The Genes–Stemness–Secretome Interplay in Malignant Pleural Mesothelioma: Molecular Dynamics and Clinical Hints
Abstract
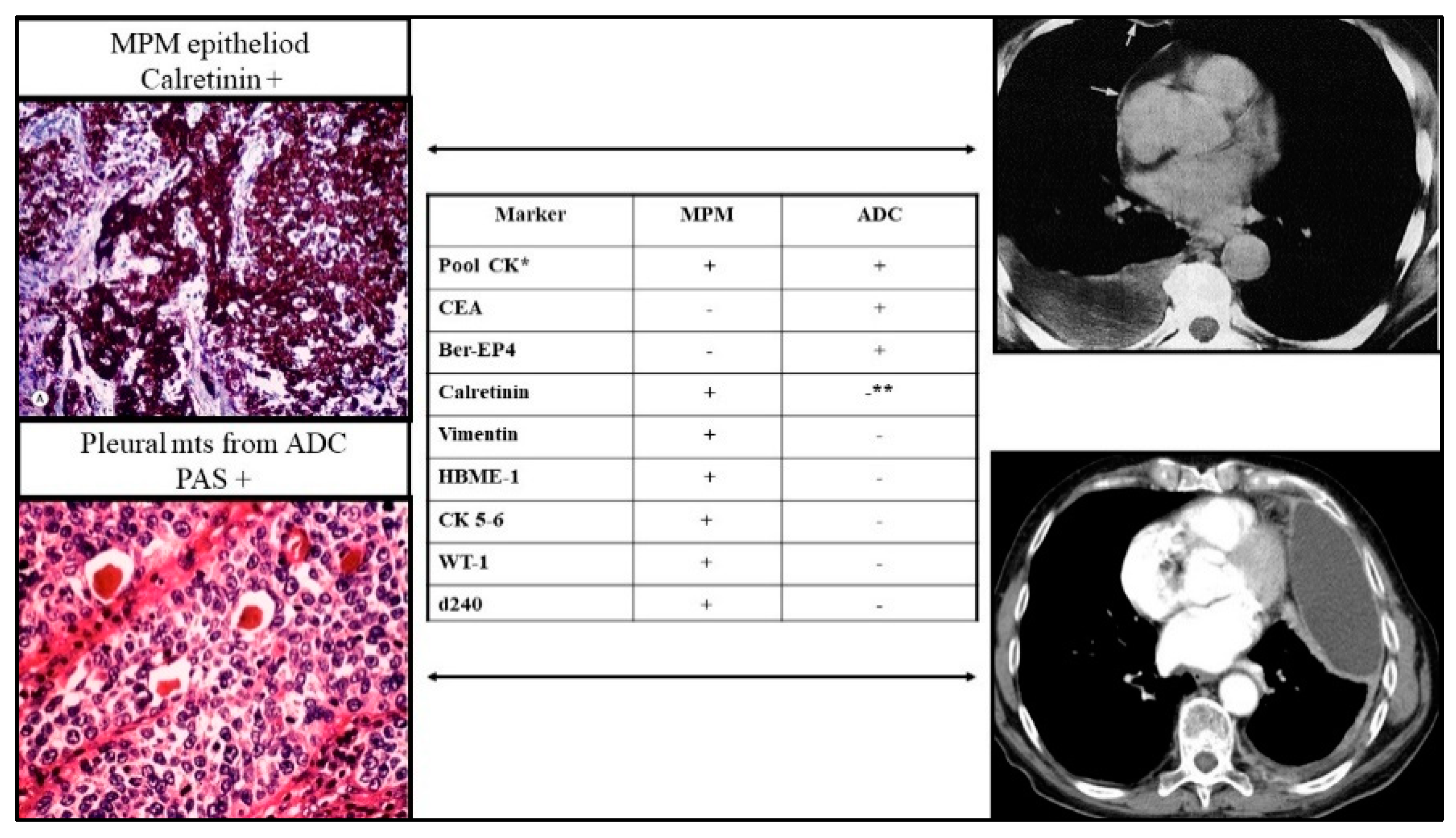
1. Introduction
2. Genetic Alterations and Disease Stage
3. The Role of Microenvironmental Hypoxia
4. The Hypoxia and Stemness Interconnection in MPM
5. Trancriptome and Secretome Profile of MPM
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Røe, O.D.; Stella, G.M. Malignant pleural mesothelioma: History, controversy and future of a manmade epidemic. Eur. Respir. Rev. 2015, 24, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Gregory, S.N.; Sarvestani, A.L.; Blakely, A.M. Malignant peritoneal mesothelioma literature review: Past, present, and future. Dig. Med. Res. 2022, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- van Kooten, J.P.; Belderbos, R.A.; von der Thüsen, J.H.; Aarts, M.J.; Verhoef, C.; Burgers, J.A.; Baas, P.; Aalbers, A.G.J.; Maat, A.P.W.M.; Aerts, J.G.J.V.; et al. Incidence, treatment and survival of malignant pleural and peritoneal mesothelioma: A population-based study. Thorax 2022, 77, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Reid, A.; de Klerk, N.H.; Magnani, C.; Ferrante, D.; Berry, G.; Musk, A.W.; Merler, E. Mesothelioma risk after 40 years since first exposure to asbestos: A pooled analysis. Thorax 2014, 69, 843–850. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Madison, R.W.; Lennerz, J.K.; Chen, K.T.; Hopkins, J.F.; Schrock, A.B.; Ritterhouse, L.L.; Lester, A.; Wharton, K.A., Jr.; Mino-Kenudson, M.; et al. Molecular Characterization of Mesothelioma: Impact of Histologic Type and Site of Origin on Molecular Landscape. JCO Precis. Oncol. 2022, 6, e2100422. [Google Scholar] [CrossRef]
- Dacic, S. Pleural mesothelioma classification-update and challenges. Mod. Pathol. 2022, 35 (Suppl. 1), 51–56. [Google Scholar] [CrossRef]
- Bianco, A.; Valente, T.; De Rimini, M.L.; Sica, G.; Fiorelli, A. Clinical diagnosis of malignant pleural mesothelioma. J. Thorac. Dis. 2018, 10 (Suppl. 2), S253–S261. [Google Scholar] [CrossRef]
- Stella, G.M.; Senetta, R.; Cassenti, A.; Ronco, M.; Cassoni, P. Cancers of unknown primary origin: Current perspectives and future therapeutic strategies. J. Transl. Med. 2012, 10, 12. [Google Scholar] [CrossRef]
- Sauter, J.L.; Dacic, S.; Galateau-Salle, F.; Attanoos, R.L.; Butnor, K.J.; Churg, A.; Husain, A.N.; Kadota, K.; Khoor, A.; Nicholson, A.G.; et al. The 2021 WHO Classification of Tumors of the Pleura: Advances Since the 2015 Classification. J. Thorac. Oncol. 2022, 17, 608–622. [Google Scholar] [CrossRef]
- Lettieri, S.; Bortolotto, C.; Agustoni, F.; Lococo, F.; Lancia, A.; Comoli, P.; Corsico, A.G.; Stella, G.M. The Evolving Landscape of the Molecular Epidemiology of Malignant Pleural Mesothelioma. J. Clin. Med. 2021, 10, 1034. [Google Scholar] [CrossRef]
- Yamada, K.M.; Cukierman, E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Tertemiz, K.C.; Ozgen Alpaydin, A.; Gurel, D.; Savas, R.; Gulcu, A.; Akkoclu, A. Multiple distant metastases in a case of malignant pleural mesothelioma. Respir. Med. Case Rep. 2014, 13, 16–18. [Google Scholar] [CrossRef]
- Taşçı, E.S.; Akın, G.; Öneç, B.; Eşbah, O. Malignant pleural mesothelioma with rarely seen metastases. J. Oncol. Sci. 2017, 3, 133–134. [Google Scholar] [CrossRef]
- Mitsimponas, N.; Petounis, A. Pancreatic metastasis from malignant pleural mesothelioma. An extremely rare site of metastasis in a patient with a very prolonged survival of seven years. Curr. Probl. Cancer Case Rep. 2021, 4, 100077. [Google Scholar] [CrossRef]
- Betti, M.; Aspesi, A.; Sculco, M.; Matullo, G.; Magnani, C.; Dianzani, I. Genetic predisposition for malignant mesothelioma: A concise review. Mutat. Res. Rev. 2019, 781, 1–10. [Google Scholar] [CrossRef]
- Levpuscek, K.; Goricar, K.; Kovac, V.; Dolzan, V.; Franko, A. The influence of genetic variability of DNA repair mechanisms on the risk of malignant mesothelioma. Radiol. Oncol. 2019, 53, 206–212. [Google Scholar] [CrossRef]
- Senk, B.; Goricar, K.; Kovac, V.; Dolzan, V.; Franko, A. Genetic polymorphisms in aquaporin 1 as risk factors for malignant mesothelioma and biomarkers of response to cisplatin treatment. Radiol. Oncol. 2019, 53, 96–104. [Google Scholar] [CrossRef]
- Betti, M.; Ferrante, D.; Padoan, M.; Guarrera, S.; Giordano, M.; Aspesi, A.; Mirabelli, D.; Casadio, C.; Ardissone, F.; Ruffini, E.; et al. XRCC1 and ERCC1 variants modify malignant mesothelioma risk: A case–control study. Mutat. Res. 2011, 708, 11–20. [Google Scholar] [CrossRef]
- Franko, A.; Kotnik, N.; Goricar, K.; Kovac, V.; Dodic-Fikfak, M.; Dolzan, V. The influence of genetic variability on the risk of developing malignant mesothelioma. Radiol. Oncol. 2018, 52, 105–111. [Google Scholar] [CrossRef]
- Malakoti, F.; Targhazeh, N.; Abadifard, E.; Zarezadeh, R.; Samemaleki, S.; Asemi, Z.; Younesi, S.; Mohammadnejad, R.; Hossini, S.H.; Karimian, A.; et al. DNA repair and damage pathways in mesothelioma development and therapy. Cancer Cell Int. 2022, 22, 176. [Google Scholar] [CrossRef] [PubMed]
- Baumann, F.; Flores, E.; Napolitano, A.; Kanodia, S.; Taioli, E.; Pass, H.; Yang, H.; Carbone, M. Mesothelioma patients with germline BAP1 mutations have 7-fold improved long-term survival Mesothelioma patients with germline BAP1 mutations have 7-fold improved long-term survival. Carcinogenesis 2015, 36, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Carbone, M.; Yang, H.; Pass, H.; Krausz, T.; Testa, J.R.; Gaudino, G. BAP1 and cancer. Nat. Rev. Cancer 2013, 13, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Righi, L.; Duregon, E.; Vatrano, S.; Izzo, S.; Giorcelli, J.; Rondón-Lagos, M.; Ascoli, V.; Ruffini, E.; Ventura, L.; Volante, M.; et al. BRCA1-Associated Protein 1 (BAP1) Immunohistochemical Expression as a Diagnostic Tool in Malignant Pleural Mesothelioma Classification: A Large Retrospective Study. J. Thorac. Oncol. 2016, 11, 2006–2017. [Google Scholar] [CrossRef] [PubMed]
- Crovella, S.; Moura, R.R.; Brandão, L.; Vita, F.; Schneider, M.; Zanconati, F.; Finotto, L.; Zacchi, P.; Zabucchi, G.; Borelli, V. Variant Enrichment Analysis to Explore Pathways Disruption in a Necropsy Series of Asbestos-Exposed Shipyard Workers. Int. J. Mol. Sci. 2022, 23, 13628. [Google Scholar] [CrossRef]
- Crovella, S.; Bianco, A.M.; Vuch, J.; Zupin, L.; Moura, R.R.; Trevisan, E.; Schneider, M.; Brollo, A.; Nicastro, E.M.; Cosenzi, A.; et al. Iron signature in asbestos-induced malignant pleural mesothelioma: A population-based autopsy study. J. Toxicol. Environ. Health A 2016, 79, 129–141. [Google Scholar] [CrossRef]
- Celsi, F.; Crovella, S.; Moura, R.R.; Schneider, M.; Vita, F.; Finotto, L.; Zabucchi, G.; Zacchi, P.; Borelli, V. Pleural mesothelioma and lung cancer: The role of asbestos exposure and genetic variants in selected iron metabolism and inflammation genes. J. Toxicol. Environ. Health A 2019, 82, 1088–1102. [Google Scholar] [CrossRef]
