Google Scholar] [CrossRef] [Green Version]
- Burger, H.; van Tol, H.; Boersma, A.W.M.; Brok, M.; Wiemer, E.A.C.; Stoter, G.; Nooter, K. Imatinib Mesylate (STI571) Is a Substrate for the Breast Cancer Resistance Protein (BCRP)/ABCG2 Drug Pump. Blood 2004, 104, 2940–2942. [Google Scholar] [CrossRef]
- Dalle Fratte, C.; Gagno, S.; Roncato, R.; Polesel, J.; Zanchetta, M.; Buzzo, M.; Posocco, B.; De Mattia, E.; Borsatti, R.; Puglisi, F.; et al. CYP2D6 and CYP2C8 Pharmacogenetics and Pharmacological Interactions to Predict Imatinib Plasmatic Exposure in GIST Patients. Brit. J. Clin. Pharma 2022, 1–10. [Google Scholar] [CrossRef]
- Rajamani, B.M.; Benjamin, E.S.B.; Abraham, A.; Ganesan, S.; Lakshmi, K.M.; Anandan, S.; Karathedath, S.; Varatharajan, S.; Mohanan, E.; Janet, N.B.; et al. Plasma Imatinib Levels and ABCB1 Polymorphism Influences Early Molecular Response and Failure-Free Survival in Newly Diagnosed Chronic Phase CML Patients. Sci. Rep. 2020, 10, 20640. [Google Scholar] [CrossRef]
- Adeagbo, B.A.; Bolaji, O.O.; Olugbade, T.A.; Durosinmi, M.A.; Bolarinwa, R.A.; Masimirembwa, C. Influence of CYP3A5*3 and ABCB1 C3435T on Clinical Outcomes and Trough Plasma Concentrations of Imatinib in Nigerians with Chronic Myeloid Leukaemia. J. Clin. Pharm. Ther. 2016, 41, 546–551. [Google Scholar] [CrossRef]
- Belohlavkova, P.; Vrbacky, F.; Voglova, J.; Racil, Z.; Zackova, D.; Hrochova, K.; Malakova, J.; Mayer, J.; Zak, P. The Significance of Enzyme and Transporter Polymorphisms for Imatinib Plasma Levels and Achieving an Optimal Response in Chronic Myeloid Leukemia Patients. Arch. Med. Sci. 2018, 14, 1416–1423. [Google Scholar] [CrossRef]
- Francis, J.; Dubashi, B.; Sundaram, R.; Pradhan, S.C.; Chandrasekaran, A. A Study to Explore the Correlation of ABCB1, ABCG2, OCT1 Genetic Polymorphisms and Trough Level Concentration with Imatinib Mesylate-Induced Thrombocytopenia in Chronic Myeloid Leukemia Patients. Cancer. Chemother. Pharm. 2015, 76, 1185–1189. [Google Scholar] [CrossRef]
- Harivenkatesh, N.; Kumar, L.; Bakhshi, S.; Sharma, A.; Kabra, M.; Velpandian, T.; Gogia, A.; Shastri, S.S.; Biswas, N.R.; Gupta, Y.K. Influence of MDR1 and CYP3A5 Genetic Polymorphisms on Trough Levels and Therapeutic Response of Imatinib in Newly Diagnosed Patients with Chronic Myeloid Leukemia. Pharmacol. Res 2017, 120, 138–145. [Google Scholar] [CrossRef]
- Seong, S.J.; Lim, M.; Sohn, S.K.; Moon, J.H.; Oh, S.-J.; Kim, B.S.; Ryoo, H.M.; Chung, J.S.; Joo, Y.D.; Bang, S.M.; et al. Influence of Enzyme and Transporter Polymorphisms on Trough Imatinib Concentration and Clinical Response in Chronic Myeloid Leukemia Patients. Ann. Oncol. 