Application of Machine Learning Models in Systemic Lupus Erythematosus
Abstract
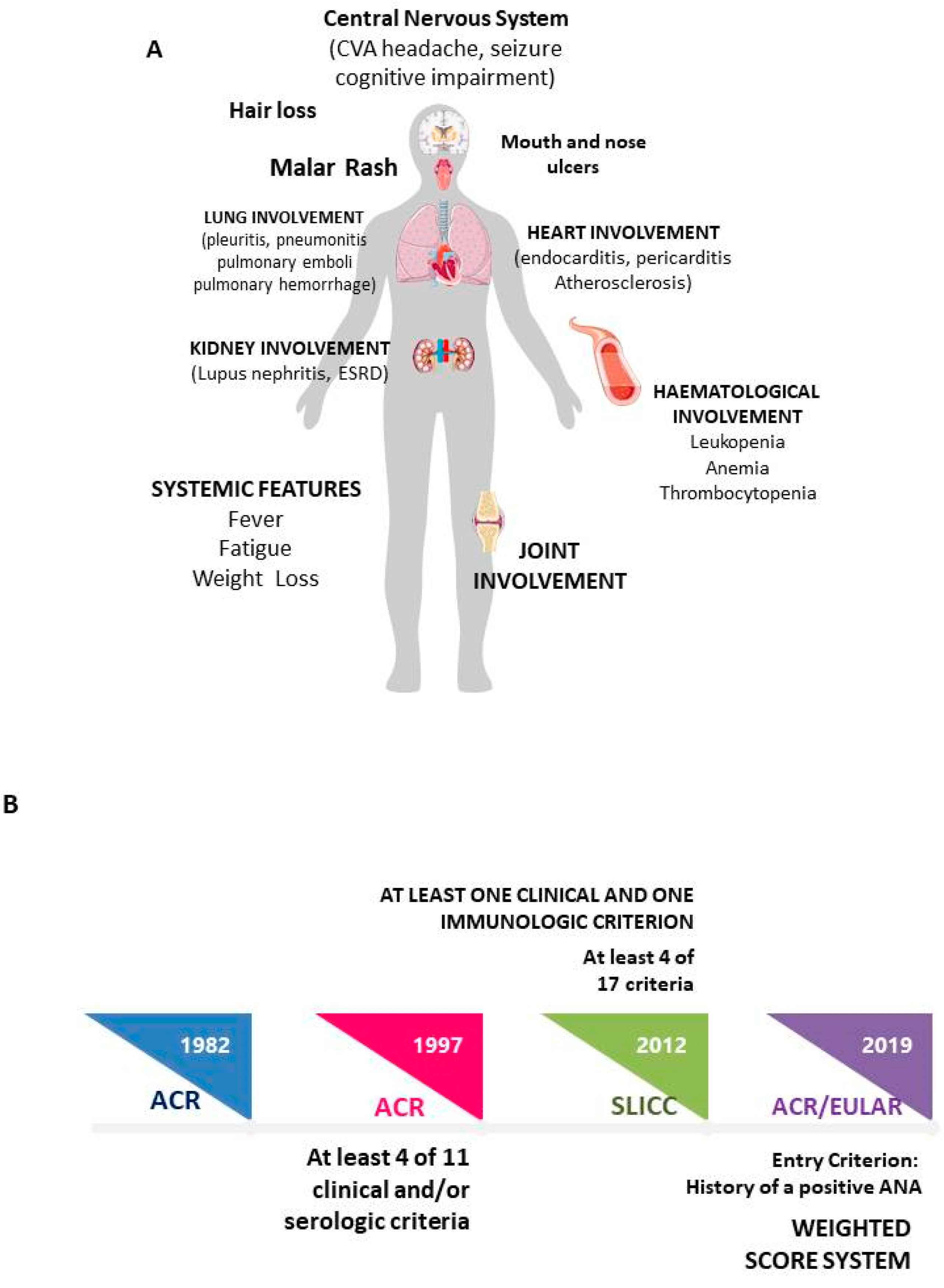
1. Introduction
Methods
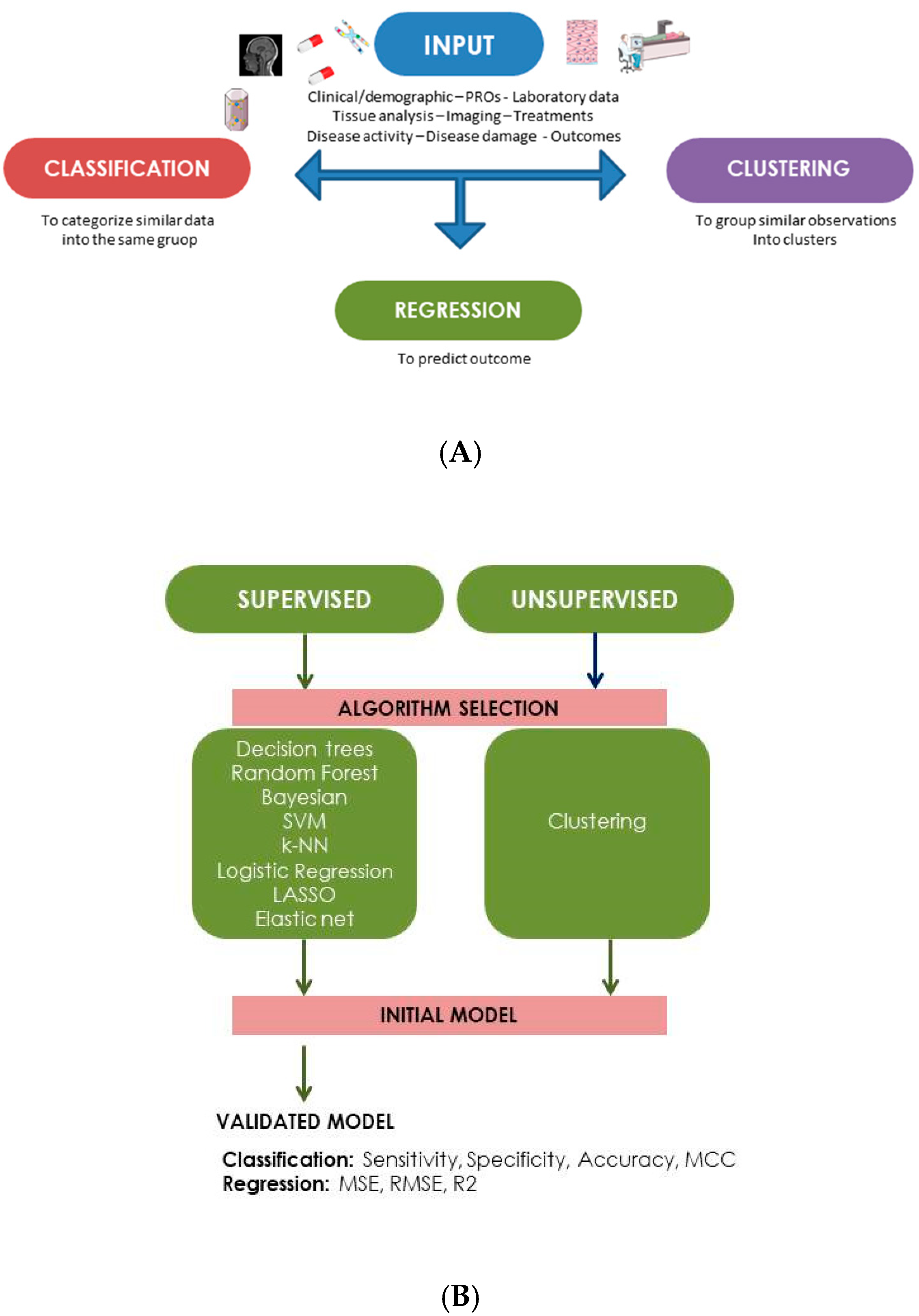
2. Machine Learning Models: General Concepts
3. Machine Learning Models in SLE Cohorts
3.1. Diagnosis
3.2. Disease Features
| Study | Disease Feature | MLM | Input Data | Results |
|---|---|---|---|---|
| Tang, 2011 [46] | Lupus Nephritis | Classification trees Logistic Regression Artificial Neural Network | Demographic, clinical, laboratory data; treatment, data about transplantation, comorbidity | Model to predict the probability of 3-year allograft survival after renal transplantation. LR, AUC = 0.74 Classification trees, AUC = 0.70 95% CI: 0.67–0.72) ANN, AUC = 0.71 |
| Chen, 2021 [47] | Lupus Nephritis | XGBoost SR-SPM | Clinical, laboratory and histological data | Development of a model to evaluate the risk of renal flare 5 years after remission. Good performance (XGBoost, C-index = 0.819) (SR-SPM, C-index = 0.746) |
| Wang, 2022 [48] | Lupus Nephritis | LASSO Support Vector Machine | LN gene expression datasets downloaded from the GEO database | Possible role as diagnostic biomarkers for C1QA (AUC = 0.741), C1QB (AUC = 0.758), MX1 (AUC = 0.865), RORC (AUC = 0.911), CD177 (AUC = 0.855), DEFA4 (AUC = 0.843), HERC5 (AUC = 0.880) |
| Stojanowski, 2022 [49] | Lupus Nephritis | Multi-layer perceptron | Demographic and laboratory features | Development of a predictive models for complete remission, (accuracy = 91.67%, AUC 0.9375) |
| Wang, 2022 [50] | Lupus Nephritis | HMFO Support Vector Machine | Demographic and laboratory features | Development of a model distinguishing between ISN/RPS pure class V and classes III ± V or IV ± V |
| Ayoub, 2022 [51] | Lupus Nephritis | Logistic Regression Random Forest Support Vector Machine Artificial Neural Network | Clinical data, urine biomarkers | Development of a model to predict 1-year treatment response (AUC = 0.7) |
| Yang, 2022 [52] | Lupus Nephritis | Mask R-CNN LSTM | Human kidney biopsy samples | Good accuracies (up to 0.940) on recognizing different glomerular diseases based on H&E whole slide images (AUC = 0.947) |
| Gu, 2021 [54] | NPSLE | LASSO Random Forest XGBoost | Clinical data, flow cytometry data on T-cell subsets, Self-Rating Anxiety/Depression Scale and Beck Depression Inventory | Identification of difference in T-cell subsets in SLE patients with or without anxiety Best performer XGBoost (AUC = 0.922) |
| Rumetshofer, 2022 [55] | NPSLE | Cluster analysis | White matter hyperintensities on MRI | Identification of five distinct clusters with predominant involvement of different areas. |
| Tan, 2022 [56] | NPSLE | Support Vector Machine Feature selection | Proton magnetic resonance spectroscopy | Diagnostic model with 94.9% accuracy, 91.3% sensitivity, 100% specificity and 0.87 cross-validation score |
| Barraclough, 2022 [57] | NPSLE | Network fusion | Cognitive assessment using the ACR Neuropsychological Battery (ACR-NB) | Identification of two subtypes with different performance in cognitive function (p < 0.03) |
| Ceccarelli, 2018 [59] | Joint involvement | Logistic Regression Forward Wrapper method Feature selection | Clinical and laboratory data, Ultrasound assessment | Good performance to identify patients with erosive arthritis (AUC = 0.806). |
| Ceccarelli, 2022 [60] | Joint involvement | Cluster analysis | Clinical and laboratory data, Ultrasound assessment | Identification of four clusters. Erosive arthritis was located in a cluster including renal and NPSLE. |
3.3. Disease Activity and Damage
3.4. Treatment
3.5. Pregnancy
3.6. Other Possible Application
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dörner, T.; Furie, R. Novel paradigms in systemic lupus erythematosus. Lancet 2019, 393, 2344–2358. [Google Scholar] [CrossRef]
- Kent, T.; Davidson, A.; Newman, D.; Buck, G.; D’Cruz, D. Burden of illness in systemic lupus erythematosus: Results from a UK patient and carer online survey. Lupus 2017, 26, 1095–1100. [Google Scholar] [CrossRef]
