BRCA Mutations—The Achilles Heel of Breast, Ovarian and Other Epithelial Cancers
Abstract
1. Introduction
2. Epidemiology of Cancer and BRCA1,2 Mutations
2.1. Pan-Cancer Overview
2.2. Ovarian Cancer
2.3. Breast Cancer
2.4. Pancreatic Cancer
2.5. Prostate Cancer
2.6. Mutations and the Founder Effect
3. Molecular Evolution of BRCA and Links to Human Cancers
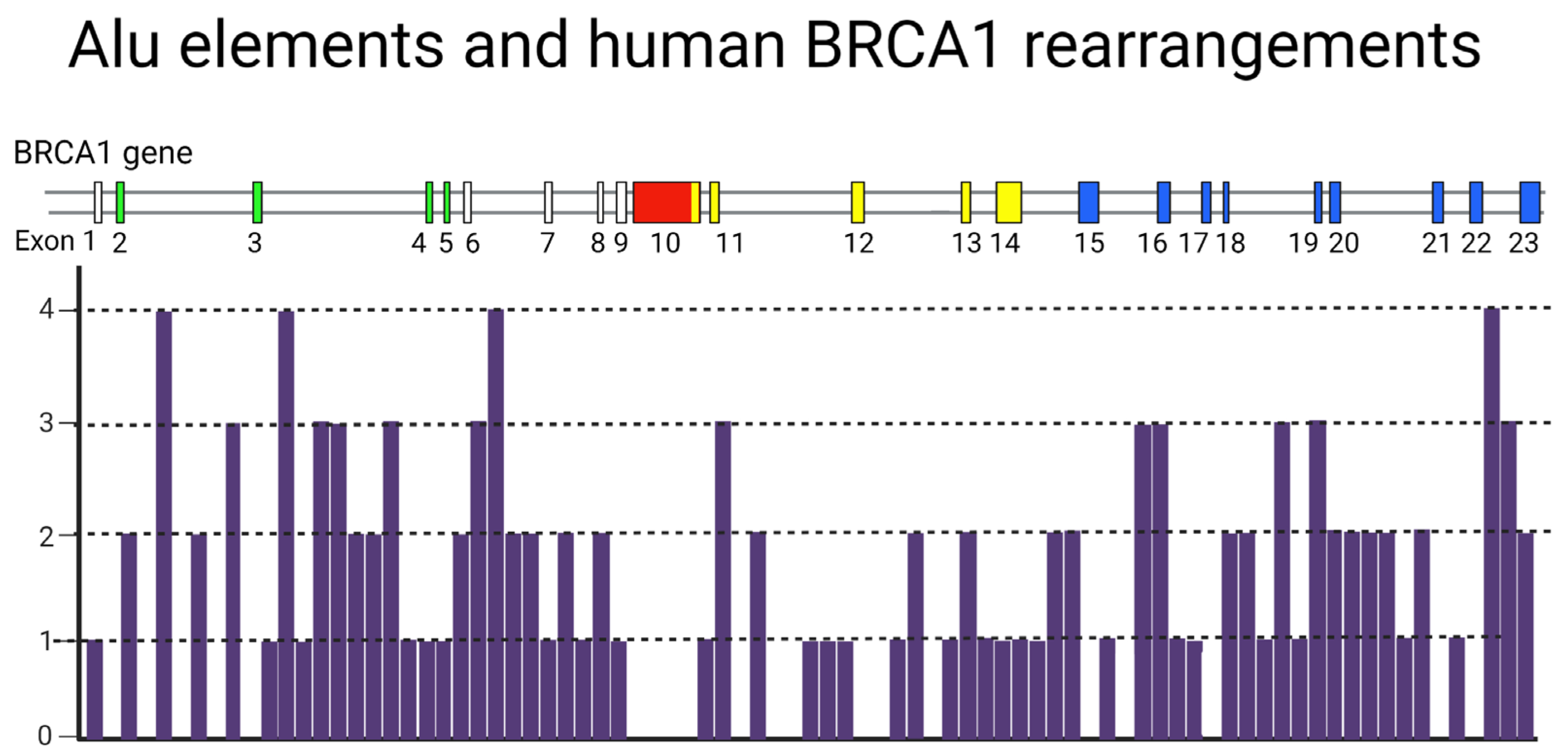
4. A Potential Mechanism for Enrichment of Mutations in the BRCA Genes
A Hypothetical Role of Transposable Elements (TEs) in BRCA-Associated Carcinogenesis
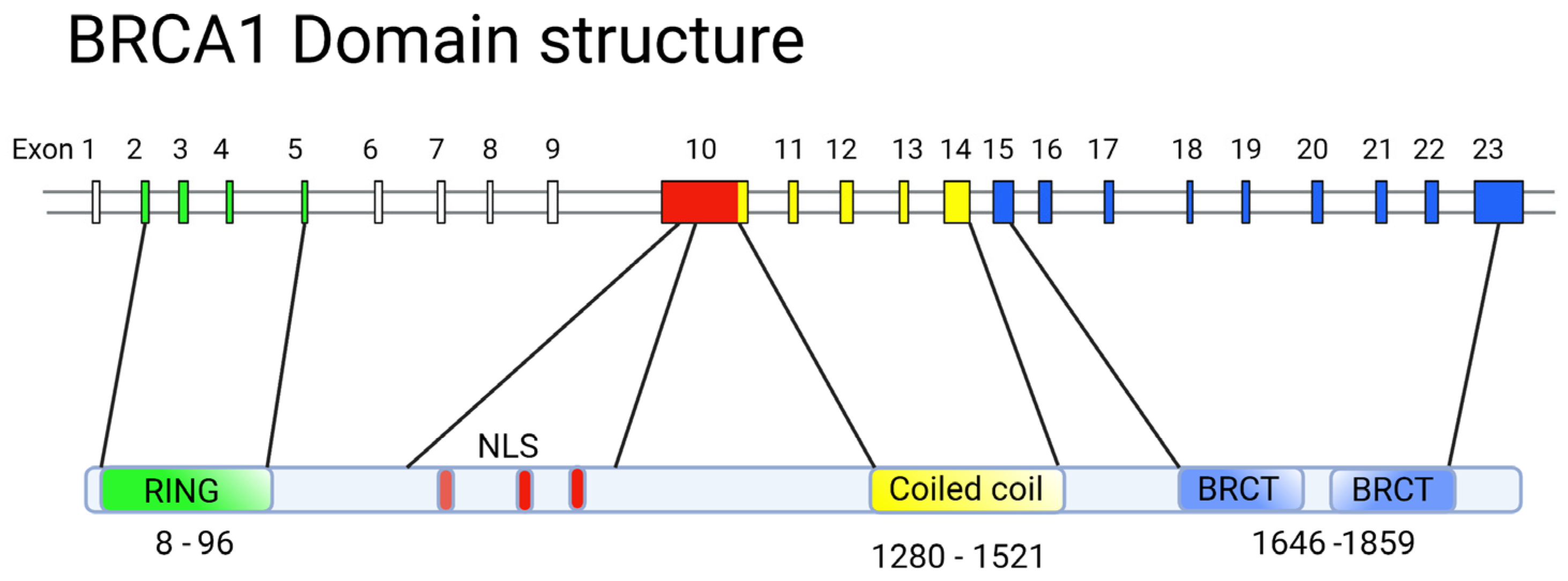
5. Structure-Function Analysis of Human BRCA1
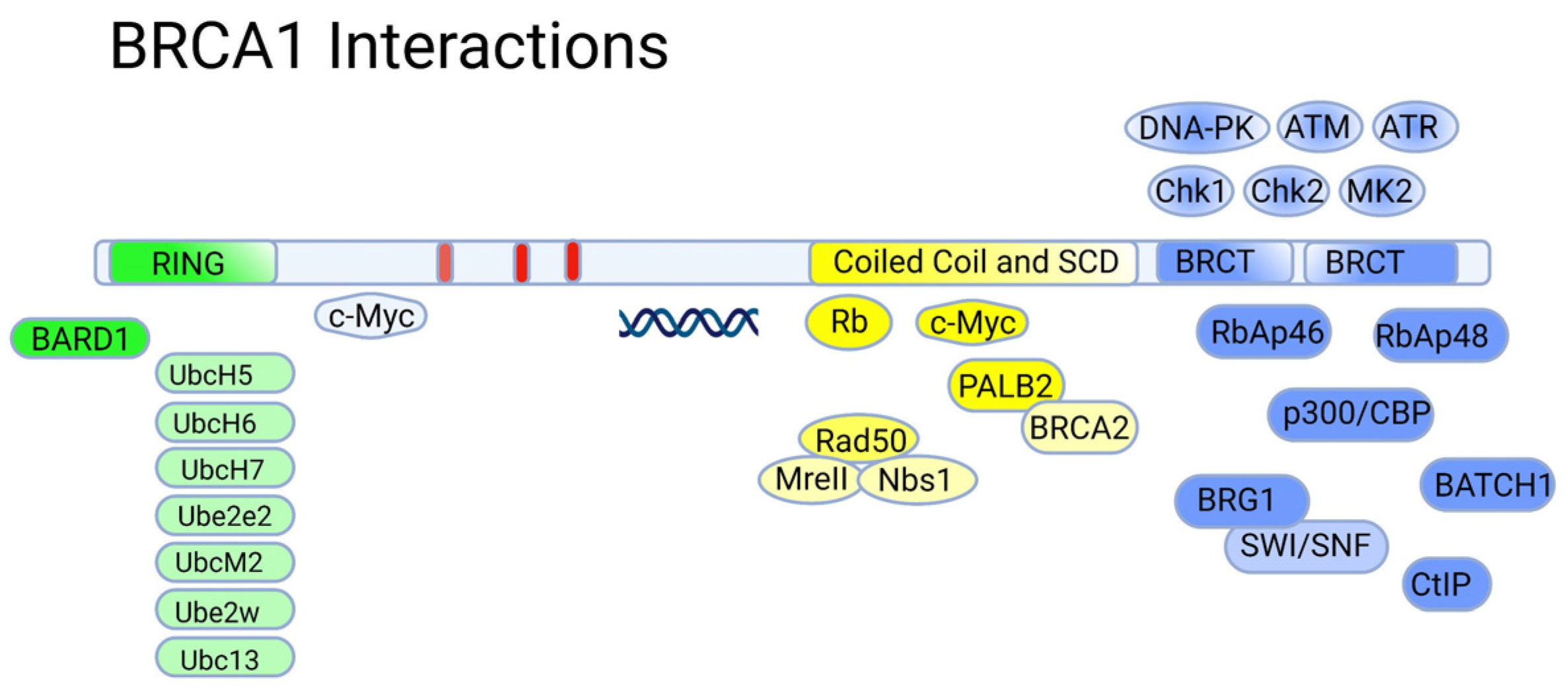
5.1. The RING Domain
5.2. The BRCT Domain
5.3. BRCA1 and p53
5.4. BRCA1 and BRCA2—A Summary on Normal Functions in Healthy Tissues
6. Survival of BRCA-Mutated Cancer Cells: Role of Tissue Microenvironment
Hypothesis: Role of Breast Adipocytes in Early Progression of BRCA1/2 Mutated Microtumors
7. Vulnerabilities of BRCA-Mutated Cancer Cells
7.1. Platinum Complexes
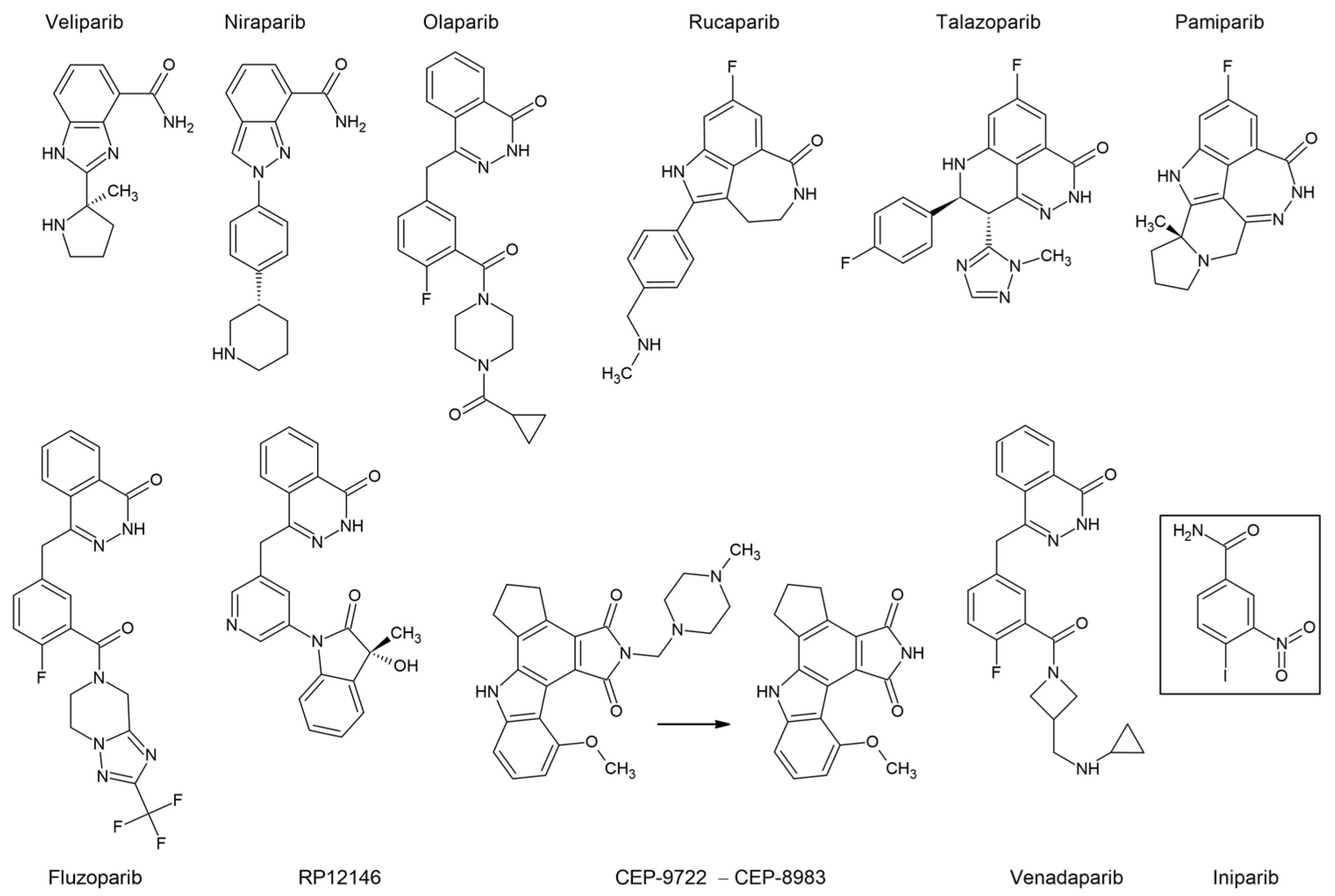
7.2. PARP Inhibitors
7.3. Boosting Synthetic Lethality by Drug Combinations
8. Future Perspectives
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hall, J.M.; Lee, M.K.; Newman, B.; Morrow, J.E.; Anderson, L.A.; Huey, B.; King, M.C. Linkage of Early-Onset Familial Breast Cancer to Chromosome 17q21. Science 1990, 250, 1684–1689. [Google Scholar] [CrossRef]
- Wooster, R.; Neuhausen, S.L.; Mangion, J.; Quirk, Y.; Ford, D.; Collins, N.; Nguyen, K.; Seal, S.; Tran, T.; Averill, D. Localization of a Breast Cancer Susceptibility Gene, BRCA2, to Chromosome 13q12-13. Science 1994, 265, 2088–2090. [Google Scholar] [CrossRef]
- King, T.A.; Li, W.; Brogi, E.; Yee, C.J.; Gemignani, M.L.; Olvera, N.; Levine, D.A.; Norton, L.; Robson, M.E.; Offit, K.; et al. Heterogenic Loss of the Wild-Type BRCA Allele in Human Breast Tumorigenesis. Ann. Surg. Oncol. 2007, 14, 2510–2518. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.C.; De, S.; Almendro, V.; Gönen, M.; Park, S.Y.; Blum, J.L.; Herlihy, W.; Ethington, G.; Schnitt, S.J.; Tung, N.; et al. Evolutionary Pathways in BRCA1-Associated Breast Tumors. Cancer Discov. 2012, 2, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Kotoula, V.; Fostira, F.; Papadopoulou, K.; Apostolou, P.; Tsolaki, E.; Lazaridis, G.; Manoussou, K.; Zagouri, F.; Pectasides, D.; Vlachos, I.; et al. The Fate of BRCA1-Related Germline Mutations in Triple-Negative Breast Tumors. Am. J. Cancer Res. 2017, 7, 98–114. [Google Scholar]
- De Talhouet, S.; Peron, J.; Vuilleumier, A.; Friedlaender, A.; Viassolo, V.; Ayme, A.; Bodmer, A.; Treilleux, I.; Lang, N.; Tille, J.-C.; et al. Clinical Outcome of Breast Cancer in Carriers of BRCA1 and BRCA2 Mutations according to Molecular Subtypes. Sci. Rep. 2020, 10, 7073. [Google Scholar] [CrossRef]
- Incorvaia, L.; Fanale, D.; Bono, M.; Calò, V.; Fiorino, A.; Brando, C.; Corsini, L.R.; Cutaia, S.; Cancelliere, D.; Pivetti, A.; et al. BRCA1/2 Pathogenic Variants in Triple-Negative versus Luminal-like Breast Cancers: Genotype-Phenotype Correlation in a Cohort of 531 Patients. Ther. Adv. Med. Oncol. 2020, 12, 1758835920975326. [Google Scholar] [CrossRef]
- Petrucelli, N.; Daly, M.B.; Pal, T. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer; University of Washington: Seattle, WA, USA, 2022. [Google Scholar]
- Marchetti, C.; Ataseven, B.; Cassani, C.; Sassu, C.M.; Congedo, L.; D’Indinosante, M.; Cappuccio, S.; Rhiem, K.; Hahnen, E.; Lucci Cordisco, E.; et al. Ovarian Cancer Onset across Different BRCA Mutation Types: A View to a More Tailored Approach for BRCA Mutated Patients. Int. J. Gynecol. Cancer 2023, 33, 257–262. [Google Scholar] [CrossRef]
- Yang, Q.; Yoshimura, G.; Nakamura, M.; Nakamura, Y.; Suzuma, T.; Umemura, T.; Mori, I.; Sakurai, T.; Kakudo, K. BRCA1 in Non-Inherited Breast Carcinomas (Review). Oncol. Rep. 2002, 9, 1329–1333. [Google Scholar] [CrossRef]
- Wilson, C.A.; Ramos, L.; Villaseñor, M.R.; Anders, K.H.; Press, M.F.; Clarke, K.; Karlan, B.; Chen, J.J.; Scully, R.; Livingston, D.; et al. Localization of Human BRCA1 and Its Loss in High-Grade, Non-Inherited Breast Carcinomas. Nat. Genet. 1999, 21, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Neff, R.T.; Senter, L.; Salani, R. BRCA Mutation in Ovarian Cancer: Testing, Implications and Treatment Considerations. Ther. Adv. Med. Oncol. 2017, 9, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-C.; Lee, Y.-L.; Li, C.-Y. BRCA Genes and Related Cancers: A Meta-Analysis from Epidemiological Cohort Studies. Medicina 2021, 57, 905. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Lee, Y.-C.; Li, C.-Y.; Lee, Y.-L.; Chen, B.-L. BRCA1 and BRCA2 Gene Mutations and Lung Cancer Sisk: A Meta-Analysis. Medicina 2020, 56, 212. [Google Scholar] [CrossRef]
- Baretta, Z.; Mocellin, S.; Goldin, E.; Olopade, O.I.; Huo, D. Effect of BRCA Germline Mutations on Breast Cancer Prognosis: A Systematic Review and Meta-Analysis. Medicina 2016, 95, e4975. [Google Scholar] [CrossRef] [PubMed]
- Gilks, C.B.; Prat, J. Ovarian Carcinoma Pathology and Genetics: Recent Advances. Hum. Pathol. 2009, 40, 1213–1223. [Google Scholar] [CrossRef]
- McCluggage, W.G. Morphological Subtypes of Ovarian Carcinoma: A Review with Emphasis on New Developments and Pathogenesis. Pathology 2011, 43, 420–432. [Google Scholar] [CrossRef]
- Kim, S.I.; Lee, M.; Kim, H.S.; Chung, H.H.; Kim, J.-W.; Park, N.H.; Song, Y.-S. Effect of BRCA Mutational Status on Survival Outcome in Advanced-Stage High-Grade Serous Ovarian Cancer. J. Ovarian Res. 2019, 12, 40. [Google Scholar] [CrossRef]