- Crovella, S.; Revelant, A.; Muraro, E.; Moura, R.R.; Brandão, L.; Trovò, M.; Steffan, A.; Zacchi, P.; Zabucchi, G.; Minatel, E.; et al. Biological Pathways Associated with the Development of Pulmonary Toxicities in Mesothelioma Patients Treated with Radical Hemithoracic Radiation Therapy: A Preliminary Study. Front. Oncol. 2021, 11, 784081. [Google Scholar] [CrossRef]
- Girardelli, M.; Maestri, I.; Rinaldi, R.R.; Tognon, M.; Boldorini, R.; Bovenzi, M.; Crovella, S.; Comar, M. NLRP1 polymorphisms in patients with asbestos-associated mesothelioma. Infect. Agent Cancer 2012, 7, 25. [Google Scholar] [CrossRef]
- Borelli, V.; Moura, R.R.; Trevisan, E.; Crovella, S. NLRP1 and NLRP3 polymorphisms in mesothelioma patients and asbestos exposed individuals a population-based autopsy study from North East Italy. Infect. Agent Cancer 2015, 10, 26. [Google Scholar] [CrossRef]
- Richards, W.G. Malignant pleural mesothelioma: Predictors and staging. Ann. Transl. Med. 2017, 5, 243. [Google Scholar] [CrossRef]
- Berzenji, L.; Van Schil, P. Multimodality treatment of malignant pleural mesothelioma. F1000Research 2018, 7, 1681. [Google Scholar] [CrossRef]
- Betti, M.; Casalone, E.; Ferrante, D.; Aspesi, A.; Morleo, G.; Biasi, A.; Sculco, M.; Mancuso, G.; Guarrera, S.; Righi, L.; et al. Germline mutations in DNA repair genes predispose asbestos-exposed patients to malignant pleural mesothelioma. Cancer Lett. 2017, 405, 38–45. [Google Scholar] [CrossRef]
- Trassl, L.; Stathopoulos, G.T. KRAS Pathway Alterations in Malignant Pleural Mesothelioma: An Underestimated Player. Cancers 2022, 14, 4303. [Google Scholar] [CrossRef]
- Shukla, A.; Hillegass, J.M.; Macpherson, M.B.; Beuschel, S.L.; Vacek, P.M.; Butnor, K.J.; Pass, H.I.; Carbone, M.; Testa, J.R.; Heintz, N.H.; et al. ERK2 is essential for the growth of human epithelioid malignant mesotheliomas. Int. J. Cancer 2011, 129, 1075–1086. [Google Scholar] [CrossRef]
- Ivanov, S.V.; Miller, J.; Lucito, R.; Tang, C.; Ivanova, A.V.; Pei, J.; Carbone, M.; Cruz, C.; Beck, A.; Webb, C.; et al. Genoic events associated with progression of pleural malignant mesothelioma. Int. J. Cancer 2009, 124, 589–599. [Google Scholar] [CrossRef]
- Stella, G.M.; Benvenuti, S.; Comoglio, P.M. Targeting the MET oncogene in cancer and metastases. Expert Opin. Investig. Drugs 2010, 19, 1381–1394. [Google Scholar] [CrossRef]
- Jagadeeswaran, R.; Ma, P.C.; Seiwert, T.Y.; Jagadeeswaran, S.; Zumba, O.; Nallasura, V.; Ahmed, S.; Filiberti, R.; Paganuzzi, M.; Runtoni, R.; et al. Functional analysis of c-Met/hepatocyte growth factor pathway in malignant pleural mesothelioma. Cancer Res. 2006, 66, 352–361. [Google Scholar] [CrossRef]
- Santoni-Rugiu, E.; Lü, M.J.S.; Jakobsen, J.N.; Melchior, L.C.; Ravn, J.; Sørensen, J.B. Correlation of MET-Receptor Overexpression with MET Gene Amplification and Patient Outcome in Malignant Mesothelioma. Int. J. Mol. Sci. 2021, 22, 12868. [Google Scholar] [CrossRef]
- Bois, M.C.; Mansfield, A.S.; Sukov, W.R.; Jenkins, S.M.; Moser, J.C.; Sattler, C.A.; Smith, C.Y.; Molina, J.R.; Peikert, T.; Roden, A.C. c-Met expression and MET amplification in malignant pleural mesothelioma. Ann. Diagn. Pathol. 2016, 23, 1–7. [Google Scholar] [CrossRef]
- Varesano, S.; Salvi, S.; Boccardo, S.; Ravetti, J.L.; Pistillo, M.P.; Canessa, P.A.; Ferro, P.; Fedeli, F.; Roncella, S. MET Gene Status in Malignant Mesothelioma Using Fluorescent In Situ Hybridization. J. Thorac. Oncol. 2016, 11, 28–30. [Google Scholar] [CrossRef] [PubMed]
- Ramundo, V.; Zanirato, G.; Aldieri, E. The Epithelial-to-Mesenchymal Transition (EMT) in the Development and Metastasis of Malignant Pleural Mesothelioma. Int. J. Mol. Sci. 2021, 22, 12216. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Vizán, P.; Das, D.; Chakravarty, P.; Vogt, J.; Rogers, K.W.; Müller, P.; Hinck, A.P.; Sapkota, G.P.; Hill, C.S. TGF-β uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. eLife 2018, 7, e31756. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-β-induced epithelial to mesenchymal transition. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef]
- Kanteti, R.; Dhanasingh, I.; Kawada, I.; Lennon, F.E.; Arif, Q.; Bueno, R.; Hasina, R.; Husain, A.N.; Vigneswaran, W.; Seiwert, T.; et al. MET and PI3K/mTOR as a Potential Combinatorial Therapeutic Target in Malignant Pleural Mesothelioma. PLoS ONE 2014, 9, e105919. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, L.; Li, H.; Eilers, G.; Kuang, Y.; Shi, S.; Yan, Z.; Li, X.; Corson, J.M.; Meng, F.; et al. Multipoint targeting of the PI3K/mTOR pathway in mesothelioma. Br. J. Cancer 2014, 110, 2479–2488. [Google Scholar] [CrossRef]
- Cova, E.; Pandolfi, L.; Colombo, M.; Frangipane, V.; Inghilleri, S.; Morosini, M.; Mrakic-Sposta, S.; Moretti, S.; Monti, M.; Pignochino, Y.; et al. Pemetrexed-loaded nanoparticles targeted to malignant pleural mesothelioma cells: An in vitro study. Int. J. Nanomed. 2019, 14, 773–785. [Google Scholar] [CrossRef]
- Echeverry, N.; Ziltener, G.; Barbone, D.; Weder, W.; Stahel, R.A.; Broaddus, V.C.; Felley-Bosco, E. Inhibition of autophagy sensitizes malignant pleural mesothelioma cells to dual PI3K/mTOR inhibitors. Cell Death Dis. 2015, 6, e1757. [Google Scholar] [CrossRef]
- Miyoshi, S.; Hamada, H.; Hamaguchi, N.; Kato, A.; Katayama, H.; Irifune, K.; Ito, R.; Miyazaki, T.; Okura, T.; Higaki, J. Antitumor activity of MEK and PI3K inhibitors against malignant pleural mesothelioma cells in vitro and in vivo. Int. J. Oncol. 2012, 41, 449–456. [Google Scholar] [CrossRef]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciplocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef]
- Hu, T.; Wolfram, J.; Srivastava, S. Extracellular Vesicles in Cancer Detection: Hopes and Hypes. Trends Cancer 2021, 7, 122–133. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.; McCombie, W. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 7, 333–351. [Google Scholar] [CrossRef]
- Mardis, E.R. The Impact of Next-Generation Sequencing on Cancer Genomics: From Discovery to Clinic. Cold Spring Harb. Perspect. Med. 2019, 9, a036269. [Google Scholar] [CrossRef]
- Ugurluer, G.; Chang, K.; Gamez, M.E.; Arnett, A.L.; Jayakrishnan, R.; Miller, R.C.; Sio, T.T. Genome-based Mutational Analysis by Next Generation Sequencing in Patients with Malignant Pleural and Peritoneal Mesothelioma. Anticancer. Res. 2016, 36, 2331–2338. [Google Scholar] [CrossRef]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef]
- De Reyniès, A.; Jaurand, M.C.; Renier, A.; Couchy, G.; Hysi, I.; Elarouci, N.; Galateau-Sallé, F.; Copin, M.C.; Hofman, P.; Cazes, A.; et al. Molecular classification of malignant pleural mesothelioma: Identification of a poor prognosis subgroup linked to the epithelial-to-mesenchymal transition. Clin. Cancer Res. 2014, 20, 1323–1334. [Google Scholar] [CrossRef]