- Al Sawah, S.; Zhang, X.; Zhu, B.; Magder, L.S.; Foster, S.A.; Iikuni, N.; Petri, M. Effect of corticosteroid use by dose on the risk of developing organ damage over time in systemic lupus erythematosus-the Hopkins Lupus Cohort. Lupus Sci. Med. 2015, 2, e000066. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997, 40, 1725. [Google Scholar] [CrossRef]
- Petri, M.; Orbai, A.M.; Alarcón, G.S.; Gordon, C.; Merrill, J.T.; Fortin, P.R.; Bruce, I.N.; Isenberg, D.; Wallace, D.J.; Nived, O.; et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012, 64, 2677–2686. [Google Scholar] [CrossRef] [PubMed]
- Aringer, M.; Costenbader, K.; Daikh, D.; Brinks, R.; Mosca, M.; Ramsey-Goldman, R.; Smolen, J.S.; Wofsy, D.; Boumpas, D.T.; Kamen, D.L.; et al. 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Systemic Lupus Erythematosus. Arthritis Rheumatol. 2019, 71, 1400–1412. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, F.; Perricone, C.; Borgiani, P.; Ciccacci, C.; Rufini, S.; Cipriano, E.; Alessandri, C.; Spinelli, F.R.; Sili Scavalli, A.; Novelli, G.; et al. Genetic Factors in Systemic Lupus Erythematosus: Contribution to Disease Phenotype. J. Immunol. Res. 2015, 2015, 745647. [Google Scholar] [CrossRef]
- Arbuckle, M.R.; McClain, M.T.; Rubertone, M.V.; Scofield, R.H.; Dennis, G.J.; James, J.A.; Harley, J.B. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N. Eng. J. Med. 2003, 349, 1526–1533. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Natalucci, F.; Olivieri, G.; Pirone, C.; Picciariello, L.; Orefice, V.; Truglia, S.; Spinelli, F.R.; Alessandri, C.; Chistolini, A.; et al. Development of Systemic Autoimmune Diseases in Healthy Subjects Persistently Positive for Antiphospholipid Antibodies: Long-Term Follow-Up Study. Biomolecules 2022, 12, 1088. [Google Scholar] [CrossRef]
- Arnaud, L.; Tektonidou, M.G. Long-term outcomes in systemic lupus erythematosus: Trends over time and major contributors. Rheumatology 2020, 59, v29–v38. [Google Scholar] [CrossRef]
- Conti, F.; Ceccarelli, F.; Perricone, C.; Leccese, I.; Massaro, L.; Pacucci, V.A.; Truglia, S.; Miranda, F.; Spinelli, F.R.; Alessandri, C.; et al. The chronic damage in systemic lupus erythematosus is driven by flares, glucocorticoids and antiphospholipid antibodies: Results from a monocentric cohort. Lupus 2016, 25, 719–726. [Google Scholar] [CrossRef]
- Fanouriakis, A.; Tziolos, N.; Bertsias, G.; Boumpas, D.T. Update οn the diagnosis and management of systemic lupus erythematosus. Ann. Rheum. Dis. 2021, 80, 14–25. [Google Scholar] [CrossRef]
- Fanouriakis, A.; Kostopoulou, M.; Alunno, A.; Aringer, M.; Bajema, I.; Boletis, J.N.; Cervera, R.; Doria, A.; Gordon, C.; Govoni, M.; et al. 2019 update of the EULAR recommendations for the management of systemic lupus erythematosus. Ann. Rheum. Dis. 2019, 78, 736–745. [Google Scholar] [CrossRef]
- Nelson, A.E.; Arbeeva, L. Narrative Review of Machine Learning in Rheumatic and Musculoskeletal Diseases for Clinicians and Researchers: Biases, Goals, and Future Directions. J. Rheumatol. 2022, 49, 1191–1200. [Google Scholar] [CrossRef]
- Kingsmore, K.M.; Puglisi, C.E.; Grammar, A.C.; Lipsky, P.E. An introduction to machine learning and analysis of its use in rheumatic diseases. Nat. Rev. Rheumatol. 2021, 17, 710–730. [Google Scholar] [CrossRef]
- Kohavi, R.; Provost, F. Glossary of terms. Machine Learning—Special Issue on Applications of Machine Learning and the Knowledge Discovery Process. Mach. Learn. 1998, 30, 271–274. [Google Scholar]
- Zhu, X.; Goldberg, A.B. Introduction to Semi-Supervised Learning. Synth. Lect. Artif. Intell. Mach. Learn. 2009, 3, 1–130. [Google Scholar]
- Lever, J.; Krzywinski, M.; Altman, N. Principal component analysis. Nat. Methods 2017, 14, 641–642. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 27–46. [Google Scholar]
- Krogh, A. What are artificial neural networks? Nat. Biotechnol. 2008, 26, 195–197. [Google Scholar] [CrossRef]
- Cross, S.S.; Harrison, R.F.; Kennedy, R.L. Introduction to neural networks. Lancet 1995, 346, 1075–1079. [Google Scholar] [CrossRef]