2013, 24, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Cargnin, S.; Ravegnini, G.; Soverini, S.; Angelini, S.; Terrazzino, S. Impact of SLC22A1 and CYP3A5 Genotypes on Imatinib Response in Chronic Myeloid Leukemia: A Systematic Review and Meta-Analysis. Pharmacol. Res. 2018, 131, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Dulucq, S.; Bouchet, S.; Turcq, B.; Lippert, E.; Etienne, G.; Reiffers, J.; Molimard, M.; Krajinovic, M.; Mahon, F.-X. Multidrug Resistance Gene (MDR1) Polymorphisms Are Associated with Major Molecular Responses to Standard-Dose Imatinib in Chronic Myeloid Leukemia. Blood 2008, 112, 2024–2027. [Google Scholar] [CrossRef]
- Bazeos, A.; Marin, D.; Reid, A.G.; Gerrard, G.; Milojkovic, D.; May, P.C.; de Lavallade, H.; Garland, P.; Rezvani, K.; Apperley, J.F.; et al. HOCT1 Transcript Levels and Single Nucleotide Polymorphisms as Predictive Factors for Response to Imatinib in Chronic Myeloid Leukemia. Leukemia 2010, 24, 1243–1245. [Google Scholar] [CrossRef]
- Koo, D.-H.; Ryu, M.-H.; Ryoo, B.-Y.; Beck, M.Y.; Na, Y.-S.; Shin, J.-G.; Lee, S.S.; Kim, E.-Y.; Kang, Y.-K. Association of ABCG2 Polymorphism with Clinical Efficacy of Imatinib in Patients with Gastrointestinal Stromal Tumor. Cancer. Chemother. Pharm. 2015, 75, 173–182. [Google Scholar] [CrossRef]
- Demetri, G.D.; Wang, Y.; Wehrle, E.; Racine, A.; Nikolova, Z.; Blanke, C.D.; Joensuu, H.; von Mehren, M. Imatinib Plasma Levels Are Correlated with Clinical Benefit in Patients with Unresectable/Metastatic Gastrointestinal Stromal Tumors. J. Clin. Oncol. 2009, 27, 3141–3147. [Google Scholar] [CrossRef]
- Picard, S.; Titier, K.; Etienne, G.; Teilhet, E.; Ducint, D.; Bernard, M.-A.; Lassalle, R.; Marit, G.; Reiffers, J.; Begaud, B.; et al. Trough Imatinib Plasma Levels Are Associated with Both Cytogenetic and Molecular Responses to Standard-Dose Imatinib in Chronic Myeloid Leukemia. Blood 2006, 109, 3496–3499. [Google Scholar] [CrossRef]
- von Mehren, M.; Widmer, N. Correlations between Imatinib Pharmacokinetics, Pharmacodynamics, Adherence, and Clinical Response in Advanced Metastatic Gastrointestinal Stromal Tumor (GIST): An Emerging Role for Drug Blood Level Testing? Cancer Treat Rev. 2011, 37, 291–299. [Google Scholar] [CrossRef]
- Gardner, E.; Burger, H.; Vanschaik, R.; Vanoosterom, A.; Debruijn, E.; Guetens, G.; Prenen, H.; Dejong, F.; Baker, S.; Bates, S. Association of Enzyme and Transporter Genotypes with the Pharmacokinetics of Imatinib. Clin. Pharmacol. Ther. 2006, 80, 192–201. [Google Scholar] [CrossRef]
- Togashi, M.; Niioka, T.; Komatsuda, A.; Nara, M.; Okuyama, S.; Omokawa, A.; Abumiya, M.; Wakui, H.; Takahashi, N.; Miura, M. Effect of CYP3A5 and ABCB1 Polymorphisms on the Interaction between Tacrolimus and Itraconazole in Patients with Connective Tissue Disease. Eur. J. Clin. Pharm. 2015, 71, 1091–1097. [Google Scholar] [CrossRef]