- Lo Iacono, M.; Monica, V.; Righi, L.; Grosso, F.; Libener, R.; Vatrano, S.; Bironzo, P.; Novello, S.; Musmeci, L.; Volante, M.; et al. Targeted next-generation sequencing of cancer genes in advanced stage malignant pleural mesothelioma: A retrospective study. J. Thorac. Oncol. 2015, 10, 492–499. [Google Scholar] [CrossRef]
- Sculco, M.; La Vecchia, M.; Aspesi, A.; Pinton, G.; Clavenna, M.G.; Casalone, E.; Allione, A.; Grosso, F.; Libener, R.; Muzio, A.; et al. Malignant pleural mesothelioma: Germline variants in DNA repair genes may steer tailored treatment. Eur. J. Cancer 2022, 163, 44–54. [Google Scholar] [CrossRef]
- Hiltbrunner, S.; Fleischmann, Z.; Sokol, E.S.; Zoche, M.; Felley-Bosco, E.; Curioni-Fontecedro, A. Genomic landscape of pleural and peritoneal mesothelioma tumours. Br. J. Cancer 2022, 127, 1997–2005. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, X.; Hou, J.; Liang, X.; Ruan, Z.; Guo, H.; Nan, K.; Jiang, L. Identification of a Five-Gene Signature for Predicting Survival in Malignant Pleural Mesothelioma Patients. Front. Genet. 2020, 11, 899. [Google Scholar] [CrossRef]
- Creaney, J.; Patch, A.M.; Addala, V.; Sneddon, S.A.; Nones, K.; Dick, I.M.; Lee, Y.C.G.; Newell, F.; Rouse, E.J.; Naeini, M.M.; et al. Comprehensive genomic and tumour immune profiling reveals potential therapeutic targets in malignant pleural mesothelioma. Genome Med. 2022, 14, 58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Luo, J.L.; Sun, Q.; Harber, J.; Dawson, A.G.; Nakas, A.; Busacca, S.; Sharkey, A.J.; Waller, D.; Sheaff, M.T.; et al. Clonal architecture in mesothelioma is prognostic and shapes the tumour microenvironment. Nat. Commun. 2021, 12, 1751. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Wilson, W.R. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 2004, 4, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.R.; Hay, M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. [Google Scholar] [CrossRef]
- Singleton, D.C.; Macann, A.; Wilson, W.R. Therapeutic targeting of the hypoxic tumour microenvironment. Nat. Rev. Clin. Oncol. 2021, 18, 751–772. [Google Scholar] [CrossRef]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef]
- Nowak, A.K.; Brosseau, S.; Cook, A.; Zalcman, G. Antiangiogeneic Strategies in Mesothelioma. Front. Oncol. 2020, 10, 126. [Google Scholar] [CrossRef]
- Nabavi, N.; Bennewith, K.L.; Churg, A.; Wang, Y.; Collins, C.C.; Mutti, L. Switching off malignant mesothelioma: Exploiting the hypoxic microenvironment. Genes Cancer 2016, 7, 340–354. [Google Scholar] [CrossRef]
- Kim, M.C.; Hwang, S.H.; Kim, N.Y.; Lee, H.-S.; Ji, S.; Yang, Y.; Kim, Y. Hypoxia promotes acquisition of aggressive phenotypes in human malignant mesothelioma. BMC Cancer 2018, 18, 819. [Google Scholar] [CrossRef]
- Kim, J.W.; Tchernyshyov, I.; Semenza, G.L.; Dang, C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef]
- Kaira, K.; Serizawa, M.; Koh, Y.; Takahashi, T.; Hanaoka, H.; Oriuchi, N.; Endo, M.; Kondo, H.; Nakajima, T.; Yamamoto, N. Relationship between 18F-FDG uptake on positron emission tomography and molecular biology in malignant pleural mesothelioma. Eur. J. Cancer 2012, 48, 1244–1254. [Google Scholar] [CrossRef]
- Taralli, S.; Giancipoli, R.G.; Caldarella, C.; Scolozzi, V.; Ricciardi, S.; Cardillo, G.; Calcagni, M.L. The Prognostic Value of 18F-FDG PET Imaging at Staging in Patients with Malignant Pleural Mesothelioma: A Literature Review. J. Clin. Med. 2021, 11, 33. [Google Scholar] [CrossRef]
- Klabatsa, A.; Chicklore, S.; Barrington, S.F.; Goh, V.; Lang-Lazdunski, L.; Cook, G.J. The association of 18F-FDG PET/CT parameters with survival in malignant pleural mesothelioma. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 276–282. [Google Scholar] [CrossRef]
- Pennacchietti, S.; Michieli, P.; Galluzzo, M.; Mazzone, M.; Giordano, S.; Comoglio, P.M. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell 2003, 3, 347–361. [Google Scholar] [CrossRef]
- Shukuya, T.; Oyanagi, J.; Serizawa, M.; Watanabe, M.; Yamamoto, N.; Koh, Y. Hypoxia Inducible Factor-1α Inhibition in Von Hippel Lindau-mutant Malignant Pleural Mesothelioma Cells. Anticancer. Res. 2020, 40, 1867–1874. [Google Scholar] [CrossRef]
- Ravenna, L.; Cardillo, I.; Curzio, G.; Baldi, A.; Mattioni, M.; Vincenzi, B.; Russo, M.A.; Soddu, S.; Verdina, A. Mesothelioma and Hypoxia: Modulation of the Inflammation-Related Phenotype and Identification of Prognostic Markers. J. Cancer Sci. Ther. 2014, 6, 9. [Google Scholar] [CrossRef]
- Liu, Z.; Tu, K.; Wang, Y.; Yao, B.; Li, Q.; Wang, L.; Dou, C.; Liu, Q.; Zheng, X. Hypoxia Accelerates Aggressiveness of Hepatocellular Carcinoma Cells Involving Oxidative Stress, Epithelial-Mesenchymal Transition and Non-Canonical Hedgehog Signaling. Cell. Physiol. Biochem. 2017, 44, 1856–1868. [Google Scholar] [CrossRef]
- De Santi, C.; Melaiu, O.; Bonotti, A.; Cascione, L.; Di Leva, G.; Foddis, R.; Cristaudo, A.; Lucchi, M.; Mora, M.; Truini, A.; et al. Deregulation of miRNAs in malignant pleural mesothelioma is associated with prognosis and suggests an alteration of cell metabolism. Sci. Rep. 2017, 7, 3140. [Google Scholar] [CrossRef]
- Li Petri, G.; El Hassouni, B.; Sciarrillo, R.; Funel, N.; Mantini, G.; van der Laan, E.A.Z.; Cascioferro, S.; Avan, A.; Zucali, P.A.; Zaffaroni, N.; et al. Impact of hypoxia on chemoresistance of mesothelioma mediated by the proton-coupled folate transporter, and preclinical activity of new anti-LDH-A compounds. Br. J. Cancer 2020, 123, 644–656. [Google Scholar] [CrossRef]
- Endoh, D.; Ishii, K.; Kohno, K.; Virgona, N.; Miyakoshi, Y.; Yano, T.; Ishida, T. Chemoresistance related to hypoxia Adaptation in mesothelioma cells from tumor spheroids. Exp. Oncol. 2022, 44, 121–125. [Google Scholar] [CrossRef]
- Klabatsa, A.; Sheaff, M.T.; Steele, J.P.; Evans, M.T.; Rudd, R.M.; Fennell, D.A. Expression and prognostic significance of hypoxia-inducible factor 1alpha (HIF-1alpha) in malignant pleural mesothelioma (MPM). Lung Cancer 2006, 51, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Blau, H.M.; Brazelton, T.R.; Weimann, J.M. The evolving concept of a stem cell: Entity or function? Cell 2001, 105, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.M.; Zhang, M.; Wood, C.G.; Pisters, L.L. Stem Cell Theory of Cancer: Origin of Tumor Heterogeneity and Plasticity. Cancers 2021, 13, 4006. [Google Scholar] [CrossRef] [PubMed]
- Lytle, N.K.; Barber, A.G.; Reya, T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer 2018, 18, 669–680. [Google Scholar] [CrossRef]