- Kumar, R.; Indrayan, A. Receiver operating characteristic (ROC) curve for medical researchers. Indian Pediatr. 2011, 48, 277–287. [Google Scholar] [CrossRef]
- Hodson, T.O. Root-mean-square error (RMSE) or mean absolute error (MAE): When to use them or not. Geosci. Model Dev. 2022, 15, 5481–5487. [Google Scholar] [CrossRef]
- Bastanlar, Y.; Özuysal, M. Introduction to machine learning. Methods Mol. Biol. 2014, 1107, 105–128. [Google Scholar]
- Libbrecht, M.; Noble, W. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Li, Y.; Ma, C.; Liao, S.; Qi, S.; Meng, S.; Cai, W.; Dai, W.; Cao, R.; Dong, X.; Krämer, B.K.; et al. Combined proteomics and single cell RNA-sequencing analysis to identify biomarkers of disease diagnosis and disease exacerbation for systemic lupus erythematosus. Front Immunol. 2022, 13, 969509. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhang, W.; Hong, X.; Zeng, Z.; Chen, Y.; Liao, S.; Cai, W.; Xu, Y.; Wang, G.; Liu, D.; et al. Screening Biomarkers for Systemic Lupus Erythematosus Based on Machine Learning and Exploring Their Expression Correlations With the Ratios of Various Immune Cells. Front Immunol. 2022, 13, 873787. [Google Scholar] [CrossRef]
- Jiang, Z.; Shao, M.; Dai, X.; Pan, Z.; Liu, D. Identification of Diagnostic Biomarkers in Systemic Lupus Erythematosus Based on Bioinformatics Analysis and Machine Learning. Front Genet. 2022, 13, 865559. [Google Scholar] [CrossRef]
- Ma, W.; Lau, Y.L.; Yang, W.; Wang, Y.F. Random forests algorithm boosts genetic risk prediction of systemic lupus erythematosus. Front Genet. 2022, 13, 902793. [Google Scholar] [CrossRef]
- Martorell-Marugán, J.; Chierici, M.; Jurman, G.; Alarcón-Riquelme, M.E.; Carmona-Sáez, P. Differential diagnosis of systemic lupus erythematosus and Sjögren’s syndrome using machine learning and multi-omics data. Comput. Biol Med. 2022, 152, 106373. [Google Scholar] [CrossRef]
- Barnado, A.; Eudy, A.M.; Blaske, A.; Wheless, L.; Kirchoff, K.; Oates, J.C.; Clowse, M.E.B. Developing and Validating Methods to Assemble Systemic Lupus Erythematosus Births in the Electronic Health Record. Arthritis Care Res. 2022, 74, 849–857. [Google Scholar] [CrossRef]
- Matthiesen, R.; Lauber, C.; Sampaio, J.L.; Domingues, N.; Alves, L.; Gerl, M.J.; Almeida, M.S.; Rodrigues, G.; Araújo Gonçalves, P.; Ferreira, J.; et al. Shotgun mass spectrometry-based lipid profiling identifies and distinguishes between chronic inflammatory diseases. EBioMedicine 2021, 70, 103504. [Google Scholar] [CrossRef]
- Adamichou, C.; Genitsaridi, I.; Nikolopoulos, D.; Nikoloudaki, M.; Repa, A.; Bortoluzzi, A.; Fanouriakis, A.; Sidiropoulos, P.; Boumpas, D.T.; Bertsias, G.K. Lupus or not? SLE Risk Probability Index (SLERPI): A simple, clinician-friendly machine learning-based model to assist the diagnosis of systemic lupus erythematosus. Ann. Rheum. Dis. 2021, 80, 758–766. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Lapucci, M.; Olivieri, G.; Sortino, A.; Natalucci, F.; Spinelli, F.R.; Alessandri, C.; Sciandrone, M.; Conti, F. Can machine learning models support physicians in systemic lupus erythematosus diagnosis? Results from a monocentric cohort. Jt. Bone Spine 2022, 89, 105292. [Google Scholar] [CrossRef]
- Park, J.; Jang, W.; Park, H.S.; Park, K.H.; Kwok, S.K.; Park, S.H.; Oh, E.J. Cytokine clusters as potential diagnostic markers of disease activity and renal involvement in systemic lupus erythematosus. J. Int. Med. Res. 2020, 48, 300060520926882. [Google Scholar] [CrossRef]
- Guthridge, J.M.; Lu, R.; Tran, L.T.; Arriens, C.; Aberle, T.; Kamp, S.; Munroe, M.E.; Dominguez, N.; Gross, T.; DeJager, W.; et al. Adults with systemic lupus exhibit distinct molecular phenotypes in a cross-sectional study. EClinicalMedicine 2020, 20, 100291. [Google Scholar] [CrossRef]
- Jorge, A.; Castro, V.M.; Barnado, A.; Gainer, V.; Hong, C.; Cai, T.; Cai, T.; Carroll, R.; Denny, J.C.; Crofford, L.; et al. Identifying lupus patients in electronic health records: Development and validation of machine learning algorithms and application of rule-based algorithms. Semin. Arthritis Rheum. 2019, 49, 84–90. [Google Scholar] [CrossRef]