- Risch, H.A.; McLaughlin, J.R.; Cole, D.E.; Rosen, B.; Bradley, L.; Kwan, E.; Jack, E.; Vesprini, D.J.; Kuperstein, G.; Abrahamson, J.L.; et al. Prevalence and Penetrance of Germline BRCA1 and BRCA2 Mutations in a Population Series of 649 Women with Ovarian Cancer. Am. J. Hum. Genet. 2001, 68, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Royer, R.; Li, S.; McLaughlin, J.R.; Rosen, B.; Risch, H.A.; Fan, I.; Bradley, L.; Shaw, P.A.; Narod, S.A. Frequencies of BRCA1 and BRCA2 Mutations among 1,342 Unselected Patients with Invasive Ovarian Cancer. Gynecol. Oncol. 2011, 121, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Ramus, S.J.; Gayther, S.A. The Contribution of BRCA1 and BRCA2 to Ovarian Cancer. Mol. Oncol. 2009, 3, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.-A.; Mooij, T.M.; Roos-Blom, M.-J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef]
- Soegaard, M.; Kjaer, S.K.; Cox, M.; Wozniak, E.; Høgdall, E.; Høgdall, C.; Blaakaer, J.; Jacobs, I.J.; Gayther, S.A.; Ramus, S.J. BRCA1 and BRCA2 Mutation Prevalence and Clinical Characteristics of a Population-Based Series of Ovarian Cancer Cases from Denmark. Clin. Cancer Res. 2008, 14, 3761–3767. [Google Scholar] [CrossRef]
- Alsop, K.; Fereday, S.; Meldrum, C.; deFazio, A.; Emmanuel, C.; George, J.; Dobrovic, A.; Birrer, M.J.; Webb, P.M.; Stewart, C.; et al. BRCA Mutation Frequency and Patterns of Treatment Response in BRCA Mutation-Positive Women with Ovarian Cancer: A Report from the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 2012, 30, 2654–2663. [Google Scholar] [CrossRef] [PubMed]
- Norquist, B.M.; Brady, M.F.; Harrell, M.I.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Burger, R.A.; Tewari, K.S.; et al. Mutations in Homologous Recombination Genes and Outcomes in Ovarian Carcinoma Patients in GOG 218: An NRG Oncology/Gynecologic Oncology Group Study. Clin. Cancer Res. 2018, 24, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.S.P.; Rothermundt, C.; Thomas, K.; Bancroft, E.; Eeles, R.; Shanley, S.; Ardern-Jones, A.; Norman, A.; Kaye, S.B.; Gore, M.E. “BRCAness” Syndrome in Ovarian Cancer: A Case-Control Study Describing the Clinical Features and Outcome of Patients with Epithelial Ovarian Cancer Associated with BRCA1 and BRCA2 Mutations. J. Clin. Oncol. 2008, 26, 5530–5536. [Google Scholar] [CrossRef]
- Eoh, K.J.; Kim, H.M.; Lee, J.-Y.; Kim, S.; Kim, S.W.; Kim, Y.T.; Nam, E.J. Mutation Landscape of Germline and Somatic BRCA1/2 in Patients with High-Grade Serous Ovarian Cancer. BMC Cancer 2020, 20, 204. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, B.T.J.; Timms, K.M.; Carey, M.S.; Gutin, A.; Meyer, L.A.; Flake, D.D.; Abkevich, V.; Potter, J.; Pruss, D.; Glenn, P.; et al. Somatic Mutations in BRCA1 and BRCA2 Could Expand the Number of Patients That Benefit from Poly (ADP Ribose) Polymerase Inhibitors in Ovarian Cancer. J. Clin. Oncol. 2010, 28, 3570–3576. [Google Scholar] [CrossRef]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib Maintenance Therapy in Platinum-Sensitive Relapsed Ovarian Cancer. N. Engl. J. Med. 2012, 366, 1382–1392. [Google Scholar] [CrossRef]
- Pennington, K.P.; Walsh, T.; Harrell, M.I.; Lee, M.K.; Pennil, C.C.; Rendi, M.H.; Thornton, A.; Norquist, B.M.; Casadei, S.; Nord, A.S.; et al. Germline and Somatic Mutations in Homologous Recombination Genes Predict Platinum Response and Survival in Ovarian, Fallopian Tube, and Peritoneal Carcinomas. Clin. Cancer Res. 2014, 20, 764–775. [Google Scholar] [CrossRef]
- Moschetta, M.; George, A.; Kaye, S.B.; Banerjee, S. BRCA Somatic Mutations and Epigenetic BRCA Modifications in Serous Ovarian Cancer. Ann. Oncol. 2016, 27, 1449–1455. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011, 474, 609–615. [CrossRef] [PubMed]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.L.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib Maintenance Therapy in Patients with Platinum-Sensitive Relapsed Serous Ovarian Cancer: A Preplanned Retrospective Analysis of Outcomes by BRCA Status in a Randomised Phase 2 Trial. Lancet Oncol. 2014, 15, 852–861. [Google Scholar] [CrossRef]
- Mehrgou, A.; Akouchekian, M. The Importance of BRCA1 and BRCA2 Genes Mutations in Breast Cancer Development. Med. J. Islam. Repub. Iran 2016, 30, 369. [Google Scholar] [PubMed]
- Aysola, K.; Desai, A.; Welch, C.; Xu, J.; Qin, Y.; Reddy, V.; Matthews, R.; Owens, C.; Okoli, J.; Beech, D.J.; et al. Triple Negative Breast Cancer—An Overview. Hered. Genet. 2013, 2013, 001. [Google Scholar] [CrossRef]
- Kotsopoulos, J. BRCA Mutations and Breast Cancer Prevention. Cancers 2018, 10, 524. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Brusco, L.; Daniels, M.; Wathoo, C.; Bailey, A.M.; Strong, L.; Shaw, K.; Lu, K.; Qi, Y.; Zhao, H.; et al. Incidental Germline Variants in 1000 Advanced Cancers on a Prospective Somatic Genomic Profiling Protocol. Ann. Oncol. 2016, 27, 795–800. [Google Scholar] [CrossRef]
- Tutt, A.; Tovey, H.; Cheang, M.C.U.; Kernaghan, S.; Kilburn, L.; Gazinska, P.; Owen, J.; Abraham, J.; Barrett, S.; Barrett-Lee, P.; et al. Carboplatin in BRCA1/2-Mutated and Triple-Negative Breast Cancer BRCAness Subgroups: The TNT Trial. Nat. Med. 2018, 24, 628–637. [Google Scholar] [CrossRef]
- Winter, C.; Nilsson, M.P.; Olsson, E.; George, A.M.; Chen, Y.; Kvist, A.; Törngren, T.; Vallon-Christersson, J.; Hegardt, C.; Häkkinen, J.; et al. Targeted Sequencing of BRCA1 and BRCA2 across a Large Unselected Breast Cancer Cohort Suggests That One-Third of Mutations Are Somatic. Ann. Oncol. 2016, 27, 1532–1538. [Google Scholar] [CrossRef]
- den Brok, W.D.; Schrader, K.A.; Sun, S.; Tinker, A.V.; Zhao, E.Y.; Aparicio, S.; Gelmon, K.A. Homologous Recombination Deficiency in Breast Cancer: A Clinical Review. JCO Precis. Oncol. 2017, 1, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bodily, W.R.; Shirts, B.H.; Walsh, T.; Gulsuner, S.; King, M.-C.; Parker, A.; Roosan, M.; Piccolo, S.R. Effects of Germline and Somatic Events in Candidate BRCA-like Genes on Breast-Tumor Signatures. PLoS ONE 2020, 15, e0239197. [Google Scholar] [CrossRef]
- Turner, N.; Tutt, A.; Ashworth, A. Hallmarks of “BRCAness” in Sporadic Cancers. Nat. Rev. Cancer 2004, 4, 814–819. [Google Scholar] [CrossRef]
- Rechsteiner, M.; Dedes, K.; Fink, D.; Pestalozzi, B.; Sobottka, B.; Moch, H.; Wild, P.; Varga, Z. Somatic BRCA1 Mutations in Clinically Sporadic Breast Cancer with Medullary Histological Features. J. Cancer Res. Clin. Oncol. 2018, 144, 865–874. [Google Scholar] [CrossRef]