- Zipori, D. The nature of stem cells: State rather than entity. Nat. Rev. Genet. 2004, 5, 873–878. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Scadden, D.T. The stem-cell niche as an entity of action. Nature 2006, 441, 1075–1079. [Google Scholar] [CrossRef]
- Crane, G.M.; Jeffery, E.; Morrison, S.J. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 2017, 17, 573–590. [Google Scholar] [CrossRef]
- Li, L.; Neaves, W.B. Normal stem cells and cancer stem cells: The niche matters. Cancer Res. 2006, 66, 4553–4557, Erratum in Cancer Res. 2006, 66, 6458. [Google Scholar] [CrossRef]
- Jones, D.L.; Wagers, A.J. No place like home: Anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 2008, 9, 11–21. [Google Scholar] [CrossRef]
- Calvi, L.M.; Link, D.C. The hematopoietic stem cell niche in homeostasis and disease. Blood 2015, 126, 2443–2451. [Google Scholar] [CrossRef]
- Yao, J.C.; Link, D.C. Concise Review: The Malignant Hematopoietic Stem Cell Niche. Stem Cells 2017, 35, 3–8. [Google Scholar] [CrossRef]
- Sánchez-Aguilera, A.; Méndez-Ferrer, S. The hematopoietic stem-cell niche in health and leukemia. Cell. Mol. Life Sci. 2017, 74, 579–590. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef]
- Bristow, R.G.; Hill, R.P. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer 2008, 8, 180–192. [Google Scholar] [CrossRef]
- Abbott, D.M.; Bortolotto, C.; Benvenuti, S.; Lancia, A.; Filippi, A.R.; Stella, G.M. Malignant Pleural Mesothelioma: Genetic and Microenviromental Heterogeneity as an Unexpected Reading Frame and Therapeutic Challenge. Cancers 2020, 12, 1186. [Google Scholar] [CrossRef]
- Wu, L.; Blum, W.; Zhu, C.Q.; Yun, Z.; Pecze, L.; Kohno, M.; Chan, M.L.; Zhao, Y.; Felley-Bosco, E.; Schwaller, B.; et al. Putative cancer stem cells may be the key target to inhibit cancer cell repopulation between the intervals of chemoradiation in murine mesothelioma. BMC Cancer 2018, 18, 471. [Google Scholar] [CrossRef]
- Li, Z.; Bao, S.; Wu, Q.; Wang, H.; Eyler, C.; Sathornsumetee, S.; Shi, Q.; Cao, Y.; Lathia, J.; McLendon, R.E.; et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 2009, 15, 501–513. [Google Scholar] [CrossRef]
- Civenni, G.; Malek, A.; Albino, D.; Garcia-Escudero, R.; Napoli, S.; Di Marco, S.; Pinton, S.; Sarti, M.; Carbone, G.M.; Catapano, C.V. RNAi-mediated silencing of Myc transcription inhibits stem-like cell maintenance and tumorigenicity in prostate cancer. Cancer Res. 2013, 73, 6816–6827. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Chu, G.J.; van Zandwijk, N.; Rasko, J.E.J. The Immune Microenvironment in Mesothelioma: Mechanisms of Resistance to Immunotherapy. Front. Oncol. 2019, 9, 1366. [Google Scholar] [CrossRef] [PubMed]
- Scherpereel, A.; Mazieres, J.; Greillier, L.; Lantuejoul, S.; Dô, P.; Bylicki, O.; Monnet, I.; Corre, R.; Audigier-Valette, C.; Locatelli-Sanchez, M.; et al. Nivolumab or nivolumab plus ipilimumab in patients with relapsed malignant pleural mesothelioma (IFCT-1501 MAPS2): A multicentre, open-label, randomised, non-comparative, phase 2 trial. Lancet Oncol. 2019, 20, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Metro, G.; Signorelli, D.; Pizzutilo, E.G.; Giannetta, L.; Cerea, G.; Garaffa, M.; Friedlaender, A.; Addeo, A.; Mandarano, M.; Bellezza, G.; et al. Immune checkpoint inhibitors for unresectable malignant pleural mesothelioma. Hum. Vaccines Immunother. 2021, 17, 2972–2980. [Google Scholar] [CrossRef] [PubMed]
- Assié, J.B.; Crépin, F.; Grolleau, E.; Canellas, A.; Geier, M.; Grébert-Manuardi, A.; Akkache, N.; Renault, A.; Hauss, P.A.; Sabatini, M.; et al. Immune-Checkpoint Inhibitors for Malignant Pleural Mesothelioma: A French, Multicenter, Retrospective Real-World Study. Cancers 2022, 14, 1498. [Google Scholar] [CrossRef]
- Marcq, E.; Waele, J.; Audenaerde, J.V.; Lion, E.; Santermans, E.; Hens, N.; Pauwels, P.; van Meerbeeck, J.P.; Smits, E.L.J. Abundant expression of TIM-3, LAG-3, PD-1 and PD-L1 as immunotherapy checkpoint targets in effusions of mesothelioma patients. Oncotarget 2017, 8, 89722–89735. [Google Scholar] [CrossRef]
- Cedrés, S.; Ponce-Aix, S.; Zugazagoitia, J.; Sansano, I.; Enguita, A.; Navarro-Mendivil, A.; Martinez-Marti, A.; Martinez, P.; Felip, E. Analysis of expression of programmed cell death 1 ligand 1 (PD-L1) in malignant pleural mesothelioma (MPM). PLoS ONE 2015, 10, e0121071. [Google Scholar] [CrossRef]
- Jin, L.; Gu, W.; Li, X.; Xie, L.; Wang, L.; Chen, Z. PD-L1 and prognosis in patients with malignant pleural mesothelioma: A meta-analysis and bioinformatics study. Ther. Adv. Med. Oncol. 2020, 12, 1758835920962362. [Google Scholar] [CrossRef]
- Bertino, P.; Premeaux, T.A.; Fujita, T.; Haun, B.K.; Marciel, M.P.; Hoffmann, F.W.; Garcia, A.; Yiang, H.; Pastorino, S.; Carbone, M.; et al. Targeting the C-terminus of galectin-9 induces mesothelioma apoptosis and M2 macrophage depletion. Oncoimmunology 2019, 8, 1601482. [Google Scholar] [CrossRef]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef]
- Das, M.; Zhu, C.; Kuchroo, V.K. Tim-3 and its role in regulating anti-tumor immunity. Immunol. Rev. 2017, 276, 97–111. [Google Scholar] [CrossRef]
- Amarnath, S.; Mangus, C.W.; Wang, J.C.; Wei, F.; He, A.; Kapoor, V.; Foley, J.E.; Massey, P.R.; Felizardo, T.C.; Riley, J.L.; et al. The PDL1-PD1 axis converts human TH1 cells into regulatory T cells. Sci. Transl. Med. 2011, 3, 111ra120. [Google Scholar] [CrossRef]
- Hsu, J.M.; Xia, W.; Hsu, Y.H.; Chan, L.C.; Yu, W.H.; Cha, J.H.; Chen, C.T.; Liao, H.W.; Kuo, C.W.; Khoo, K.H.; et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat. Comm. 2018, 9, 1908. [Google Scholar] [CrossRef]
- Darvin, P.; Sasidharan Nair, V.; Elkord, E. PD-L1 Expression in Human Breast Cancer Stem Cells Is Epigenetically Regulated through Posttranslational Histone Modifications. J. Oncol. 2019, 2019, 3958908. [Google Scholar] [CrossRef]
- Bronte, G.; Procopio, A.D.; Graciotti, L. The application of cancer stem cell model in malignant mesothelioma. Crit. Rev. Oncol. Hematol. 2022, 174, 103698. [Google Scholar] [CrossRef]
- Ghani, F.I.; Yamazaki, H.; Iwata, S.; Okamoto, T.; Aoe, K.; Okabe, K.; Mimura, Y.; Fujimoto, N.; Kishimoto, T.; Yamada, T.; et al. Identification of cancer stem cell markers in human malignant mesothelioma cells. Biochem. Biophys. Res. Comm. 2011, 404, 735–742. [Google Scholar] [CrossRef]
- Yamazaki, H.; Naito, M.; Ghani, F.I.; Dang, N.H.; Iwata, S.; Morimoto, C. Characterization of cancer stem cell properties of CD24 and CD26-positive human malignant mesothelioma cells. Biochem. Biophys. Res. Comm. 2012, 419, 529–536. [Google Scholar] [CrossRef]
- Cortes-Dericks, L.; Carboni, G.L.; Schmid, R.A.; Karoubi, G. Putative cancer stem cells in malignant pleural mesothelioma show resistance to cisplatin and pemetrexed. Int. J. Oncol. 2010, 37, 437–444. [Google Scholar] [CrossRef]