- Murray, S.G.; Avati, A.; Schmajuk, G.; Yazdany, J. Automated and flexible identification of complex disease: Building a model for systemic lupus erythematosus using noisy labeling. J. Am. Med. Inform. Assoc. 2019, 26, 61–65. [Google Scholar] [CrossRef]
- Turner, C.A.; Jacobs, A.D.; Marques, C.K.; Oates, J.C.; Kamen, D.L.; Anderson, P.E.; Obeid, J.S. Word2Vec inversion and traditional text classifiers for phenotyping lupus. BMC Med. Inform. Decis. Mak. 2017, 17, 126. [Google Scholar] [CrossRef]
- Dai, Y.; Hu, C.; Wang, L.; Huang, Y.; Zhang, L.; Xiao, X.; Tan, Y. Serum peptidome patterns of human systemic lupus erythematosus based on magnetic bead separation and MALDI-TOF mass spectrometry analysis. Scand. J. Rheumatol. 2010, 3, 240–246. [Google Scholar] [CrossRef]
- Huang, Z.; Shi, Y.; Cai, B.; Wang, L.; Wu, Y.; Ying, B.; Qin, L.; Hu, C.; Li, Y. MALDI-TOF MS combined with magnetic beads for detecting serum protein biomarkers and establishment of boosting decision tree model for diagnosis of systemic lupus erythematosus. Rheumatology 2009, 48, 626–631. [Google Scholar] [CrossRef]
- Diaz-Gallo, L.M.; Oke, V.; Lundström, E.; Elvin, K.; Ling Wu, Y.; Eketjäll, S.; Zickert, A.; Gustafsson, J.T.; Jönsen, A.; Leonard, D.; et al. Four Systemic Lupus Erythematosus Subgroups, Defined by Autoantibodies Status, Differ Regarding HLA-DRB1 Genotype Associations and Immunological and Clinical Manifestations. ACR Open Rheumatol. 2022, 4, 27–39. [Google Scholar] [CrossRef]
- Lu, Z.; Li, W.; Tang, Y.; Da, Z.; Li, X. Lymphocyte subset clustering analysis in treatment-naive patients with systemic lupus erythematosus. Clin. Rheumatol. 2021, 40, 1835–1842. [Google Scholar] [CrossRef]
- Reynolds, J.A.; McCarthy, E.M.; Haque, S.; Ngamjanyaporn, P.; Sergeant, J.C.; Lee, E.; Lee, E.; Kilfeather, S.A.; Parker, B.; Bruce, I.N. Cytokine profiling in active and quiescent SLE reveals distinct patient subpopulations. Arthritis Res. Ther. 2018, 20, 173. [Google Scholar] [CrossRef]
- Anders, H.J.; Saxena, R.; Zhao, M.H.; Parodis, I.; Salmon, J.E.; Mohan, C. Lupus nephritis. Nat. Rev. Dis. Primers. 2020, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Poynton, M.R.; Hurdle, J.F.; Baird, B.C.; Koford, J.K.; Goldfarb-Rumyantzev, A.S. Predicting three-year kidney graft survival in recipients with systemic lupus erythematosus. ASAIO J. 2011, 57, 300–309. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, S.; Chen, T.; Liang, D.; Yang, J.; Zeng, C.; Li, X.; Xie, G.; Liu, Z. Machine Learning for Prediction and Risk Stratification of Lupus Nephritis Renal Flare. Am. J. Nephrol. 2021, 52, 152–160. [Google Scholar] [CrossRef]
- Wang, L.; Yang, Z.; Yu, H.; Lin, W.; Wu, R.; Yang, H.; Yang, K. Predicting diagnostic gene expression profiles associated with immune infiltration in patients with lupus nephritis. Front. Immunol. 2022, 13, 839197. [Google Scholar] [CrossRef]
- Stojanowski, J.; Konieczny, A.; Rydzyńska, K.; Kasenberg, I.; Mikołajczak, A.; Gołębiowski, T.; Krajewska, M.; Kusztal, M. Artificial neural network—An effective tool for predicting the lupus nephritis outcome. BMC Nephrol. 2022, 23, 381. [Google Scholar] [CrossRef]
- Wang, M.; Liang, Y.; Hu, Z.; Chen, S.; Shi, B.; Heidari, A.A.; Zhang, Q.; Chen, H.; Chen, X. Lupus nephritis diagnosis using enhanced moth flame algorithm with support vector machines. Comput. Biol. Med. 2022, 145, 105435. [Google Scholar] [CrossRef]
- Ayoub, I.; Wolf, B.J.; Geng, L.; Song, H.; Khatiwada, A.; Tsao, B.P.; Oates, J.C.; Rovin, B.H. Prediction models of treatment response in lupus nephritis. Kidney Int. 2022, 101, 379–389. [Google Scholar] [CrossRef]
- Yang, C.K.; Lee, C.Y.; Wang, H.S.; Huang, S.C.; Liang, P.I.; Chen, J.S.; Kuo, C.F.; Tu, K.H.; Yeh, C.Y.; Chen, T.D. Glomerular disease classification and lesion identification by machine learning. Biomed. J. 2022, 4, 675–685. [Google Scholar] [CrossRef]
- Carrión-Barberà, I.; Salman-Monte, T.C.; Vílchez-Oya, F.; Monfort, J. Neuropsychiatric involvement in systemic lupus erythematosus: A review. Autoimmun Rev. 2021, 20, 102780. [Google Scholar] [CrossRef]