- Greer, J.B.; Whitcomb, D.C. Role of BRCA1 and BRCA2 Mutations in Pancreatic Cancer. Gut 2007, 56, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Kowalewski, A.; Szylberg, Ł.; Saganek, M.; Napiontek, W.; Antosik, P.; Grzanka, D. Emerging Strategies in BRCA-Positive Pancreatic Cancer. J. Cancer Res. Clin. Oncol. 2018, 144, 1503–1507. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Lu, Y.; Jin, K.; Cheng, H.; Guo, M.; Liu, Z.; Long, J.; Liu, C.; Ni, Q.; Yu, X. Pancreatic Cancer: BRCA Mutation and Personalized Treatment. Expert Rev. Anticancer Ther. 2015, 15, 1223–1231. [Google Scholar] [CrossRef]
- Castro, E.; Eeles, R. The Role of BRCA1 and BRCA2 in Prostate Cancer. Asian J. Androl. 2012, 14, 409–414. [Google Scholar] [CrossRef]
- Messina, C.; Cattrini, C.; Soldato, D.; Vallome, G.; Caffo, O.; Castro, E.; Olmos, D.; Boccardo, F.; Zanardi, E. BRCA Mutations in Prostate Cancer: Prognostic and Predictive Implications. J. Oncol. 2020, 2020, 4986365. [Google Scholar] [CrossRef]
- Narod, S.A.; Neuhausen, S.; Vichodez, G.; Armel, S.; Lynch, H.T.; Ghadirian, P.; Cummings, S.; Olopade, O.; Stoppa-Lyonnet, D.; Couch, F.; et al. Rapid Progression of Prostate Cancer in Men with a BRCA2 Mutation. Br. J. Cancer 2008, 99, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Pal, T.; Vadaparampil, S.; Betts, J.; Miree, C.; Li, S.; Narod, S.A. BRCA1/2 in High-Risk African American Women with Breast Cancer: Providing Genetic Testing through Various Recruitment Strategies. Genet. Test. 2008, 12, 401–407. [Google Scholar] [CrossRef]
- Ferla, R.; Calò, V.; Cascio, S.; Rinaldi, G.; Badalamenti, G.; Carreca, I.; Surmacz, E.; Colucci, G.; Bazan, V.; Russo, A. Founder Mutations in BRCA1 and BRCA2 Genes. Ann. Oncol. 2007, 18 (Suppl. 6), vi93–vi98. [Google Scholar] [CrossRef]
- Roa, B.B.; Boyd, A.A.; Volcik, K.; Richards, C.S. Ashkenazi Jewish Population Frequencies for Common Mutations in BRCA1 and BRCA2. Nat. Genet. 1996, 14, 185–187. [Google Scholar] [CrossRef] [PubMed]
- Janavičius, R. Founder BRCA1/2 Mutations in the Europe: Implications for Hereditary Breast-Ovarian Cancer Prevention and Control. EPMA J. 2010, 1, 397–412. [Google Scholar] [CrossRef]
- Sokolenko, A.P.; Sokolova, T.N.; Ni, V.I.; Preobrazhenskaya, E.V.; Iyevleva, A.G.; Aleksakhina, S.N.; Romanko, A.A.; Bessonov, A.A.; Gorodnova, T.V.; Anisimova, E.I.; et al. Frequency and Spectrum of Founder and Non-Founder BRCA1 and BRCA2 Mutations in a Large Series of Russian Breast Cancer and Ovarian Cancer Patients. Breast Cancer Res. Treat. 2020, 184, 229–235. [Google Scholar] [CrossRef]
- Suspitsin, E.N.; Sherina, N.Y.; Ponomariova, D.N.; Sokolenko, A.P.; Iyevleva, A.G.; Gorodnova, T.V.; Zaitseva, O.A.; Yatsuk, O.S.; Togo, A.V.; Tkachenko, N.N.; et al. High Frequency of BRCA1, but Not CHEK2 or NBS1 (NBN), Founder Mutations in Russian Ovarian Cancer Patients. Hered. Cancer Clin. Pract. 2009, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Day, M.; Rappas, M.; Ptasinska, K.; Boos, D.; Oliver, A.W.; Pearl, L.H. BRCT Domains of the DNA Damage Checkpoint Proteins TOPBP1/Rad4 Display Distinct Specificities for Phosphopeptide Ligands. Elife 2018, 7, e39979. [Google Scholar] [CrossRef]
- Koonin, E.V.; Altschul, S.F.; Bork, P. BRCA1 Protein Products... Functional Motifs. Nat. Genet. 1996, 13, 266–268. [Google Scholar] [CrossRef]
- Fabian, D.; Flatt, T. The Evolution of Aging; Nature Education Knowledge; Cambridge University Press: Cambridge, UK, 2011; Volume 3. [Google Scholar]
- Ben-Aharon, I.; Levi, M.; Margel, D.; Yerushalmi, R.; Rizel, S.; Perry, S.; Sharon, E.; Hasky, N.; Abir, R.; Fisch, B.; et al. Premature Ovarian Aging in BRCA Carriers: A Prototype of Systemic Precocious Aging? Oncotarget 2018, 9, 15931–15941. [Google Scholar] [CrossRef] [PubMed]
- Kępczyński, Ł.; Połatyńska, K.; Nykel, A.; Sałamunia, J.; Kałużewski, T.; Kużawczyk, A.; Gach, A. Age of Natural Menopause Onset in BRCA1/2 Carriers—Systematic Review and Meta-Analysis. Prz. Menopauzalny 2020, 19, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Drechsel, K.C.E.; van Tilborg, T.C.; Eijkemans, M.J.C.; Lentjes, E.G.W.M.; Homminga, I.; Goddijn, M.; van Golde, R.J.T.; Verpoest, W.; Lichtenbelt, K.; Broekmans, J.M.; et al. The Impact of BRCA1- and BRCA2 Mutations on Ovarian Reserve Status. Reprod. Sci. 2022, 30, 270–282. [Google Scholar] [CrossRef]
- Semmler, L.; Reiter-Brennan, C.; Klein, A. BRCA1 and Breast Cancer: A Review of the Underlying Mechanisms Resulting in the Tissue-Specific Tumorigenesis in Mutation Carriers. J. Breast Cancer 2019, 22, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different Roles in a Common Pathway of Genome Protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.H.; Serrano, L. Cell Type-Specific Properties and Environment Shape Tissue Specificity of Cancer Genes. Sci. Rep. 2016, 6, 20707. [Google Scholar] [CrossRef] [PubMed]
- Ade, C.; Roy-Engel, A.M.; Deininger, P.L. Alu Elements: An Intrinsic Source of Human Genome Instability. Curr. Opin. Virol. 2013, 3, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Welcsh, P.L.; King, M.C. BRCA1 and BRCA2 and the Genetics of Breast and Ovarian Cancer. Hum. Mol. Genet. 2001, 10, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.M.; Lee, M.K.; Szabo, C.I.; Jerome, N.; McEuen, M.; Taylor, M.; Hood, L.; King, M.C. Complete Genomic Sequence and Analysis of 117 Kb of Human DNA Containing the Gene BRCA1. Genome Res. 1996, 6, 1029–1049. [Google Scholar] [CrossRef]
- Montagna, M.; Santacatterina, M.; Torri, A.; Menin, C.; Zullato, D.; Chieco-Bianchi, L.; D’Andrea, E. Identification of a 3 Kb Alu-Mediated BRCA1 Gene Rearrangement in Two Breast/Ovarian Cancer Families. Oncogene 1999, 18, 4160–4165. [Google Scholar] [CrossRef] [PubMed]