- Pasdar, E.A.; Smits, M.; Stapelberg, M.; Bajzikova, M.; Stantic, M.; Goodwin, J.; Yan, B.; Stursa, J.; Kovarova, J.; Sachaphibulkij, K.; et al. Characterisation of mesothelioma-initiating cells and their susceptibility to anti-cancer agents. PLoS ONE 2015, 10, e0119549. [Google Scholar] [CrossRef]
- Garber, K. Cancer stem cell pipeline flounders. Nat. Rev. Drug Discov. 2018, 17, 771–773. [Google Scholar] [CrossRef]
- Fan, H.; Guan, J.L. Compensatory function of Pyk2 protein in the promotion of focal adhesion kinase (FAK)-null mammary cancer stem cell tumorigenicity and metastatic activity. J. Biol. Chem. 2011, 286, 18573–18582. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.S.; Owen, K.A.; Conger, K.; Llewellyn, R.A.; Bouton, A.H.; Casanova, J.E. Non-redundant functions of FAK and Pyk2 in intestinal epithelial repair. Sci. Rep. 2019, 9, 4497. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.L. Integrin signaling through FAK in the regulation of mammary stem cells and breast cancer. IUBMB Life 2010, 2, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Fennell, D.A.; Baas, P.; Taylor, P.; Nowak, A.K.; Gilligan, D.; Nakano, T.; Pachter, J.A.; Weaver, D.T.; Scherpereel, A.; Pavlakis, N.; et al. Maintenance Defactinib Versus Placebo after First-Line Chemotherapy in Patients with Merlin-Stratified Pleural Mesothelioma: COMMAND-A Double-Blind, Randomized, Phase II Study. J. Clin. Oncol. 2019, 37, 790–798. [Google Scholar] [CrossRef]
- Loving, H.S.; Underbakke, E.S. Conformational Dynamics of FERM-Mediated Autoinhibition in Pyk2 Tyrosine Kinase. Biochemistry 2019, 58, 3767–3776. [Google Scholar] [CrossRef]
- Sekido, Y.; Pass, H.I.; Bader, S.; Mew, D.J.; Christman, M.F.; Gazdar, A.F.; Minna, J.D. Neurofibromatosis type 2 (NF2) gene is somatically mutated in mesothelioma but not in lung cancer. Cancer Res. 1995, 55, 1227–1231. [Google Scholar]
- Sato, T.; Sekido, Y. NF2/Merlin Inactivation and Potential Therapeutic Targets in Mesothelioma. Int. J. Mol. Sci. 2018, 19, 988. [Google Scholar] [CrossRef]
- Trofatter, J.A.; MacCollin, M.M.; Rutter, J.L.; Murrell, J.R.; Duyao, M.P.; Parry, D.M.; Eldridge, R.; Kley, N.; Menon, A.G.; Pulaski, K.; et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 1993, 72, 791–800. [Google Scholar] [CrossRef]
- Pignochino, Y.; Dell’Aglio, C.; Inghilleri, S.; Zorzetto, M.; Basiricò, M.; Capozzi, F.; Canta, M.; Piloni, D.; Cemmi, F.; Sangiolo, D.; et al. The combination of sorafenib and everolimus shows antitumor activity in preclinical models of malignant pleural mesothelioma. BMC Cancer 2015, 15, 374. [Google Scholar] [CrossRef]
- Sekido, Y. Inactivation of Merlin in malignant mesothelioma cells and the Hippo signaling cascade dysregulation. Pathol. Int. 2011, 61, 331–344. [Google Scholar] [CrossRef]
- Janse van Rensburg, H.J.; Yang, X. Essential signalling in NF2 loss-related tumours: The therapeutic potential of CRL4DCAF1 and mTOR combined inhibition. J. Thorac. Dis. 2017, 9, 3533–3536. [Google Scholar] [CrossRef]
- Larsson, J.; Ohishi, M.; Garrison, B.; Aspling, M.; Janzen, V.; Adams, G.B.; Curto, M.; McClatchey, A.I.; Schipani, E.; Scadden, D.T. Nf2/merlin regulates hematopoietic stem cell behavior by altering microenvironmental architecture. Cell Stem Cell 2008, 3, 221–227. [Google Scholar] [CrossRef]
- Blum, W.; Pecze, L.; Felley-Bosco, E.; Wu, L.; de Perrot, M.; Schwaller, B. (Stem Cell Factor-Based Identification and Functional Properties of In Vitro-Selected Subpopulations of Malignant Mesothelioma Cells. Stem Cell Rep. 2017, 8, 1005–1017. [Google Scholar] [CrossRef]
- Chasse, M.H.; Johnson, B.K.; Boguslawski, E.A.; Sorensen, K.M.; Rosien, J.E.; Kang, M.H.; Reynolds, C.P.; Heo, L.; Madaj, Z.B.; Beddows, I.; et al. Mithramycin induces promoter reprogramming and differentiation of rhabdoid tumor. EMBO Mol. Med. 2021, 13, e12640. [Google Scholar] [CrossRef]
- Rao, M.; Atay, S.M.; Shukla, V.; Hong, Y.; Upham, T.; Ripley, R.T.; Hong, J.A.; Zhang, M.; Reardon, E.; Fetsch, P.; et al. Mithramycin Depletes Specificity Protein 1 and Activates p53 to Mediate Senescence and Apoptosis of Malignant Pleural Mesothelioma Cells. Clin. Cancer Res. 2016, 22, 1197–1210. [Google Scholar] [CrossRef]
- Macharia, L.W.; Wanjiru, C.M.; Mureithi, M.W.; Pereira, C.M.; Ferrer, V.P.; Moura-Neto, V. MicroRNAs, Hypoxia and the Stem-Like State as Contributors to Cancer Aggressiveness. Front. Genet. 2019, 10, 125. [Google Scholar] [CrossRef]
- Kartha, R.V.; Subramanian, S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front. Genet. 2014, 5, 8. [Google Scholar] [CrossRef]
- Alexander, R.P.; Fang, G.; Rozowsky, J.; Snyder, M.; Gerstein, M.B. Annotating non-coding regions of the genome. Nat. Rev. Genet. 2010, 11, 559–571. [Google Scholar] [CrossRef]
- Moriondo, G.; Scioscia, G.; Soccio, P.; Tondo, P.; De Pace, C.C.; Sabato, R.; Foschino Barbaro, M.P.; Lacedonia, D. Effect of Hypoxia-Induced Micro-RNAs Expression on Oncogenesis. Int. J. Mol. Sci. 2022, 23, 6294. [Google Scholar] [CrossRef]
- Sawai, S.; Wong, P.F.; Ramasamy, T.S. Hypoxia-regulated microRNAs: The molecular drivers of tumor progression. Crit. Rev. Biochem. Mol. Biol. 2022, 57, 351–376. [Google Scholar] [CrossRef]
- Shen, G.; Li, X.; Jia, Y.F.; Piazza, G.A.; Xi, Y. Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol. Sin. 2013, 34, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Gee, H.E.; Ivan, C.; Calin, G.A.; Ivan, M. HypoxamiRs and cancer: From biology to targeted therapy. Antioxidants Redox Signal. 2014, 21, 1220–1238. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Lv, Q.; Ye, W.; Wong, C.K.; Cai, G.; Gu, D.; Ji, Y.; Zhao, C.; Wang, J.; Yang, B.B.; et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE 2006, 1, e116. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Johnson, T.G.; van Zandwijk, N. Manipulating microRNAs for the Treatment of Malignant Pleural Mesothelioma: Past, Present and Future. Front. Oncol. 2020, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- Fennell, D. miR-16: Expanding the range of molecular targets in mesothelioma. Lancet Oncol. 2017, 18, 1296–1297. [Google Scholar] [CrossRef]
- Viteri, S.; Rosell, R. An innovative mesothelioma treatment based on miR-16 mimic loaded EGFR targeted minicells (TargomiRs). Transl. Lung Cancer Res. 2018, 7 (Suppl. 1), S1–S4. [Google Scholar] [CrossRef]
- Caruso, S.; Bazan, V.; Rolfo, C.; Insalaco, L.; Fanale, D.; Bronte, G.; Corsini, L.R.; Rizzo, S.; Cicero, G.; Russo, A. MicroRNAs in colorectal cancer stem cells: New regulators of cancer stemness? Oncogenesis 2012, 1, e32. [Google Scholar] [CrossRef]
- Ali Hosseini Rad, S.M.; Bavarsad, M.S.; Arefian, E.; Jaseb, K.; Shahjahani, M.; Saki, N. The Role of microRNAs in Stemness of Cancer Stem Cells. Oncol. Rev. 2013, 7, e8. [Google Scholar] [CrossRef]
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415. [Google Scholar] [CrossRef]