- Gu, X.X.; Jin, Y.; Fu, T.; Zhang, X.M.; Li, T.; Yang, Y.; Li, R.; Zhou, W.; Guo, J.X.; Zhao, R.; et al. Relevant Characteristics Analysis Using Natural Language Processing and Machine Learning Based on Phenotypes and T-Cell Subsets in Systemic Lupus Erythematosus Patients with Anxiety. Front. Psychiatry 2021, 12, 793505. [Google Scholar] [CrossRef] [PubMed]
- Rumetshofer, T.; Inglese, F.; de Bresser, J.; Mannfolk, P.; Strandberg, O.; Jönsen, A.; Bengtsson, A.; Nilsson, M.; Knutsson, L.; Lätt, J.; et al. Tract-based white matter hyperintensity patterns in patients with systemic lupus erythematosus using an unsupervised machine learning approach. Sci. Rep. 2022, 12, 21376. [Google Scholar] [CrossRef]
- Tan, G.; Huang, B.; Cui, Z.; Dou, H.; Zheng, S.; Zhou, T. A noise-immune reinforcement learning method for early diagnosis of neuropsychiatric systemic lupus erythematosus. Math. Biosci. Eng. 2022, 19, 2219–2239. [Google Scholar] [CrossRef]
- Barraclough, M.; Erdman, L.; Diaz-Martinez, J.P.; Knight, A.; Bingham, K.; Su, J.; Kakvan, M.; Muñoz Grajales, C.; Tartaglia, M.C.; Ruttan, L.; et al. Systemic lupus erythematosus phenotypes formed from machine learning with a specific focus on cognitive impairment. Rheumatology 2022, 17, keac653. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Perricone, C.; Cipriano, E.; Massaro, L.; Natalucci, F.; Capalbo, G.; Leccese, I.; Bogdanos, D.; Spinelli, F.R.; Alessandri, C.; et al. Joint involvement in systemic lupus erythematosus: From pathogenesis to clinical assessment. Semin. Arthritis Rheum. 2017, 47, 53–64. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Sciandrone, M.; Perricone, C.; Galvan, G.; Cipriano, E.; Galligari, A.; Levato, T.; Colasanti, T.; Massaro, L.; Natalucci, F.; et al. Biomarkers of erosive arthritis in systemic lupus erythematosus: Application of machine learning models. PLoS ONE 2018, 13, e0207926. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Natalucci, F.; Pirone, C.; Olivieri, G.; Colasanti, T.; Picciariello, L.; Spinelli, F.R.; Alessandri, C.; Conti, F. Erosive arthritis in systemic lupus erythematosus: Application of cluster analysis. Clin. Exp. Rheumatol. 2022, 40, 2175–2178. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Natalucci, F.; Olivieri, G.; Perricone, C.; Pirone, C.; Spinelli, F.R.; Alessandri, C.; Conti, F. Erosive arthritis in systemic lupus erythematosus: Not only Rhupus. Lupus 2021, 30, 2029–2041. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, Y.; Zhou, Y.; Tang, X.; Tang, L.; Wang, J. Identification of crucial genes for predicting the risk of atherosclerosis with system lupus erythematosus based on comprehensive bioinformatics analysis and machine learning. Comput. Biol. Med. 2023, 152, 106388. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, Z.; Xiao, Y.; Wan, W.; Yang, X. The shared biomarkers and pathways of systemic lupus erythematosus and metabolic syndrome analyzed by bioinformatics combining machine learning algorithm and single-cell sequencing analysis. Front. Immunol. 2022, 13, 1015882. [Google Scholar] [CrossRef] [PubMed]
- Isenberg, D.A.; Merrill, J.T. Why, why, why de-lupus (does so badly in clinical trials). Expert Rev. Clin. Immunol. 2016, 12, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Toro-Domínguez, D.; Martorell-Marugán, J.; Goldman, D.; Petri, M.; Carmona-Sáez, P.; Alarcón-Riquelme, M.E. Stratification of Systemic Lupus Erythematosus Patients Into Three Groups of Disease Activity Progression According to Longitudinal Gene Expression. Arthritis Rheumatol. 2018, 70, 2025–2035. [Google Scholar] [CrossRef]
- Alves, P.; Bandaria, J.; Leavy, M.B.; Gliklich, B.; Boussios, C.; Su, Z.; Curhan, G. Validation of a machine learning approach to estimate Systemic Lupus Erythematosus Disease Activity Index score categories and application in a real-world dataset. RMD Open 2021, 7, e001586. [Google Scholar] [CrossRef] [PubMed]
- Kegerreis, B.; Catalina, M.D.; Bachali, P.; Geraci, N.S.; Labonte, A.C.; Zeng, C.; Stearrett, N.; Crandall, K.A.; Lipsky, P.E.; Grammar, A.C. Machine learning approaches to predict lupus disease activity from gene expression data. Sci. Rep. 2019, 9, 9617. [Google Scholar] [CrossRef]