- Petain, A.; Kattygnarath, D.; Azard, J.; Chatelut, E.; Delbaldo, C.; Geoerger, B.; Barrois, M.; Séronie-Vivien, S.; LeCesne, A.; Vassal, G.; et al. Population Pharmacokinetics and Pharmacogenetics of Imatinib in Children and Adults. Clin. Cancer Res. 2008, 14, 7102–7109. [Google Scholar] [CrossRef]
- Maekawa, K.; Yamamura, M.; Matsuki, A.; Ishikawa, T.; Hirai, T.; Yamaguchi, Y.; Saito, Y.; Kanda, T. Impacts of SNPs on Adverse Events and Trough Concentration of Imatinib in Patients with Gastrointestinal Stromal Tumors. Drug. Metab. Pharm. 2022, 43, 100441. [Google Scholar] [CrossRef]
- Dessilly, G.; Panin, N.; Elens, L.; Haufroid, V.; Demoulin, J.-B. Impact of ABCB1 1236C > T-2677G > T-3435C > T Polymorphisms on the Anti-Proliferative Activity of Imatinib, Nilotinib, Dasatinib and Ponatinib. Sci. Rep. 2016, 6, 29559. [Google Scholar] [CrossRef] [PubMed]
- Gouin-Thibault, I.; Delavenne, X.; Blanchard, A.; Siguret, V.; Salem, J.E.; Narjoz, C.; Gaussem, P.; Beaune, P.; Funck-Brentano, C.; Azizi, M.; et al. Interindividual Variability in Dabigatran and Rivaroxaban Exposure: Contribution of ABCB1 Genetic Polymorphisms and Interaction with Clarithromycin. J. Thromb. Haemost. 2017, 15, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chia, Y.L.; Nedelman, J.; Schran, H.; Mahon, F.-X.; Molimard, M. A Therapeutic Drug Monitoring Algorithm for Refining the Imatinib Trough Level Obtained at Different Sampling Times. Ther. Drug Monit. 2009, 31, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Slim, K.; Nini, E.; Forestier, D.; Kwiatkowski, F.; Panis, Y.; Chipponi, J. Methodological Index for Non-Randomized Studies (MINORS): Development and Validation of a New Instrument: Methodological Index for Non-Randomized Studies. ANZ J. Surg. 2003, 73, 712–716. [Google Scholar] [CrossRef]
- Normand, S.L. Meta-Analysis: Formulating, Evaluating, Combining, and Reporting. Stat. Med. 1999, 18, 321–359. [Google Scholar] [CrossRef]


| Patient Characteristic | N | % |
|---|---|---|
| Gender | ||
| Male | 16 | 48.5 |
| Female | 17 | 51.5 |
| Age at enrollment | ||
| Median (range) | 66 (35–83) | |
| Primary tumor site | ||
| Stomach | 15 | 45.5 |
| Intestinal | 13 | 39.4 |
| Other 1 | 5 | 15.1 |
| Imatinib setting at enrollment | ||
| Adjuvant | 9 | 27.3 |
| First line | 24 | 72.7 |
| SNP | Genotype Frequency, N (%) | Allele Frequency | HWE | |||
|---|---|---|---|---|---|---|
| Wild Type | Het | Variant | p | q | p-Value | |
| ABCB1 c.3435C>T | 9 (27.3) | 15 (45.4) | 9 (27.3) | 0.500 | 0.500 | 0.3657 |
| ABCB1 c.2677G>T | 14 (42.4) | 13 (39.4) | 6 (18.2) | 0.621 | 0.379 | 0.1050 |
| ABCB1 c.1236C>T | 14 (42.4) | 13 (39.4) | 6 (18.2) | 0.621 | 0.379 | 0.1050 |
| ABCG2 c.412C>A | 23 (69.7) | 9 (27.7) | 1 (3.0) | 0.836 | 0.164 | 0.8564 |
| Genotype | N | Imatinib Ctrough | p-Value | |
|---|---|---|---|---|