- Flores-Huerta, N.; Silva-Cázares, M.B.; Arriaga-Pizano, L.A.; Prieto-Chávez, J.L.; López-Camarillo, C. LncRNAs and microRNAs as Essential Regulators of Stemness in Breast Cancer Stem Cells. Biomolecules 2021, 11, 380. [Google Scholar] [CrossRef]
- Bari, E.; Ferrarotti, I.; Torre, M.L.; Corsico, A.G.; Perteghella, S. Mesenchymal stem/stromal cell secretome for lung regeneration: The long way through “pharmaceuticalization” for the best formulation. J. Control. Release 2019, 309, 11–24. [Google Scholar] [CrossRef]
- Crivelli, B.; Chlapanidas, T.; Perteghella, S.; Lucarelli, E.; Pascucci, L.; Brini, A.T.; Ferrero, I.; Marazzi, M.; Pessina, A.; Torre, M.L.; et al. Mesenchymal stem/stromal cell extracellular vesicles: From active principle to next generation drug delivery system. J. Control Release 2017, 262, 104–117. [Google Scholar] [CrossRef]
- Ito, F.; Kato, K.; Yanatori, I.; Murohara, T.; Toyokuni, S. Ferroptosis-dependent extracellular vesicles from macrophage contribute to asbestos-induced mesothelial carcinogenesis through loading ferritin. Redox Biol. 2021, 47, 102174. [Google Scholar] [CrossRef]
- Raposo, G.; Stahl, P.D. Extracellular vesicles: A new communication paradigm? Nat. Rev. Mol. Cell Biol. 2019, 20, 509–510. [Google Scholar] [CrossRef]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef]
- Katsuda, T.; Kosaka, N.; Ochiya, T. The roles of extracellular vesicles in cancer biology: Toward the development of novel cancer biomarkers. Proteomics 2014, 14, 412–425. [Google Scholar] [CrossRef]
- Nogues, L.; Benito-Martin, A.; Hergueta-Redondo, M.; Peinado, H. The influence of tumour-derived extracellular vesicles on local and distal metastatic dissemination. Mol. Asp. Med. 2018, 60, 15–26. [Google Scholar] [CrossRef]
- Ahmadzada, T.; Kao, S.; Reid, G.; Clarke, S.; Grau, G.E.; Hosseini-Beheshti, E. Extracellular vesicles as biomarkers in malignant pleural mesothelioma: A review. Crit. Rev. Oncol. Hematol. 2020, 150, 102949. [Google Scholar] [CrossRef]
- Munson, P.; Lam, Y.W.; Dragon, J.; MacPherson, M.; Shukla, A. Exosomes from asbestos-exposed cells modulate gene expression in mesothelial cells. FASEB J. 2018, 32, 4328–4342. [Google Scholar] [CrossRef]
- Munson, P.; Lam, Y.W.; MacPherson, M.; Beuschel, S.; Shukla, A. Mouse serum exosomal proteomic signature in response to asbestos exposure. J. Cell. Biochem. 2018, 119, 6266–6273. [Google Scholar] [CrossRef]
- Ghio, A.J.; Stonehuerner, J.; Richards, J.; Devlin, R.B. Iron Homeostasis in the lung following asbestos exposure. Antioxidants Redox Signal. 2008, 10, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Pass, H.I.; Levin, S.M.; Harbut, M.R.; Melamed, J.; Chiriboga, L.; Donington, J.; Huflejt, M.; Carbone, M.; Chia, D.; Goodglick, L.; et al. Fibulin-3 as a Blood and Effusion Biomarker for Pleural Mesothelioma. N. Engl. J. Med. 2012, 367, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.H.; Cerione, R.A.; Antonyak, M.A. Extracellular Vesicles and Their Roles in Cancer Progression. Methods Mol. Biol. 2021, 2174, 143–170. [Google Scholar] [CrossRef] [PubMed]
- Thayanithy, V.; Babatunde, V.; Dickson, E.L.; Wong, P.; Oh, S.; Ke, X.; Barlas, A.; Fujisawa, S.; Romin, Y.; Moreira, A.L.; et al. Tumor exosomes induce tunneling nanotubes in lipid raft-enriched regions of human mesothelioma cells. Exp. Cell Res. 2014, 323, 178–188. [Google Scholar] [CrossRef]
- Greening, D.W.; Ji, H.; Chen, M.S.; Robinson, B.W.S.; Dick, I.M.; Creaney, J.; Simpson, R.J. Secreted primary human malignant mesothelioma exosome signature reflects oncogenic cargo. Sci. Rep. 2016, 6, 32643. [Google Scholar] [CrossRef]
- Creaney, J.; Dick, I.M.; Leon, J.S.; Robinson, B.W.S. A Proteomic Analysis of the Malignant Mesothelioma Secretome Using iTRAQ. Cancer Genom. Proteom. 2017, 14, 103–117. [Google Scholar] [CrossRef]
- Hegmans, J.; Bard, M.P.L.; Hemmes, A.; Luider, T.M.; Kleijmeer, M.J.; Prins, J.B.; Zitvogel, L.; Burgers, S.A.; Hoogsteden, H.C.; Lambrecht, B.N. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am. J. Pathol. 2004, 164, 1807–1815. [Google Scholar] [CrossRef]
- Bard, M.P.; Hegmans, J.P.; Hemmes, A.; Luider, T.M.; Willemsen, R.; Severijnen, L.A.A.; van Meerbeeck, J.P.; Burgers, S.A.; Hoogsteden, H.C.; Lambrecht, B.N. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol. 2004, 31, 114–121. [Google Scholar] [CrossRef]
- Monaco, F.; Gaetani, S.; Alessandrini, F.; Tagliabracci, A.; Bracci, M.; Valentino, M.; Neuzil, J.; Amati, M.; Bovenzi, M.; Tomasetti, M.; et al. Exosomal transfer of miR-126 promotes the anti-tumour response in malignant mesothelioma: Role of miR-126 in cancer-stroma communication. Cancer Lett. 2019, 463, 27–36. [Google Scholar] [CrossRef]
- Munson, P.B.; Hall, E.M.; Farina, N.H.; Pass, H.I.; Shukla, A. Exosomal miR-16-5p as a target for malignant mesothelioma. Sci. Rep. 2019, 9, 11688. [Google Scholar] [CrossRef]
- Clayton, A.; Tabi, Z. Exosomes and the MICA-NKG2D system in cancer. Blood Cells Mol. Dis. 2005, 34, 206–213. [Google Scholar] [CrossRef]
- Mahaweni, N.M.; Kaijen-Lambers, M.E.; Dekkers, J.; Aerts, J.G.; Hegmans, J.P. Tumour-derived exosomes as antigen delivery carriers in dendritic cell-based immunotherapy for malignant mesothelioma. J. Extracell. Vesicles 2013, 2, 22492. [Google Scholar] [CrossRef]
- Javadi, J.; Goergens, A.; Vanky, H.; Gupta, D.; Hjerpe, A.; El-Andaloussi, S.; Hagey, D.; Dobra, K. Diagnostic and Prognostic Utility of the Extracellular Vesicles Subpopulations Present in Pleural Effusion. Biomolecules 2021, 11, 1606. [Google Scholar] [CrossRef]
- Cavalleri, T.; Angelici, L.; Favero, C.; Dioni, L.; Mensi, C.; Bareggi, C.; Palleschi, A.; Rimessi, A.; Consonni, D.; Bordini, L.; et al. Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos-exposed subjects suggest a 2-miRNA signature as potential biomarker of disease. PLoS ONE 2017, 12, e0176680. [Google Scholar] [CrossRef]
- van der Koog, L.; Gandek, T.B.; Nagelkerke, A. Liposomes and Extracellular Vesicles as Drug Delivery Systems: A Comparison of Composition, Pharmacokinetics, and Functionalization. Adv. Heal. Mater. 2022, 11, e2100639. [Google Scholar] [CrossRef]
- Zhou, X.; Miao, Y.Q.; Wang, Y.; He, S.F.; Guo, L.M.; Mao, J.S.; Chen, M.S.; Yang, Y.T.; Zhang, X.X.; Gan, Y. Tumour-derived extracellular vesicle membrane hybrid lipid nanovesicles enhance siRNA delivery by tumour-homing and intracellular freeway transportation. J. Extracell. Vesicles 2022, 11, e12198. [Google Scholar] [CrossRef]
- Chernova, T.; Grosso, S.; Sun, X.M.; Tenor, A.R.; Cabeza, J.Z.; Craxton, A.; Self, E.L.; Nakas, A.; Cain, K.; MacFarlane, M.; et al. Extracellular Vesicles Isolated from Malignant Mesothelioma Cancer-Associated Fibroblasts Induce Pro-Oncogenic Changes in Healthy Mesothelial Cells. Int. J. Mol. Sci. 2022, 23, 12469. [Google Scholar] [CrossRef]
| Gene | Location | Pathway | Variant Types |