- Yones, S.A.; Annett, A.; Stoll, P.; Diamanti, K.; Holmfeldt, L.; Barrenäs, C.F.; Meadows, J.R.S.; Komorowski, J. Interpretable machine learning identifies paediatric Systemic Lupus Erythematosus subtypes based on gene expression data. Sci. Rep. 2022, 1, 7433. [Google Scholar] [CrossRef]
- Zheng, J.; Zhu, L.; Ju, B.; Zhang, J.; Luo, J.; Wang, Y.; Lv, X.; Pu, D.; He, L.; Wang, J. Peripheral immunophenotypes associated with the flare in the systemic lupus erythematosus patients with low disease activity state. Clin. Immunol. 2022, 245, 109166. [Google Scholar] [CrossRef]
- Gladman, D.D.; Urowitz, M.B.; Goldsmith, C.H.; Fortin, P.; Ginzler, E.; Gordon, C.; Hanly, J.G.; Isenberg, D.A.; Kalunian, K.; Nived, O.; et al. The reliability of the Systemic Lupus International Collaborating Clinics/American College of Rheumatology Damage Index in patients with systemic lupus erythematosus. Arthritis Rheum. 1997, 40, 809–813. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Sciandrone, M.; Perricone, C.; Galvan, G.; Morelli, F.; Vicente, L.N.; Leccese, I.; Massaro, L.; Cipriano, E.; Spinelli, F.R.; et al. Prediction of chronic damage in systemic lupus erythematosus by using machine-learning models. PLoS ONE 2017, 3, e0174200. [Google Scholar] [CrossRef]
- Ahn, G.Y.; Lee, J.; Won, S.; Ha, E.; Kim, H.; Nam, B.; Kim, J.S.; Kang, J.; Kim, J.H.; Song, G.G.; et al. Identifying damage clusters in patients with systemic lupus erythematosus. Int. J. Rheum. Dis. 2020, 23, 84–91. [Google Scholar] [CrossRef]
- Pego-Reigosa, J.M.; Lois-Iglesias, A.; Rúa-Figueroa, Í.; Galindo, M.; Calvo-Alén, J.; de Uña-Álvarez, J.; Balboa-Barreiro, V.; Ibáñez Ruan, J.; Olivé, A.; Rodríguez-Gómez, M.; et al. Relationship between damage clustering and mortality in systemic lupus erythematosus in early and late stages of the disease: Cluster analyses in a large cohort from the Spanish Society of Rheumatology Lupus Registry. Rheumatology 2016, 55, 1243–1250. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Olivieri, G.; Sortino, A.; Dominici, L.; Arefayne, F.; Celia, A.I.; Cipriano, E.; Garufi, C.; Lapucci, M.; Mancuso, S.; et al. Comprehensive disease control in systemic lupus erythematosus. Semin. Arthritis Rheum. 2021, 51, 404–408. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Olivieri, G.; Pirone, C.; Ciccacci, C.; Picciariello, L.; Natalucci, F.; Perricone, C.; Spinelli, F.R.; Alessandri, C.; Borgiani, P.; et al. The Impacts of the Clinical and Genetic Factors on Chronic Damage in Caucasian Systemic Lupus Erythematosus Patients. J. Clin. Med. 2022, 11, 3368. [Google Scholar] [CrossRef]
- Lever, E.; Alves, M.R.; Isenberg, D.A. Towards Precision Medicine in Systemic Lupus Erythematosus. Pharmgenomics Pers. Med. 2020, 13, 39–49. [Google Scholar] [CrossRef]
- Kan, H.; Nagar, S.; Patel, J.; Wallace, D.J.; Molta, C.; Chang, D.J. Longitudinal Treatment Patterns and Associated Outcomes in Patients with Newly Diagnosed Systemic Lupus Erythematosus. Clin. Ther. 2016, 38, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.M.; Alase, A.; Wigston, Z.; Psarras, A.; Burska, A.; Sutton, E.; Md Yusof, M.Y.; Reynolds, J.A.; Masterplans Consortium; McHugh, N.; et al. Gene expression and autoantibody analysis reveals distinct ancestry-specific profiles associated with response to rituximab in refractory systemic lupus erythematosus. Arthritis Rheumatol. 2022; Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Li, Y.F.; Zhang, C.; He, S.M.; Chen, X. Predicting the effect of sirolimus on disease activity in patients with systemic lupus erythematosus using machine learning. J. Clin. Pharm. Ther. 2022, 47, 1845–1850. [Google Scholar] [CrossRef] [PubMed]
- Toro-Domínguez, D.; Martorell-Marugán, J.; Martinez-Bueno, M.; López-Domínguez, R.; Carnero-Montoro, E.; Barturen, G.; Goldman, D.; Petri, M.; Carmona-Sáez, P.; Alarcón-Riquelme, M.E. Scoring personalized molecular portraits identify Systemic Lupus Erythematosus subtypes and predict individualized drug responses, symptomatology and disease progression. Brief Bioinform. 2022, 23, bbac332. [Google Scholar] [CrossRef] [PubMed]