- Girolimetti, G.; Perrone, A.M.; Santini, D.; Barbieri, E.; Guerra, F.; Ferrari, S.; Zamagni, C.; De Iaco, P.; Gasparre, G.; Turchetti, D. BRCA-Associated Ovarian Cancer: From Molecular Genetics to Risk Management. Biomed. Res. Int. 2014, 2014, 787143. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.K.; Han, K.; Wang, J.; Lee, J.; Wang, H.; Callinan, P.A.; Dyer, M.; Cordaux, R.; Liang, P.; Batzer, M.A. Human Genomic Deletions Mediated by Recombination between Alu Elements. Am. J. Hum. Genet. 2006, 79, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Bozsik, A.; Pócza, T.; Papp, J.; Vaszkó, T.; Butz, H.; Patócs, A.; Oláh, E. Complex Characterization of Germline Large Genomic Rearrangements of the BRCA1 and BRCA2 Genes in High-Risk Breast Cancer Patients-Novel Variants from a Large National Center. Int. J. Mol. Sci. 2020, 21, 4650. [Google Scholar] [CrossRef]
- Unger, M.A.; Nathanson, K.L.; Calzone, K.; Antin-Ozerkis, D.; Shih, H.A.; Martin, A.M.; Lenoir, G.M.; Mazoyer, S.; Weber, B.L. Screening for Genomic Rearrangements in Families with Breast and Ovarian Cancer Identifies BRCA1 Mutations Previously Missed by Conformation-Sensitive Gel Electrophoresis or Sequencing. Am. J. Hum. Genet. 2000, 67, 841–850. [Google Scholar] [CrossRef]
- Nordling, M.; Karlsson, P.; Wahlström, J.; Engwall, Y.; Wallgren, A.; Martinsson, T. A Large Deletion Disrupts the Exon 3 Transcription Activation Domain of the BRCA2 Gene in a Breast/Ovarian Cancer Family. Cancer Res. 1998, 58, 1372–1375. [Google Scholar]
- Ewald, I.P.; Ribeiro, P.L.I.; Palmero, E.I.; Cossio, S.L.; Giugliani, R.; Ashton-Prolla, P. Genomic Rearrangements in BRCA1 and BRCA2: A Literature Review. Genet. Mol. Biol. 2009, 32, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-F.; Hu, Z.; Rao, N.-Y.; Song, C.-G.; Zhang, B.; Cao, M.-Z.; Su, F.-X.; Wang, Y.-S.; He, P.-Q.; Di, G.-H.; et al. The Prevalence of BRCA1 and BRCA2 Germline Mutations in High-Risk Breast Cancer Patients of Chinese Han Nationality: Two Recurrent Mutations Were Identified. Breast Cancer Res. Treat. 2008, 110, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Fachal, L.; Blanco, A.; Santamariña, M.; Carracedo, A.; Vega, A. Large Genomic Rearrangements of BRCA1 and BRCA2 among Patients Referred for Genetic Analysis in Galicia (NW Spain): Delimitation and Mechanism of Three Novel BRCA1 Rearrangements. PLoS ONE 2014, 9, e93306. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.I.; McBee, R.M.; Le, U.Q.; Stone, A.C.; Wilkerson, G.K.; Demogines, A.M.; Sawyer, S.L. Rapid Evolution of BRCA1 and BRCA2 in Humans and Other Primates. BMC Evol. Biol. 2014, 14, 155. [Google Scholar] [CrossRef] [PubMed]
- O’Donovan, P.J.; Livingston, D.M. BRCA1 and BRCA2: Breast/Ovarian Cancer Susceptibility Gene Products and Participants in DNA Double-Strand Break Repair. Carcinogenesis 2010, 31, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Hemel, D.; Domchek, S.M. Breast Cancer Predisposition Syndromes. Hematol. Oncol. Clin. N. Am. 2010, 24, 799–814. [Google Scholar] [CrossRef] [PubMed]
- Preisler-Adams, S.; Schönbuchner, I.; Fiebig, B.; Welling, B.; Dworniczak, B.; Weber, B.H.F. Gross Rearrangements in BRCA1 but Not BRCA2 Play a Notable Role in Predisposition to Breast and Ovarian Cancer in High-Risk Families of German Origin. Cancer Genet. Cytogenet. 2006, 168, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Rohlfs, E.M.; Puget, N.; Graham, M.L.; Weber, B.L.; Garber, J.E.; Skrzynia, C.; Halperin, J.L.; Lenoir, G.M.; Silverman, L.M.; Mazoyer, S. An Alu-Mediated 7.1 Kb Deletion of BRCA1 Exons 8 and 9 in Breast and Ovarian Cancer Families That Results in Alternative Splicing of Exon 10. Genes Chromosom. Cancer 2000, 28, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Agata, S.; Dalla Palma, M.; Callegaro, M.; Scaini, M.C.; Menin, C.; Ghiotto, C.; Nicoletto, O.; Zavagno, G.; Chieco-Bianchi, L.; D’Andrea, E.; et al. Large Genomic Deletions Inactivate the BRCA2 Gene in Breast Cancer Families. J. Med. Genet. 2005, 42, e64. [Google Scholar] [CrossRef]
- Karhu, R.; Laurila, E.; Kallioniemi, A.; Syrjäkoski, K. Large Genomic BRCA2 Rearrangements and Male Breast Cancer. Cancer Detect. Prev. 2006, 30, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.M.; Davis, T.A.; Silva, A.G.S.; Kirk, J.A.; Leary, J.A. kConFab Investigators Large Genomic Rearrangements of Both BRCA2 and BRCA1 Are a Feature of the Inherited Breast/Ovarian Cancer Phenotype in Selected Families. J. Med. Genet. 2005, 42, e31. [Google Scholar] [CrossRef]
- Peixoto, A.; Santos, C.; Rocha, P.; Pinto, P.; Bizarro, S.; Teixeira, M.R. Molecular Diagnosis of the Portuguese Founder Mutation BRCA2 c.156_157insAlu. Breast Cancer Res. Treat. 2009, 117, 215–217. [Google Scholar] [CrossRef]
- Duncan, J.A.; Reeves, J.R.; Cooke, T.G. BRCA1 and BRCA2 Proteins: Roles in Health and Disease. Mol. Pathol. 1998, 51, 237–247. [Google Scholar] [CrossRef]
- Brzovic, P.S.; Rajagopal, P.; Hoyt, D.W.; King, M.-C.; Klevit, R.E. Structure of a BRCA1–BARD1 Heterodimeric RING–RING Complex. Nat. Struct. Mol. Biol. 2001, 8, 833–837. [Google Scholar] [CrossRef]
- Wang, Y.; Bernhardy, A.J.; Johnson, N. Abstract A23: BRCA1 Mutations in the BRCT Domain Can Be Removed through Alternative Splicing and Induce PARP Inhibitor Resistance. Mol. Cancer Res. 2017, 15, A23. [Google Scholar] [CrossRef]
- Wu, W.; Koike, A.; Takeshita, T.; Ohta, T. The Ubiquitin E3 Ligase Activity of BRCA1 and Its Biological Functions. Cell Div. 2008, 3, 1. [Google Scholar] [CrossRef]
- Hashizume, R.; Fukuda, M.; Maeda, I.; Nishikawa, H.; Oyake, D.; Yabuki, Y.; Ogata, H.; Ohta, T. The RING Heterodimer BRCA1-BARD1 Is a Ubiquitin Ligase Inactivated by a Breast Cancer-Derived Mutation. J. Biol. Chem. 2001, 276, 14537–14540. [Google Scholar] [CrossRef] [PubMed]
- Birrane, G.; Varma, A.K.; Soni, A.; Ladias, J.A.A. Crystal Structure of the BARD1 BRCT Domains. Biochemistry 2007, 46, 7706–7712. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Xu, X.L.; Yang, M.C.; Wei, F.; Ayi, T.C.; Bowcock, A.M.; Baer, R. Cell Cycle-Dependent Colocalization of BARD1 and BRCA1 Proteins in Discrete Nuclear Domains. Proc. Natl. Acad. Sci. USA 1997, 94, 12075–12080. [Google Scholar] [CrossRef]