| Mean | SD | |||
| ABCB1 c.3435C>T | ||||
| Wild-type | 9 | 1135.9 | 530.7 | 0.4186 |
| Heterozygous | 15 | 934.8 | 514.7 | |
| Variant | 9 | 1083.6 | 620.7 | |
| ABCB1 c.2677G>T | ||||
| Wild-type | 14 | 1049.4 | 573.1 | 0.1907 |
| Heterozygous | 13 | 1160.0 | 703.8 | |
| Variant | 6 | 761.4 | 318.2 | |
| ABCB1 c.1236C>T | ||||
| Wild-type | 14 | 1038.0 | 551.6 | 0.1907 |
| Heterozygous | 13 | 1191.1 | 695.9 | |
| Variant | 6 | 716.0 | 245.3 | |
| ABCB1 Haplotype | ||||
| Wild-type | 8 | 1135.9 | 412.6 | 0.2185 |
| Heterozygous | 20 | 1059.3 | 566.0 | |
| Variant | 5 | 813.5 | 279.7 | |
| ABCG2 c.412C>A | ||||
| Wild-type | 23 | 1011.9 | 657.0 | 0.5434 |
| Heterozygous | 9 | 1030.3 | 487.8 | |
| Variant | 1 | 1792.8 | - | |
| Study | Country | Disease | Study Type | Enrollment Interval | Sample Size (Genotyped) | Genotyping Method | Imatinib Quantification Method | Analyzed SNPs | Wild-Type (n) | Heterozygous (n) | Variant (n) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adeagbo et al. [15] | Nigeria | CML | Observational, case–control, prospective | N.A. | 110 (109) | TaqMan allele discrimination assay | HPLC coupled with a diode array UV detector | ABCB1 c.3435C>T | 80 | 24 | 5 |
| Belohlavkova et al. [16] | Czech Republic | CML | Observational, retrospective | 1997–2012 | 112 | TaqMan allele discrimination assay | HPLC | ABCB1 c.3435C>T | 22 | 58 | 32 |
| ABCB1 c.1236C>T | 32 | 62 | 18 | ||||||||
| ABCG2 c.421C>A | 87 | 23 | 2 | ||||||||
| Francis et al. [17] | India | CML | Observational, prospective | 2012–2014 | 111 (73) | TaqMan allele discrimination assay | HPLC coupled with electrospray-ionization MS/MS | ABCB1 c.3435C>T | 11 | 37 | 25 |
| ABCB1 c.2677G>T | 8 | 33 | 32 | ||||||||
| ABCG2 c.421C>A | 54 | 19 | 0 | ||||||||
| Harivenkatesh et al. [18] | India | CML | Observational, prospective | 2013–2016 | 173 | Direct Sanger sequencing | HPLC coupled with MS/MS | ABCB1 c.3435C>T | 19 | 88 | 66 |
| ABCB1 c.1236C>T | 19 | 113 | 41 | ||||||||
| ABCB1 c.2677G>T | 18 | 76 | 79 | ||||||||
| Rajamani et al. [14] | India | CML | Observational, prospective | N.A. | 160 (67) | PCR restriction fragment length polymorphism (RFLP) | HPLC | ABCB1 c.3435C>T | 7 | 39 | 21 |
| ABCB1 c.1236C>T | 10 | 30 | 27 | ||||||||
| ABCB1 c.2677G>T | 6 | 35 | 26 | ||||||||
| ABCG2 c.421C>A | 45 | 17 | 5 | ||||||||
| Seong et al. [19] | Korea | CML | Observational, prospective | N.A. | 82 | TaqMan allele discrimination assay | HPLC coupled with MS/MS | ABCB1 c.3435C>T | 35 | 38 | 9 |
| ABCB1 c.1236C>T | 17 | 37 | 28 | ||||||||
| ABCG2 c.421C>A | 41 | 32 | 8 | ||||||||
| CRO-Aviano Study | Italy | GIST | Observational, prospective | 2015–2021 | 33 | Targeted NGS | HPLC coupled with MS/MS | ABCB1 c.3435C>T | 9 | 15 | 9 |
| ABCB1 c.1236C>T | 14 | 13 | 6 | ||||||||