- Kwok, L.-W.; Tam, L.-S.; Zhu, T.; Leung, Y.-Y.; Li, E. Predictors of maternal and fetal outcomes in pregnancies of patients with systemic lupus erythematosus. Lupus 2011, 20, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, L.; Bertsias, G.K.; Agmon-Levin, N.; Brown, S.; Cervera, R.; Costedoat-Chalumeau, N.; Doria, A.; Fischer-Betz, R.; Forger, F.; Moraes-Fontes, M.F.; et al. EULAR recommendations for women’s health and the management of family planning, assisted reproduction, pregnancy and menopause in patients with systemic lupus erythematosus and/or antiphospholipid syndrome. Ann. Rheum. Dis. 2017, 76, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhou, Y.; Shi, J.; Yang, J.; Huang, H.; Zhang, M.; Wang, S.; Ma, Q.; Liu, Y.; Li, B.; et al. Potential genetic biomarkers predict adverse pregnancy outcome during early and mid-pregnancy in women with systemic lupus erythematosus. Front. Endocrinol 2022, 13, 957010. [Google Scholar] [CrossRef] [PubMed]
- Fazzari, M.J.; Guerra, M.M.; Salmon, J.; Kim, M.Y. Adverse pregnancy outcomes in women with systemic lupus erythematosus: Can we improve predictions with machine learning? Lupus Sci. Med. 2022, 1, e000769. [Google Scholar] [CrossRef]
- Jorge, A.M.; Smith, D.; Wu, Z.; Chowdhury, T.; Costenbader, K.; Zhang, Y.; Choi, H.K.; Feldman, C.H.; Zhao, Y. Exploration of machine learning methods to predict systemic lupus erythematosus hospitalizations. Lupus 2022, 31, 1296–1305. [Google Scholar] [CrossRef] [PubMed]
- Margiotta, D.P.E.; Fasano, S.; Basta, F.; Pierro, L.; Riccardi, A.; Navarini, L.; Valentini, G.; Afeltra, A. Clinical features of patients with systemic lupus erythematosus according to health-related quality of life, entity of pain, fatigue and depression: A cluster analysis. Clin. Exp. Rheumatol. 2019, 37, 535–539. [Google Scholar] [PubMed]
- Margiotta, D.P.E.; Laudisio, A.; Navarini, L.; Basta, F.; Mazzuca, C.; Angeletti, S.; Ciccozzi, M.; Incalzi, R.A.; Afeltra, A. Pattern of sleep dysfunction in systemic lupus erythematosus: A cluster analysis. Clin. Rheumatol. 2019, 38, 1561–1570. [Google Scholar] [CrossRef]
| Study | MLM | Input Data | Results |
|---|---|---|---|
| Li, 2022 [26] | Random Forest | PBMC proteomics | Six-protein combination (IFIT3, MX1, TOMM40, STAT1, STAT2, and OAS3) exhibited good performance for SLE disease diagnosis (AUC= 0.723 versus HC; AUC= 0.815 versus RA). Nine-protein combination (PHACTR2, GOT2, L-selectin, CMC4, MAP2K1, CMPK2, ECPAS, SRA1, and STAT2) showed a robust performance in assessing disease exacerbation (AUC = 0.990) |
| Zhong, 2022 [27] | LASSO Support Vector Machines | Differentially expressed genes (DEGs) | Selection of six candidate diagnostic biomarkers for SLE (ABCB1, EIF2AK2, HERC6, ID3, IFI27, and PLSCR1), with AUC from 0.96 to 0.913 |
| Jiang, 2022 [28] | Logistic regression Random Forest XGBoost Support Vector Machines Artificial Neural Network | Genetic biomarkers from GSE65391 and GSE72509 datasets | IFI44 was determined to be the optimal diagnostic biomarker of SLE |
| Ma, 2022 [29] | Random Forest Support Vector Machines Artificial Neural Network | Genome-wide association studies | RF model AUC = 0.84 At the optimal cut-off, the RF predictor reached a sensitivity of 84% with a specificity of 68% in SLE classification. |
| Martorell-Marugán, 2022 [30] | XGBoost model | Biomarkers for gene expression and DNA methylation | The model is able to discriminate SLE from Sjogren Syndrome (gene expression MCC = 0.5791 ± 0.0409; methylation data MCC = 0.5546 ± 0.0484) |
| Barnado, 2022 [31] | Random Forest XGBoost model | Electonic Health Data | PPV 74–77% |
| Matthiesen, 2021 [32] | Partial Least Square | Plasma lipidomes | SLE vs CVD (Sensitivity = 0.91, Specificity = 1) IS vs SLE (Sensitivity = 1, Specificity = 0.82) |