- Wang, B.; Matsuoka, S.; Ballif, B.A.; Zhang, D.; Smogorzewska, A.; Gygi, S.P.; Elledge, S.J. Abraxas and RAP80 Form a BRCA1 Protein Complex Required for the DNA Damage Response. Science 2007, 316, 1194–1198. [Google Scholar] [CrossRef] [PubMed]
- Mittenberg, A.G.; Moiseeva, T.N.; Barlev, N.A. Role of Proteasomes in Transcription and Their Regulation by Covalent Modifications. Front. Biosci. 2008, 13, 7184–7192. [Google Scholar] [CrossRef] [PubMed]
- Paull, T.T.; Cortez, D.; Bowers, B.; Elledge, S.J.; Gellert, M. Direct DNA Binding by Brca1. Proc. Natl. Acad. Sci. USA 2001, 98, 6086–6091. [Google Scholar] [CrossRef] [PubMed]
- Starita, L.M.; Parvin, J.D. The Multiple Nuclear Functions of BRCA1: Transcription, Ubiquitination and DNA Repair. Curr. Opin. Cell Biol. 2003, 15, 345–350. [Google Scholar] [CrossRef]
- Zheng, L.; Pan, H.; Li, S.; Flesken-Nikitin, A.; Chen, P.L.; Boyer, T.G.; Lee, W.H. Sequence-Specific Transcriptional Corepressor Function for BRCA1 through a Novel Zinc Finger Protein, ZBRK1. Mol. Cell 2000, 6, 757–768. [Google Scholar] [CrossRef]
- Clark, S.L.; Rodriguez, A.M.; Snyder, R.R.; Hankins, G.D.V.; Boehning, D. Structure-Function of the Tumor Suppressor BRCA1. Comput. Struct. Biotechnol. J. 2012, 1, e201204005. [Google Scholar] [CrossRef]
- Lee, M.S.; Green, R.; Marsillac, S.M.; Coquelle, N.; Williams, R.S.; Yeung, T.; Foo, D.; Hau, D.D.; Hui, B.; Monteiro, A.N.A.; et al. Comprehensive Analysis of Missense Variations in the BRCT Domain of BRCA1 by Structural and Functional Assays. Cancer Res. 2010, 70, 4880–4890. [Google Scholar] [CrossRef]
- Williams, R.S.; Bernstein, N.; Lee, M.S.; Rakovszky, M.L.; Cui, D.; Green, R.; Weinfeld, M.; Glover, J.N.M. Structural Basis for Phosphorylation-Dependent Signaling in the DNA-Damage Response. Biochem. Cell Biol. 2005, 83, 721–727. [Google Scholar] [CrossRef]
- Zhang, J.; Willers, H.; Feng, Z.; Ghosh, J.C.; Kim, S.; Weaver, D.T.; Chung, J.H.; Powell, S.N.; Xia, F. Chk2 Phosphorylation of BRCA1 Regulates DNA Double-Strand Break Repair. Mol. Cell. Biol. 2004, 24, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef]
- Reinhardt, H.C.; Yaffe, M.B. Kinases That Control the Cell Cycle in Response to DNA Damage: Chk1, Chk2, and MK2. Curr. Opin. Cell Biol. 2009, 21, 245–255. [Google Scholar] [CrossRef]
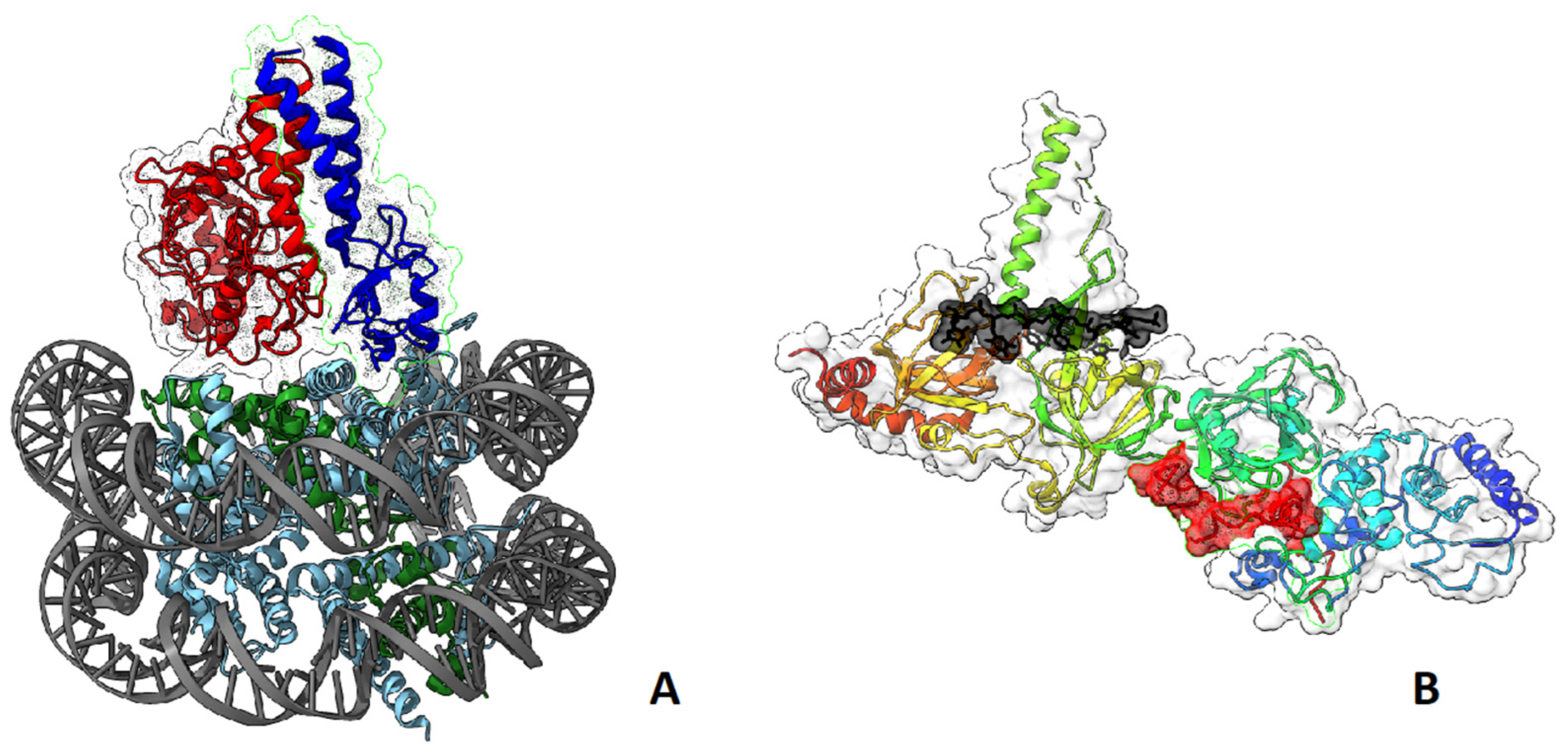
- Witus, S.R.; Burrell, A.L.; Farrell, D.P.; Kang, J.; Wang, M.; Hansen, J.M.; Pravat, A.; Tuttle, L.M.; Stewart, M.D.; Brzovic, P.S.; et al. BRCA1/BARD1 Site-Specific Ubiquitylation of Nucleosomal H2A Is Directed by BARD1. Nat. Struct. Mol. Biol. 2021, 28, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Botuyan, M.V.; Zhao, D.; Cui, G.; Mer, E.; Mer, G. Mechanisms of BRCA1-BARD1 Nucleosome Recognition and Ubiquitylation. Nature 2021, 596, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.R.; Clifford, G.; Bonnet, C.; Groth, A.; Wilson, M.D.; Chapman, J.R. BARD1 Reads H2A Lysine 15 Ubiquitination to Direct Homologous Recombination. Nature 2021, 596, 433–437. [Google Scholar] [CrossRef]
- Yang, H.; Jeffrey, P.D.; Miller, J.; Kinnucan, E.; Sun, Y.; Thoma, N.H.; Zheng, N.; Chen, P.-L.; Lee, W.-H.; Pavletich, N.P. BRCA2 Function in DNA Binding and Recombination from a BRCA2-DSS1-SsDNA Structure. Science 2002, 297, 1837–1848. [Google Scholar] [CrossRef]
- Trego, K.S.; Groesser, T.; Davalos, A.R.; Parplys, A.C.; Zhao, W.; Nelson, M.R.; Hlaing, A.; Shih, B.; Rydberg, B.; Pluth, J.M.; et al. Non-Catalytic Roles for XPG with BRCA1 and BRCA2 in Homologous Recombination and Genome Stability. Mol. Cell 2016, 61, 535–546. [Google Scholar] [CrossRef]
- Scully, R.; Livingston, D.M. In Search of the Tumour-Suppressor Functions of BRCA1 and BRCA2. Nature 2000, 408, 429–432. [Google Scholar] [CrossRef]
- Deng, C.X.; Scott, F. Role of the Tumor Suppressor Gene Brca1 in Genetic Stability and Mammary Gland Tumor Formation. Oncogene 2000, 19, 1059–1064. [Google Scholar] [CrossRef]
- Foray, N.; Marot, D.; Randrianarison, V.; Venezia, N.D.; Picard, D.; Perricaudet, M.; Favaudon, V.; Jeggo, P. Constitutive Association of BRCA1 and C-Abl and Its ATM-Dependent Disruption after Irradiation. Mol. Cell. Biol. 2002, 22, 4020–4032. [Google Scholar] [CrossRef]
- Levav-Cohen, Y.; Goldberg, Z.; Zuckerman, V.; Grossman, T.; Haupt, S.; Haupt, Y. C-Abl as a Modulator of P53. Biochem. Biophys. Res. Commun. 2005, 331, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Hantschel, O.; Rix, U.; Schmidt, U.; Bürckstümmer, T.; Kneidinger, M.; Schütze, G.; Colinge, J.; Bennett, K.L.; Ellmeier, W.; Valent, P.; et al. The Btk Tyrosine Kinase Is a Major Target of the Bcr-Abl Inhibitor Dasatinib. Proc. Natl. Acad. Sci. USA 2007, 104, 13283–13288. [Google Scholar] [CrossRef]
- Althubiti, M.; Rada, M.; Samuel, J.; Escorsa, J.M.; Najeeb, H.; Lee, K.-G.; Lam, K.-P.; Jones, G.D.D.; Barlev, N.A.; Macip, S. BTK Modulates P53 Activity to Enhance Apoptotic and Senescent Responses. Cancer Res. 2016, 76, 5405–5414. [Google Scholar] [CrossRef] [PubMed]
- Rada, M.; Barlev, N.; Macip, S. BTK: A Two-Faced Effector in Cancer and Tumour Suppression. Cell Death Dis. 2018, 9, 1064. [Google Scholar] [CrossRef]
- Xu, X.; Qiao, W.; Linke, S.P.; Cao, L.; Li, W.M.; Furth, P.A.; Harris, C.C.; Deng, C.X. Genetic Interactions between Tumor Suppressors Brca1 and P53 in Apoptosis, Cell Cycle and Tumorigenesis. Nat. Genet. 2001, 28, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Somasundaram, K.; Peng, Y.; Tian, H.; Zhang, H.; Bi, D.; Weber, B.L.; El-Deiry, W.S. BRCA1 Physically Associates with P53 and Stimulates Its Transcriptional Activity. Oncogene 1998, 16, 1713–1721. [Google Scholar] [CrossRef]
- Pietrasik, S.; Zajac, G.; Morawiec, J.; Soszynski, M.; Fila, M.; Blasiak, J. Interplay between BRCA1 and GADD45A and Its Potential for Nucleotide Excision Repair in Breast Cancer Pathogenesis. Int. J. Mol. Sci. 2020, 21, 870. [Google Scholar] [CrossRef] [PubMed]
- Evers, B.; Jonkers, J. Mouse Models of BRCA1 and BRCA2 Deficiency: Past Lessons, Current Understanding and Future Prospects. Oncogene 2006, 25, 5885–5897. [Google Scholar] [CrossRef]