| ABCB1 c.2677G>T | 14 | 13 | 6 | ||||||||
| ABCG2 c.421C>A | 23 | 9 | 1 |
| Genotype | N Studies | Ctrough | Test for Heterogeneity | Subgroup Differences | |
|---|---|---|---|---|---|
| Studies (Total Patients) | Mean (95% CI) | I2 % | p-Value | p-Value | |
| ABCB1 c.3435C>T | |||||
| CC + CT vs. TT a | 7 (649) | 1209.8 (893.6–1526.1) vs. 1378.6 (1059.3–1698.0) | 95 | <0.01 | 0.46 |
| CC vs. CT + TT b | 1088.1 (876.7–1297.5) vs. 1250.9 (790.6–1711.1) | 96 | <0.01 | 0.53 | |
| CC vs. CT vs. TT c | 1122.7 (871.1–1374.3) vs. 1194.6 (708.2–1681.0) vs. 1347.0 (1038.3–1656.1) | 95 | <0.01 | 0.54 | |
| ABCB1 c.2677G>T | |||||
| GG + GT vs. TT a | 4 (346) | 1408.7 (1196.1–1621.4) vs. 1403.0 (871.0–1936.3) | 88 | <0.01 | 0.99 |
| GG vs. GT + TT b | 1078.5 (791.7–1364.2) vs. 1475.9 (1174.2–1777.7) | 89 | <0.01 | 0.06 | |
| GG vs. GT vs. TT c | 1078.5 (791.7–1365.2) vs. 1504.7 (1329.9–1679.6) vs. 1403.9 (871.5–1936.3) | 88 | <0.01 | 0.05 | |
| ABCB1 c.1236C>T | |||||
| CC + CT vs. TT a | 5 (467) | 1307.3 (1057.7–1556.9) vs. 1295.2 (786.6–1803.9) | 89 | <0.01 | 0.97 |
| CC vs. CT + TT b | 1065.7 (760.9–1370.5) vs. 1365.4 (1049.3–1681.4) | 89 | <0.01 | 0.18 | |
| CC vs. CT vs. TT c | 1065.7 (760.9–1370.5) vs. 1394.0 (1120.6–1667.4) vs. 1295.2 (786.6–1803.9) | 87 | <0.01 | 0.29 | |
| ABCG2 c.412C>A | |||||
| CC + CA vs. AA a | 4 (293) | 1198.6 (1036.0–1361.2) vs. 1463.2 (1276.5–1649.9) | 67 | <0.01 | 0.04 |
| CC vs. CA + AA b | 1177.3 (1059.0–1295.6) vs. 1263.47 (1000.6–1526.3) | 37 | 0.14 | 0.56 | |
| CC vs. CA vs. AA c | 1177.3 (1059.0–1295.6) vs. 1228.4 (950.01–1506.8) vs. 1463.2 (1276.5–1649.9) | 56 | <0.01 | 0.04 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dalle Fratte, C.; Polesel, J.; Gagno, S.; Posocco, B.; De Mattia, E.; Roncato, R.; Orleni, M.; Puglisi, F.; Guardascione, M.; Buonadonna, A.; et al. Impact of ABCG2 and ABCB1 Polymorphisms on Imatinib Plasmatic Exposure: An Original Work and Meta-Analysis. Int. J. Mol. Sci. 2023, 24, 3303. https://doi.org/10.3390/ijms24043303
Dalle Fratte C, Polesel J, Gagno S, Posocco B, De Mattia E, Roncato R, Orleni M, Puglisi F, Guardascione M, Buonadonna A, et al. Impact of ABCG2 and ABCB1 Polymorphisms on Imatinib Plasmatic Exposure: An Original Work and Meta-Analysis. International Journal of Molecular Sciences. 2023; 24(4):3303. https://doi.org/10.3390/ijms24043303
Chicago/Turabian StyleDalle Fratte, Chiara, Jerry Polesel, Sara Gagno, Bianca Posocco, Elena De Mattia, Rossana Roncato, Marco Orleni, Fabio Puglisi, Michela Guardascione, Angela Buonadonna, and et al. 2023. "Impact of ABCG2 and ABCB1 Polymorphisms on Imatinib Plasmatic Exposure: An Original Work and Meta-Analysis" International Journal of Molecular Sciences 24, no. 4: 3303. https://doi.org/10.3390/ijms24043303