|---|---|---|---|
| BAP 1 | 3p21.1 | BRCA-associated protein-1 (ubiquitin carboxy-terminal hydrolase) (BAP1) is a gene that encodes a protein that has a high affinity for the BRCA1 protein and functions as a tumor suppressor protein | Missense mutations, nonsense mutations, silent mutations, frameshift insertions and deletions, and in-frame insertions and deletions. |
| NF2 | 22q12.2 | Neurofibromin 2 (NF2) is a gene that encodes a protein that functions by connecting cytoskeletal components with cell-surface proteins, cytoskeletal proteins, and ion transport proteins. | Fusions, missense mutations, nonsense mutations, silent mutations, frameshift deletions and insertions, and in-frame deletions and insertions. |
| CDKN2A | 9p21.3 | Cell cycle control. Cyclin-dependent kinase inhibitor 2A (CDKN2A) gene encodes several protein isoforms that function as inhibitors of CDK4 and ARF. | Missense mutations, nonsense mutations, silent mutations, in-frame deletions, frameshift deletions and insertions, and whole gene deletions. |
| CDKN2B | 9p21.3 | Cell cycle control. Cyclin-dependent kinase inhibitor 2B (CDKN2B, also known as p15) is a gene that encodes a protein that binds to CDK4 or CDK6 and inhibits their activation. | Missense and silent mutations. |
| TP53 | 17p13.1 | Cell cycle control.Tumor protein p53 (TP53) is a gene that codes for a tumor suppressor protein, cellular tumor antigen p53. The protein regulates expression of genes involved in cell cycle arrest, apoptosis, senescence, DNA repair, and changes in metabolism. | The most common alterations in TP53 are TP53 Mutation (32.56%), TP53 Missense (26.61%), TP53 c.217-c.1178 Missense (26.50%), TP53 Exon 5 Mutation (9.30%), and TP53 Exon 8 Mutation (8.49%) [3]. |
| SETD2 | 3p21.31 | Chromatin remodeling/DNA methylation. SET domain containing 2 (SETD2) is a gene that encodes a protein that is a member of a class of huntingtin interacting proteins. The protein functions as a histone methyltransferase specific for lysine-36 of histone H3. | Missense mutations, nonsense mutations, silent mutations, frameshift deletions and insertions, and in-frame deletions. |
| PBRM1 | 3p21.1 | Chromatin remodeling/DNA methylation. Polybromo 1 (PBRM1) is a gene that encodes a protein that is a member of a protein complex that functions in ligand-dependent transcriptional activation by nuclear hormone receptors. | Missense mutations, nonsense mutations, silent mutations, frameshift deletions and insertions, and in-frame deletions. |
| KMT2D | 12q13.12 | Chromatin remodeling/DNA methylation. Lysine (K)-specific methyltransferase 2D (KMT2D) is a gene that encodes a protein that functions as a histone methyltransferase that methylates the LYS-4 position of histone H3. | Missense mutations, nonsense mutations, silent mutations, frameshift deletions and insertions, and in-frame deletions. |
| FBXW7 | 4q31.3 | F-box/WD repeat-containing protein 7. F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase (FBXW7) is a gene that encodes a member of the F-box protein family. | Missense, nonsense, silent, and frameshift insertions and deletions. |
| ATM | 11q22.3 | DNA damage/repair. ATM serine/threonine kinase (ATM) is a gene that encodes a protein that is a member of the PI3/PI4-kinase family. The protein functions as a cell cycle checkpoint kinase and regulates multiple downstream effectors. | Missense mutations, nonsense mutations, silent mutations, whole gene deletions, frameshift deletions and insertions, and in-frame deletions and insertions. |
| LATS2 | 13q12.11 | Serine/threonine-protein kinase LATS2. | The most common alterations in LATS2 are LATS2 Amplification (0.33%), LATS2 Loss (0.20%), LATS2 P479_A480dup (0.02%), LATS2 R1054* (0.02%), and LATS2 A476T (0.02%). |
| CREBBP | 16p13.3 | CREB-binding protein. CREB-binding protein (CREBBP) is a gene that encodes a protein that functions in transcriptional activation and is involved in the regulation of embryonic development, growth control, and homeostasis. The protein also acetylates histone proteins. | Fusions, missense mutations, nonsense mutations, silent mutations, frameshift insertions and deletions, and in-frame. |
| ARID1B | 6q25.3 | AT-rich interactive domain-containing protein 1B. AT rich interactive domain 1B (SWI 1-like) (ARID1B) is a gene that encodes a protein that is a component of the SWI/SNF chromatin remodeling complex. The protein functions in cell-cycle activation. | Missense mutations, nonsense mutations, silent mutations, frameshift deletions and insertions, and in-frame deletions and insertions. |
| PTEN | 10q23.31 | PI3K/AKT1/MTOR. PTEN (phosphatase and tensin homolog) is a gene that encodes for phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN. This protein is a lipid/protein phosphatase that plays a role in multiple cell processes, including growth, proliferation, survival, and maintenance of genomic integrity. PTEN acts as a tumor suppressor by negatively regulating the PI3K/AKT signaling pathway. | Somatic mutations of PTEN occur in multiple malignancies. Germline mutations of PTEN lead to inherited hamartoma and Cowden syndrome. |
| TET2 | 4q24 | Chromatin remodeling/DNA methylation. Tet methylcytosine dioxygenase 2 (TET2; also known as ten-eleven translocation 2) is a gene that codes for methylcytosine dioxygenase TET2, a protein involved in epigenetic regulation of myelopoeisis. | TET2 is a tumor suppressor, and so in cancer, loss of TET2 function, which can occur via TET2 mutation, TET2 deletion, or IDH1 or IDH2 mutation, can cause myeloid or lymphoid transformations. Mutations in TET2 have been found in MDS, AML, ALL, and other hematologic malignancies. |
| DNMT3A | 2p23.3 | Chromatin remodeling/DNA methylation. DNMT3A (DNA (cytosine-5-)-methyltransferase 3 alpha) gene encodes the DNA (cytosine-5)-methyltransferase 3A protein, which is involved in epigenetic gene regulation. | DNMT3A is most frequently mutated in hematologic malignancies, but it has also been observed in other cancers, including lung cancer and MPM. The most common alterations in DNMT3A are DNMT3A Mutation (2.95%), DNMT3A R882H (0.41%), DNMT3A Nonsense (0.38%), DNMT3A R882C (0.21%), and DNMT3A Loss (0.08%). |
| KMT2A | 11q23.3 | MLL cleavage product C180. Lysine (K)-specific methyltransferase 2A (KMT2A; also known as MLL) is a gene that encodes a protein that functions as a transcriptional coactivator. The protein is involved in cellular processes including the regulation of gene expression and hematopoiesis. | Fusions, rearrangements, missense mutations, nonsense mutations, silent mutations, frameshift insertions and deletions, and in-frame deletions. |