- Clarke, C.L.; Sandle, J.; Jones, A.A.; Sofronis, A.; Patani, N.R.; Lakhani, S.R. Mapping Loss of Heterozygosity in Normal Human Breast Cells from BRCA1/2 Carriers. Br. J. Cancer 2006, 95, 515–519. [Google Scholar] [CrossRef]
- Armes, J.E.; Egan, A.J.M.; Southey, M.C.; Dite, G.S.; McCredie, M.R.E.; Giles, G.G.; Hopper, J.L.; Venter, D.J. The Histologic Phenotypes of Breast Carcinoma Occurring before Age 40 Years in Women with and without BRCA1 or BRCA2 Germline Mutations. Cancer 1998, 83, 2335–2345. [Google Scholar] [CrossRef]
- Mote, P.A.; Leary, J.A.; Avery, K.A.; Sandelin, K.; Chenevix-Trench, G.; Kirk, J.A.; Clarke, C.L. Germ-Line Mutations in BRCA1 or BRCA2 in the Normal Breast Are Associated with Altered Expression of Estrogen-Responsive Proteins and the Predominance of Progesterone Receptor A. Genes Chromosom. Cancer 2004, 39, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Ingthorsson, S.; Traustadottir, G.A.; Gudjonsson, T. Cellular Plasticity and Heterotypic Interactions during Breast Morphogenesis and Cancer Initiation. Cancers 2022, 14, 5209. [Google Scholar] [CrossRef]
- Avşar Abdik, E. Differentiated Pre-Adipocytes Promote Proliferation, Migration and Epithelial-Mesenchymal Transition in Breast Cancer Cells of Different P53 Status. Mol. Biol. Rep. 2021, 48, 5187–5198. [Google Scholar] [CrossRef]
- Choi, J.; Cha, Y.J.; Koo, J.S. Adipocyte Biology in Breast Cancer: From Silent Bystander to Active Facilitator. Prog. Lipid Res. 2018, 69, 11–20. [Google Scholar] [CrossRef]
- Takehara, M.; Sato, Y.; Kimura, T.; Noda, K.; Miyamoto, H.; Fujino, Y.; Miyoshi, J.; Nakamura, F.; Wada, H.; Bando, Y.; et al. Cancer-Associated Adipocytes Promote Pancreatic Cancer Progression through SAA1 Expression. Cancer Sci. 2020, 111, 2883–2894. [Google Scholar] [CrossRef]
- Kothari, C.; Diorio, C.; Durocher, F. The Importance of Breast Adipose Tissue in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 5760. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; He, S. Multi-faceted Role of Cancer-associated Adipocytes in the Tumor Microenvironment (Review). Mol. Med. Rep. 2021, 24, 866. [Google Scholar] [CrossRef] [PubMed]
- Jafari, N.; Kolla, M.; Meshulam, T.; Shafran, J.S.; Qiu, Y.; Casey, A.N.; Pompa, I.R.; Ennis, C.S.; Mazzeo, C.S.; Rabhi, N.; et al. Adipocyte-Derived Exosomes May Promote Breast Cancer Progression in Type 2 Diabetes. Sci. Signal. 2021, 14, eabj2807. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jung, W.H.; Koo, J.S. Adipocytes Can Induce Epithelial-Mesenchymal Transition in Breast Cancer Cells. Breast Cancer Res. Treat. 2015, 153, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, B.; Zhao, Y.; Tao, Z.; Wang, Y.; Chen, G.; Hu, X. Mammary Adipocytes Protect Triple-Negative Breast Cancer Cells from Ferroptosis. J. Hematol. Oncol. 2022, 15, 72. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; He, X.; Tong, C.; Li, H.; Xie, C.; Wu, Y.; Wang, L.; Yan, X.; Luo, D.; Tang, Y.; et al. Cancer-Associated Adipocytes Promote the Invasion and Metastasis in Breast Cancer through LIF/CXCLs Positive Feedback Loop. Int. J. Biol. Sci. 2022, 18, 1363–1380. [Google Scholar] [CrossRef]
- Jones, L.P.; Buelto, D.; Tago, E.; Owusu-Boaitey, K.E. Abnormal Mammary Adipose Tissue Environment of Brca1 Mutant Mice Show a Persistent Deposition of Highly Vascularized Multilocular Adipocytes. J. Cancer Sci. Ther. 2011. [Google Scholar] [CrossRef] [PubMed]
- Miran, I.; Scherer, D.; Ostyn, P.; Mazouni, C.; Drusch, F.; Bernard, M.; Louvet, E.; Adam, J.; Mathieu, M.-C.; Haffa, M.; et al. Adipose Tissue Properties in Tumor-Bearing Breasts. Front. Oncol. 2020, 10, 1506. [Google Scholar] [CrossRef]
- Koellensperger, E.; Bonnert, L.-C.; Zoernig, I.; Marmé, F.; Sandmann, S.; Germann, G.; Gramley, F.; Leimer, U. The Impact of Human Adipose Tissue-Derived Stem Cells on Breast Cancer Cells: Implications for Cell-Assisted Lipotransfers in Breast Reconstruction. Stem Cell Res. Ther. 2017, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; He, Y.; Yu, X. Bone Marrow Adipocyte: An Intimate Partner With Tumor Cells in Bone Metastasis. Front. Endocrinol. 2018, 9, 339. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Ceccaldi, R.; Shapiro, G.I.; D’Andrea, A.D. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015, 5, 1137–1154. [Google Scholar] [CrossRef]
- Torrisi, R.; Zuradelli, M.; Agostinetto, E.; Masci, G.; Losurdo, A.; De Sanctis, R.; Santoro, A. Platinum Salts in the Treatment of BRCA-Associated Breast Cancer: A True Targeted Chemotherapy? Crit. Rev. Oncol. Hematol. 2019, 135, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Giannone, G.; Scotto, G.; Katsaros, D.; De Giorgi, U.; Farolfi, A.; Borella, F.; Cosma, S.; Ferrero, A.; Mangiacotti, S.; Villa, M.; et al. Hypersensitivity to Platinum Salts According to BRCA Status in Ovarian Cancer: A Retrospective Analysis of Clinical Outcomes and Systematic Review of Literature. Gynecol. Oncol. 2021, 162, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Wattenberg, M.M.; Asch, D.; Yu, S.; O’Dwyer, P.J.; Domchek, S.M.; Nathanson, K.L.; Rosen, M.A.; Beatty, G.L.; Siegelman, E.S.; Reiss, K.A. Platinum Response Characteristics of Patients with Pancreatic Ductal Adenocarcinoma and a Germline BRCA1, BRCA2 or PALB2 Mutation. Br. J. Cancer 2020, 122, 333–339. [Google Scholar] [CrossRef]
- Byrski, T.; Huzarski, T.; Dent, R.; Gronwald, J.; Zuziak, D.; Cybulski, C.; Kladny, J.; Gorski, B.; Lubinski, J.; Narod, S.A. Response to Neoadjuvant Therapy with Cisplatin in BRCA1-Positive Breast Cancer Patients. Breast Cancer Res. Treat. 2009, 115, 359–363. [Google Scholar] [CrossRef]
- Maxwell, K.N.; Wubbenhorst, B.; Wenz, B.M.; De Sloover, D.; Pluta, J.; Emery, L.; Barrett, A.; Kraya, A.A.; Anastopoulos, I.N.; Yu, S.; et al. BRCA Locus-Specific Loss of Heterozygosity in Germline BRCA1 and BRCA2 Carriers. Nat. Commun. 2017, 8, 319. [Google Scholar] [CrossRef]