| Adamichou, 2020 [33] | LASSO Logistic Regression | Clinical/laboratory features according classification criteria EULAR/ACR | Accuracy = 94.8% for identifying SLE High sensitivity for early disease (93.8%), LN (97.9%), NPSLE (91.8%), SLE requiring immunosuppressives/biologics (96.4%). Development of a scoring system (>7, 94.2%) accuracy |
| Ceccarelli, 2021 [34] | ReliefF algorithm, Logistic Regression Support Vector Machines DT models | Clinical/laboratory features according classification criteria EULAR/ACR | At the ReliefF model, anti-dsDNA positivity, low C3/C4 serum levels and malar/maculopapular rash resulted the strongest predictor features. A good model’s performance was obtained already when only the three highest scoring features were considered (AUC = 0.94) |
| Park, 2020 [35] | Cluster Analysis | Serum cyotkines | Cluster analysis revealed two distinct patient groups characterized by high levels of IL8, MIP1α and MIP1β (group 1) or of IL2, IL6, IL10, IL12, IFNγ and TNF (group 2). Active disease was more common in group 1 (55.7%) than in group 2 (34.8%). More patients in group 2 had renal involvement (42/115, 36.5%) than in group 1 (22/88, 25%). |
| Guthridge, 2020 [36] | K-means clustering Random Forest | Data from plasma, serum, RNA, clinical, laboratory features | Identification of 7 SLE clusters. Inflammation and interferon modules were elevated in Clusters 1 (moderately) and 4 (strongly), with decreased T-cell modules in Cluster 4. Active clinical features were similar across clusters. Clinical SLEDAI trended highest in Clusters 3 and 4, though Cluster 3 lacked strong interferon and inflammation signatures. Renal activity was more frequent in Cluster 4, and rare in Clusters 2, 5, and 7. Serology findings were lowest in Clusters 2 and 5. |
| Jorge, 2019 [37] | Logistic Regression | Registration data according classification criteria ACR/SLICC | PPV 90% for definite SLE PPV 92% for definite/probable SLE |
| Murray, 2019 [38] | Logistic Regression | Electronic health record | AUC=0.97 to automate identification of SLE patients |
| Turner, 2017 [39] | Artificial Neural Network Random Forest Naïve Bayes model Support Vector Machines Word2Vec | Electronic health record | ICD-9 accuracy 90.00% (AUC = 0.9) Shallow neural network with CUIs accuracy 92.10% (AUC = 0.970) Random forest with BOWs accuracy 95.25% (AUC = 0.994) Random forest with CUIs accuracy 95.00% (AUC = 0.979) Word2Vec inversion accuracy 90.03% (AUC = 0.905) |
| Dai, 2010 [40] | k-nearest neighbors | Serum peptidome patterns | Blinded verification of the classification model showed 91.7% sensitivity in active SLE, 83.3% sensitivity in stable SLE, and 86.7% specificity in normal controls. |
| Huang, 2009 [41] | Decision Trees | Serum proteomic | A panel of four potential protein biomarkers could accurately recognize 25 of 32 patients with SLE, 36 of 42 patients with other autoimmune diseases and 36 of 40 healthy people. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceccarelli, F.; Natalucci, F.; Picciariello, L.; Ciancarella, C.; Dolcini, G.; Gattamelata, A.; Alessandri, C.; Conti, F. Application of Machine Learning Models in Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2023, 24, 4514. https://doi.org/10.3390/ijms24054514
Ceccarelli F, Natalucci F, Picciariello L, Ciancarella C, Dolcini G, Gattamelata A, Alessandri C, Conti F. Application of Machine Learning Models in Systemic Lupus Erythematosus. International Journal of Molecular Sciences. 2023; 24(5):4514. https://doi.org/10.3390/ijms24054514
Chicago/Turabian StyleCeccarelli, Fulvia, Francesco Natalucci, Licia Picciariello, Claudia Ciancarella, Giulio Dolcini, Angelica Gattamelata, Cristiano Alessandri, and Fabrizio Conti. 2023. "Application of Machine Learning Models in Systemic Lupus Erythematosus" International Journal of Molecular Sciences 24, no. 5: 4514. https://doi.org/10.3390/ijms24054514
APA StyleCeccarelli, F., Natalucci, F., Picciariello, L., Ciancarella, C., Dolcini, G., Gattamelata, A., Alessandri, C., & Conti, F. (2023). Application of Machine Learning Models in Systemic Lupus Erythematosus. International Journal of Molecular Sciences, 24(5), 4514. https://doi.org/10.3390/ijms24054514