| FAT1 | 4q35.2 | Protocadherin Fat 1, nuclear form. FAT atypical cadherin 1 (FAT1) is a gene that encodes a tumor suppressor protein that controls cell proliferation. | Missense mutations, synonymous mutations, nonsense mutations, frameshift deletions, and frameshift insertions. |
| MTAP | 9p21.3 | S-methyl-5’-thioadenosine phosphorylase. | The most common alterations in MTAP are MTAP Loss (4.92%), MTAP Amplification (1.28%), MTAP-RAF1 Fusion (0.04%), MTAP A191fs (0.05%), and MTAP A76V (0.04%). |
| EP300 | 22q13.2 | Histone acetyltransferase p300. E1A binding protein p300 (EP300) is a gene that encodes a protein that functions in transcriptional regulation by histone acetylation. | Fusions, missense mutations, nonsense mutations, silent mutations, frameshift insertions and deletions, and in-frame insertions and deletions. |
| PTCH1 | 9q22.32 | Hedgehog signaling. Patched 1 (PTCH1) is a gene that encodes a protein that belongs to the patched gene family. The protein functions as a receptor protein for sonic hedgehog, desert hedgehog, and indian hedgehog proteins. | Fusions, missense mutations, nonsense mutations, silent mutations, whole gene deletions, frameshift deletions and insertions, and in-frame deletions and insertions. |
| LATS1 | 6q25.1 | Serine/threonine-protein kinase LATS1. | The most common alterations in LATS1 are LATS1 Amplification (0.16%), LATS1 Loss (0.14%), LATS1 R995C (0.03%), LATS1 R670W (0.02%), and LATS1 R737* (0.03%). |
| RECQL4 | 8q24.3 | ATP-dependent DNA helicase Q4. RecQ helicase-like 4 (RECQL4) is a gene that encodes a DNA helicase that is predominantly expressed in thymus and testis. | Missense mutations, synonymous mutations, nonsense mutations, and frameshift deletions. |
| ROS1 | 6q22.1 | Kinase fusions, receptor tyrosine kinase/growth factor signaling. ROS1 (ROS proto-oncogene 1, receptor tyrosine kinase) is a gene that encodes the proto-oncogene tyrosine-protein kinase ROS protein, a receptor tyrosine kinase (RTK) of the insulin receptor family. OS1 fusions have been described in glioblastoma and colangiocarcinoma. ROS1 fusions containing an intact tyrosine kinase domain possess oncogenic activity. Signaling downstream of ROS1 fusions results in activation of cellular pathways known to be involved in cell growth and cell proliferation. | The most common alterations in ROS1 are ROS1 Mutation (3.56%), ROS1 Fusion (0.29%), ROS1 Amplification (0.17%), ROS1 Loss (0.12%), and ROS1-CD74 Fusion (0.05%). |
| WT1 | 11p13 | Beta-Catenin/WNT signaling. Wilms tumor 1 (WT1) is a gene that encodes a transcription factor that contains four zinc finger motifs and a proline/glutamine-rich DNA binding region at opposite termini. | Fusions, missense mutations, nonsense mutations, silent mutations and frameshift deletions. |
| BCOR | Xp11.4 | BCL-6 corepressor. BCL6 corepressor (BCOR) is a gene that encodes the BCL-6 corepressor protein. BCOR is a member of the ankyrin repeat domain containing gene family. The corepressor expressed by BCOR binds to BCL6, a DNA-binding protein that acts as a transcription repressor for genes involved in regulation of B cells, a type of immune cell. BCOR mutations have been observed in myelodysplastic syndromes, endometrial cancer, and other cancers. | The most common alterations in BCOR are BCOR Mutation (3.39%), BCOR Frameshift (0.63%), BCOR Nonsense (0.44%), BCOR N1459S (0.34%), and BCOR Loss (0.22%). |
| ARID2 | 12q12 | AT-rich interactive domain-containing protein 2. AT-rich interactive domain 2 (ARID; RFX-like) (ARID2) is a gene that encodes a protein that functions in a chromatin remodeling complex to promote gene transcription | Missense mutations, nonsense mutations, silent mutations, frameshift insertions and deletions, and in-frame insertions and deletions. |
| ASXL1 | 20q11.21 | Chromatin remodeling/DNA methylation. Additional sex combs like transcriptional regulator 1 (official symbol ASXL1) is a gene that encodes the putative Polycomb group protein ASXL1. Normal ASXL1 plays a role in embryonic development. | The most common alterations in ASXL1 are ASXL1 Mutation (2.62%), ASXL1 Nonsense (0.65%), ASXL1 Amplification (0.67%), ASXL1 R693* (0.10%), and ASXL1 Y591* (0.07%). |
| Location | Pathway | Variant Types | |
|---|---|---|---|
| WT-1 | 11p13 | Beta-Catenin/WNT signaling | Expression |
| PMS2 Deficient Expression | 7p22.1 | Deficient Expression | |
| MSLN Overexpression | 16p13.3 | Overexpression | |
| MSLN Expression | 16p13.3 | Expression | |
| MSH6 Deficient Expression | 2p16.3 | Chromatin remodeling/DNA methylation | Deficient Expression |
| MSH2 Deficient Expression | 2p21-p16.3 | Chromatin remodeling/DNA methylation | Deficient Expression |
| MLH1 Deficient Expression | 3p22.2 | DNA damage/repair | Deficient Expression |
| HLA-A*02:01 Positive | |||
| Deficient DNA Mismatch Repair (dMMR) | Predictive biomarker for use of nivolumab, pembrolizumab, dostarlimab, fluorouracil, and ipilimumab in patients | ||
| BRCA2 Mutation | 13q13.1 | DNA damage/repair | Missense mutations, nonsense mutations, silent mutations, whole gene deletions, frameshift deletions and insertions, and in-frame deletions |
| BRCA2 Loss | 13q13.1 | DNA damage/repair | Loss |
| BRCA1 | 17q21.31 | DNA damage/repair | Missense mutations, nonsense mutations, silent mutations, frameshift deletions and insertions, and in-frame deletions |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stella, G.M.; Marchiò, C.; Bari, E.; Ferrarotti, I.; Bertuccio, F.R.; Di Gennaro, A.; Abbott, D.M.; Putignano, P.; Campo, I.; Torre, M.L.; et al. The Genes–Stemness–Secretome Interplay in Malignant Pleural Mesothelioma: Molecular Dynamics and Clinical Hints. Int. J. Mol. Sci. 2023, 24, 3496. https://doi.org/10.3390/ijms24043496
Stella GM, Marchiò C, Bari E, Ferrarotti I, Bertuccio FR, Di Gennaro A, Abbott DM, Putignano P, Campo I, Torre ML, et al. The Genes–Stemness–Secretome Interplay in Malignant Pleural Mesothelioma: Molecular Dynamics and Clinical Hints. International Journal of Molecular Sciences. 2023; 24(4):3496. https://doi.org/10.3390/ijms24043496
Chicago/Turabian StyleStella, Giulia M., Caterina Marchiò, Elia Bari, Ilaria Ferrarotti, Francesco R. Bertuccio, Antonella Di Gennaro, David Michael Abbott, Paola Putignano, Ilaria Campo, Maria Luisa Torre, and et al. 2023. "The Genes–Stemness–Secretome Interplay in Malignant Pleural Mesothelioma: Molecular Dynamics and Clinical Hints" International Journal of Molecular Sciences 24, no. 4: 3496. https://doi.org/10.3390/ijms24043496
APA StyleStella, G. M., Marchiò, C., Bari, E., Ferrarotti, I., Bertuccio, F. R., Di Gennaro, A., Abbott, D. M., Putignano, P., Campo, I., Torre, M. L., & Corsico, A. G. (2023). The Genes–Stemness–Secretome Interplay in Malignant Pleural Mesothelioma: Molecular Dynamics and Clinical Hints. International Journal of Molecular Sciences, 24(4), 3496. https://doi.org/10.3390/ijms24043496