- Afghahi, A.; Timms, K.M.; Vinayak, S.; Jensen, K.C.; Kurian, A.W.; Carlson, R.W.; Chang, P.-J.; Schackmann, E.; Hartman, A.-R.; Ford, J.M.; et al. Tumor BRCA1 Reversion Mutation Arising during Neoadjuvant Platinum-Based Chemotherapy in Triple-Negative Breast Cancer Is Associated with Therapy Resistance. Clin. Cancer Res. 2017, 23, 3365–3370. [Google Scholar] [CrossRef] [PubMed]
- Pilié, P.G.; Tang, C.; Mills, G.B.; Yap, T.A. State-of-the-Art Strategies for Targeting the DNA Damage Response in Cancer. Nat. Rev. Clin. Oncol. 2019, 16, 81–104. [Google Scholar] [CrossRef]
- Mateo, J.; Lord, C.J.; Serra, V.; Tutt, A.; Balmaña, J.; Castroviejo-Bermejo, M.; Cruz, C.; Oaknin, A.; Kaye, S.B.; de Bono, J.S. A Decade of Clinical Development of PARP Inhibitors in Perspective. Ann. Oncol. 2019, 30, 1437–1447. [Google Scholar] [CrossRef]
- Bredow, K.; Blümcke, B.; Schneider, S.; Püsken, M.; Schmutzler, R.; Rhiem, K. Long-Term Survival of a BRCA2 Mutation Carrier Following Second Ovarian Cancer Relapse Using PARPi Therapy: A Case Report. Mol. Clin. Oncol. 2022, 17, 137. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yu, X. Comparison between Talazoparib and Conventional Chemotherapy in the Treatment of HER2-Positive Breast Cancer Patients: A Retrospective Study. Front. Immunol. 2022, 13, 901636. [Google Scholar] [CrossRef] [PubMed]
- Zong, D.; Adam, S.; Wang, Y.; Sasanuma, H.; Callén, E.; Murga, M.; Day, A.; Kruhlak, M.J.; Wong, N.; Munro, M.; et al. BRCA1 Haploinsufficiency Is Masked by RNF168-Mediated Chromatin Ubiquitylation. Mol. Cell 2019, 73, 1267–1281.e7. [Google Scholar] [CrossRef]
- Sullivan-Reed, K.; Bolton-Gillespie, E.; Dasgupta, Y.; Langer, S.; Siciliano, M.; Nieborowska-Skorska, M.; Hanamshet, K.; Belyaeva, E.A.; Bernhardy, A.J.; Lee, J.; et al. Simultaneous Targeting of PARP1 and RAD52 Triggers Dual Synthetic Lethality in BRCA-Deficient Tumor Cells. Cell Rep. 2018, 23, 3127–3136. [Google Scholar] [CrossRef]
- Kayumov, M.; Jia, L.; Pardaev, A.; Song, S.-S.; Mirzaakhmedov, S.; Ding, C.; Cheng, Y.-J.; Zhang, R.I.; Bao, X.; Miao, Z.-H.; et al. Design, Synthesis and Pharmacological Evaluation of New PARP1 Inhibitors by Merging Pharmacophores of Olaparib and the Natural Product Alantolactone. Eur. J. Med. Chem. 2022, 240, 114574. [Google Scholar] [CrossRef]
- Sinha, S.; Chatterjee, S.; Paul, S.; Das, B.; Dash, S.R.; Das, C.; Kundu, C.N. Olaparib Enhances the Resveratrol-Mediated Apoptosis in Breast Cancer Cells by Inhibiting the Homologous Recombination Repair Pathway. Exp. Cell Res. 2022, 420, 113338. [Google Scholar] [CrossRef]
- Kim, H.; Xu, H.; George, E.; Hallberg, D.; Kumar, S.; Jagannathan, V.; Medvedev, S.; Kinose, Y.; Devins, K.; Verma, P.; et al. Combining PARP with ATR Inhibition Overcomes PARP Inhibitor and Platinum Resistance in Ovarian Cancer Models. Nat. Commun. 2020, 11, 3726. [Google Scholar] [CrossRef]
- Xu, H.; Hurley, L.H. A First-in-Class Clinical G-Quadruplex-Targeting Drug. The Bench-to-Bedside Translation of the Fluoroquinolone QQ58 to CX-5461 (Pidnarulex). Bioorg. Med. Chem. Lett. 2022, 77, 129016. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.M.; Moldovan, G.-L. Mechanisms of PARP1 Inhibitor Resistance and Their Implications for Cancer Treatment. NAR Cancer 2022, 4, zcac042. [Google Scholar] [CrossRef] [PubMed]
- Mustafina, O.E. The Possible Roles of Human Alu Elements in Aging. Front. Genet. 2013, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.E.; White, T.B.; Streva, V.A.; DeFreece, C.B.; Hedges, D.J.; Deininger, P.L. The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair. PLoS Genet. 2015, 11, e1005016. [Google Scholar] [CrossRef]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Petit, S.A.; Ficarro, S.B.; Toomire, K.J.; Xie, A.; Lim, E.; Cao, S.A.; Park, E.; Eck, M.J.; Scully, R.; et al. PARP1-Driven Poly-ADP-Ribosylation Regulates BRCA1 Function in Homologous Recombination-Mediated DNA Repair. Cancer Discov. 2014, 4, 1430–1447. [Google Scholar] [CrossRef] [PubMed]
- Lodovichi, S.; Quadri, R.; Sertic, S.; Pellicioli, A. PARylation of BRCA1 Limits DNA Break Resection through BRCA2 and EXO1. Cell Rep. 2023, 42, 112060. [Google Scholar] [CrossRef]
- Tookman, L.A.; Browne, A.K.; Connell, C.M.; Bridge, G.; Ingemarsdotter, C.K.; Dowson, S.; Shibata, A.; Lockley, M.; Martin, S.A.; McNeish, I.A. RAD51 and BRCA2 Enhance Oncolytic Adenovirus Type 5 Activity in Ovarian Cancer. Mol. Cancer Res. 2016, 14, 44–55. [Google Scholar] [CrossRef]
- Reisländer, T.; Lombardi, E.P.; Groelly, F.J.; Miar, A.; Porru, M.; Di Vito, S.; Wright, B.; Lockstone, H.; Biroccio, A.; Harris, A.; et al. BRCA2 Abrogation Triggers Innate Immune Responses Potentiated by Treatment with PARP Inhibitors. Nat. Commun. 2019, 10, 3143. [Google Scholar] [CrossRef]
- Loizzi, V.; Dellino, M.; Cerbone, M.; Arezzo, F.; Cazzato, G.; Damiani, G.R.; Pinto, V.; Silvestris, E.; Kardhashi, A.; Cicinelli, E.; et al. The Role of Hormonal Replacement Therapy in BRCA Mutated Patients: Lights and Shadows. Int. J. Mol. Sci. 2023, 24, 764. [Google Scholar] [CrossRef]
- Singer, C.F. Nonsurgical Prevention Strategies in BRCA1 and BRCA2 Mutation Carriers. Breast Care 2021, 16, 144–148. [Google Scholar] [CrossRef]
- Nehme, R.; Diab-Assaf, M.; Decombat, C.; Delort, L.; Caldefie-Chezet, F. Targeting Adiponectin in Breast Cancer. Biomedicines 2022, 10, 2958. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of P53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef]
- Williamson, C.T.; Kubota, E.; Hamill, J.D.; Klimowicz, A.; Ye, R.; Muzik, H.; Dean, M.; Tu, L.; Gilley, D.; Magliocco, A.M.; et al. Enhanced Cytotoxicity of PARP Inhibition in Mantle Cell Lymphoma Harbouring Mutations in Both ATM and P53. EMBO Mol. Med. 2012, 4, 515–527. [Google Scholar] [CrossRef]
- Smeby, J.; Kryeziu, K.; Berg, K.C.G.; Eilertsen, I.A.; Eide, P.W.; Johannessen, B.; Guren, M.G.; Nesbakken, A.; Bruun, J.; Lothe, R.A.; et al. Molecular Correlates of Sensitivity to PARP Inhibition beyond Homologous Recombination Deficiency in Pre-Clinical Models of Colorectal Cancer Point to Wild-Type TP53 Activity. EBioMedicine 2020, 59, 102923. [Google Scholar] [CrossRef]
- Hong, T.; Lei, G.; Chen, X.; Li, H.; Zhang, X.; Wu, N.; Zhao, Y.; Zhang, Y.; Wang, J. PARP Inhibition Promotes Ferroptosis via Repressing SLC7A11 and Synergizes with Ferroptosis Inducers in BRCA-Proficient Ovarian Cancer. Redox Biol. 2021, 42, 101928. [Google Scholar] [CrossRef]
- Feng, Z.; Kachnic, L.; Zhang, J.; Powell, S.N.; Xia, F. DNA Damage Induces P53-Dependent BRCA1 Nuclear Export. J. Biol. Chem. 2004, 279, 28574–28584. [Google Scholar] [CrossRef]
- Parfenyev, S.; Singh, A.; Fedorova, O.; Daks, A.; Kulshreshtha, R.; Barlev, N.A. Interplay between P53 and Non-Coding RNAs in the Regulation of EMT in Breast Cancer. Cell Death Dis. 2021, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Barlev, N.A.; Sayan, B.S.; Candi, E.; Okorokov, A.L. The MicroRNA and P53 Families Join Forces against Cancer. Cell Death Differ. 2010, 17, 373–375. [Google Scholar] [CrossRef] [PubMed]
- Lezina, L.; Aksenova, V.; Fedorova, O.; Malikova, D.; Shuvalov, O.; Antonov, A.V.; Tentler, D.; Garabadgiu, A.V.; Melino, G.; Barlev, N.A. KMT Set7/9 Affects Genotoxic Stress Response via the Mdm2 Axis. Oncotarget 2015, 6, 25843–25855. [Google Scholar] [CrossRef]
- Portman, N.; Milioli, H.H.; Alexandrou, S.; Coulson, R.; Yong, A.; Fernandez, K.J.; Chia, K.M.; Halilovic, E.; Segara, D.; Parker, A.; et al. MDM2 Inhibition in Combination with Endocrine Therapy and CDK4/6 Inhibition for the Treatment of ER-Positive Breast Cancer. Breast Cancer Res. 2020, 22, 87. [Google Scholar] [CrossRef] [PubMed]
- Bulatov, E.; Sayarova, R.; Mingaleeva, R.; Miftakhova, R.; Gomzikova, M.; Ignatyev, Y.; Petukhov, A.; Davidovich, P.; Rizvanov, A.; Barlev, N.A. Isatin-Schiff Base-Copper (II) Complex Induces Cell Death in P53-Positive Tumors. Cell Death Discov. 2018, 4, 103. [Google Scholar] [CrossRef]
- Davidovich, P.; Aksenova, V.; Petrova, V.; Tentler, D.; Orlova, D.; Smirnov, S.; Gurzhiy, V.; Okorokov, A.L.; Garabadzhiu, A.; Melino, G.; et al. Discovery of Novel Isatin-Based P53 Inducers. ACS Med. Chem. Lett. 2015, 6, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, O.; Daks, A.; Petrova, V.; Petukhov, A.; Lezina, L.; Shuvalov, O.; Davidovich, P.; Kriger, D.; Lomert, E.; Tentler, D.; et al. Novel Isatin-Derived Molecules Activate P53 via Interference with Mdm2 to Promote Apoptosis. Cell Cycle 2018, 17, 1917–1930. [Google Scholar] [CrossRef]
- Gorodnova, T.V.; Sokolenko, A.P.; Kotiv, K.B.; Sokolova, T.N.; Ivantsov, A.O.; Guseynov, K.D.; Nekrasova, E.A.; Smirnova, O.A.; Berlev, I.V.; Imyanitov, E.N. Neoadjuvant Therapy of BRCA1-Driven Ovarian Cancer by Combination of Cisplatin, Mitomycin C and Doxorubicin. Hered. Cancer Clin. Pract. 2021, 19, 14. [Google Scholar] [CrossRef] [PubMed]
- Fasching, P.A.; Loibl, S.; Hu, C.; Hart, S.N.; Shimelis, H.; Moore, R.; Schem, C.; Tesch, H.; Untch, M.; Hilfrich, J.; et al. BRCA1/2 Mutations and Bevacizumab in the Neoadjuvant Treatment of Breast Cancer: Response and Prognosis Results in Patients With Triple-Negative Breast Cancer From the GeparQuinto Study. J. Clin. Oncol. 2018, 36, 2281–2287. [Google Scholar] [CrossRef] [PubMed]


| PARP Inhibitor | Cancer Type | Co-Target | Co-Treatment | Phase | Register Number |
|---|---|---|---|---|---|
| Olaparib | BC, OC, FTC, EndA, UCC | mTORC1/2 or AKT | Vistusertib or Capivasertib | 1b | NCT02208375 |
| Olaparib | OC, FTC, PPC | CTLA-4 | Tremelimumab | 2 | NCT02571725 |
| Talazoparib | TNBC | mTOR/PI3K | Gedatolisib | 2 | NCT03911973 |
| Olaparib | BC | CDK4,6 and HR | Palbociclib, Fulvestrant | 1 | NCT03685331 |
| Niraparib | FTC, OC, EndA, PPC | PI3K | Copanlisib | 1 | NCT03586661 |
| Olaparib | TNBC | PD-L1 | Durvalumab | 2 | NCT05209529 |
| Niraparib | PanC | PD-1 | Dostarlimab | 2 | NCT04493060 |
| Talazoparib | melanoma | PD-1 | Nivolumab | 2 | NCT04187833 |
| Niraparib | rare tumors | PD-1 | Sintilimab | 2 | NCT04423185 |
| Olaparib | BC | VEGFR or ATR | Cediranib or Ceralasertib | 2 | NCT04090567 |
| Fluzoparib | HER2- BC | VEGFR | Apatinib | 3 | NCT04296370 |
| Olaparib | OC, FTC, genital neoplasms | multiple receptor tyrosine kinases | Anlotinib | 2 | NCT04566952 |
| Olaparib | serous OC | ATR | Ceralasertib | 2 | NCT03462342 |
| Rucaparib | mesothelioma | — | — | 2 | NCT03654833 |
| Olaparib | Pt-resistant OC | CDK4,6 | Abemaciclib | 1/1b | NCT04633239 |
| Olaparib | prostate cancer | LRHL | Leuprolide | 2 | NCT05498272 |
| Talazoparib | OC and other | BRD2,3,4 | ZEN-3694 | 2 | NCT05327010 |
| Veliparib | BC | — | Temozolomide | 2 | NCT01009788 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loboda, A.P.; Adonin, L.S.; Zvereva, S.D.; Guschin, D.Y.; Korneenko, T.V.; Telegina, A.V.; Kondratieva, O.K.; Frolova, S.E.; Pestov, N.B.; Barlev, N.A. BRCA Mutations—The Achilles Heel of Breast, Ovarian and Other Epithelial Cancers. Int. J. Mol. Sci. 2023, 24, 4982. https://doi.org/10.3390/ijms24054982
Loboda AP, Adonin LS, Zvereva SD, Guschin DY, Korneenko TV, Telegina AV, Kondratieva OK, Frolova SE, Pestov NB, Barlev NA. BRCA Mutations—The Achilles Heel of Breast, Ovarian and Other Epithelial Cancers. International Journal of Molecular Sciences. 2023; 24(5):4982. https://doi.org/10.3390/ijms24054982
Chicago/Turabian StyleLoboda, Anna P., Leonid S. Adonin, Svetlana D. Zvereva, Dmitri Y. Guschin, Tatyana V. Korneenko, Alexandra V. Telegina, Olga K. Kondratieva, Sofia E. Frolova, Nikolay B. Pestov, and Nick A. Barlev. 2023. "BRCA Mutations—The Achilles Heel of Breast, Ovarian and Other Epithelial Cancers" International Journal of Molecular Sciences 24, no. 5: 4982. https://doi.org/10.3390/ijms24054982
APA StyleLoboda, A. P., Adonin, L. S., Zvereva, S. D., Guschin, D. Y., Korneenko, T. V., Telegina, A. V., Kondratieva, O. K., Frolova, S. E., Pestov, N. B., & Barlev, N. A. (2023). BRCA Mutations—The Achilles Heel of Breast, Ovarian and Other Epithelial Cancers. International Journal of Molecular Sciences, 24(5), 4982. https://doi.org/10.3390/ijms24054982







