Long Non-Coding RNA lncWOX11a Suppresses Adventitious Root Formation of Poplar by Regulating the Expression of PeWOX11a
Abstract
1. Introduction
2. Results
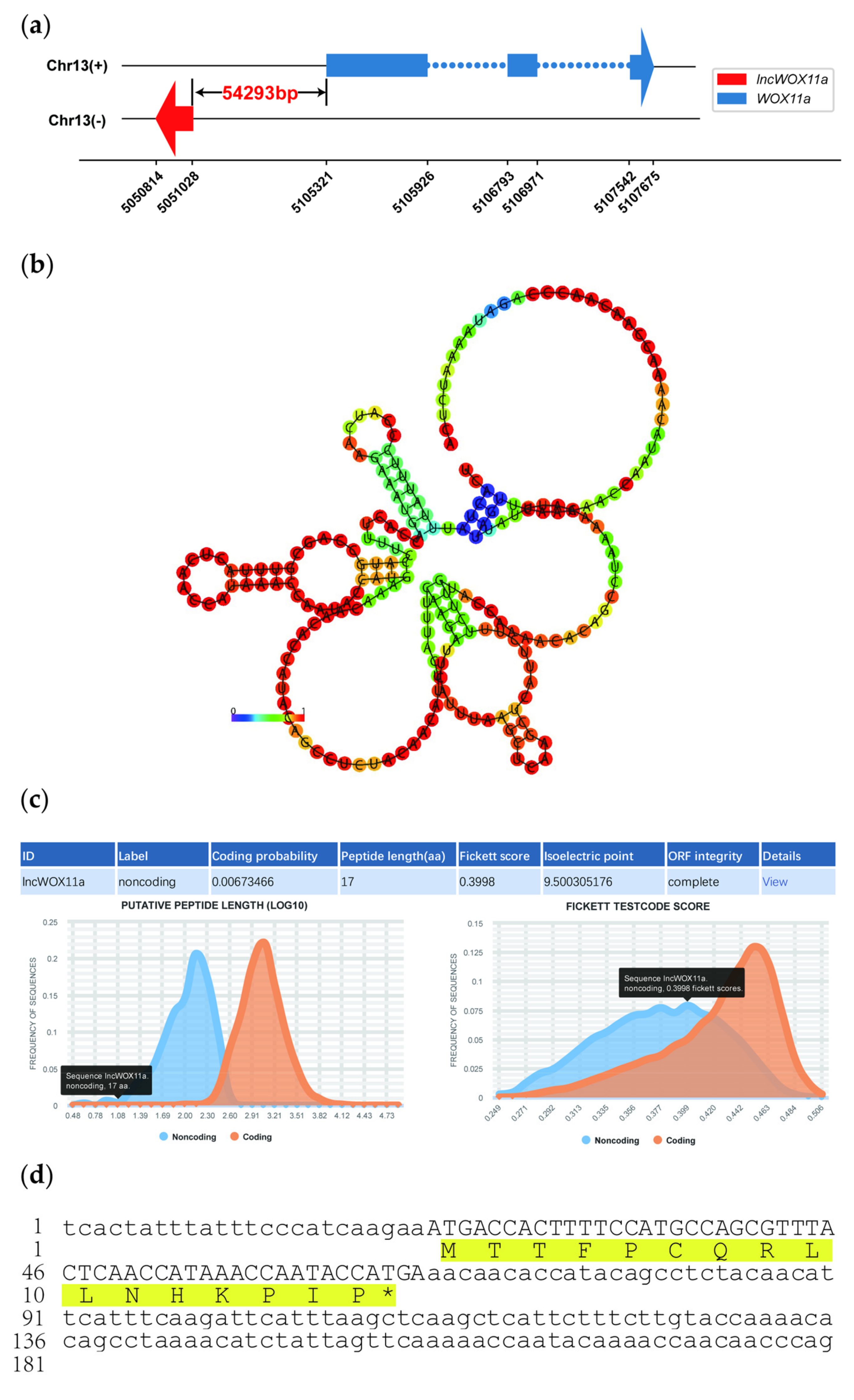
2.1. Isolation and Sequence Analysis of lncWOX11a
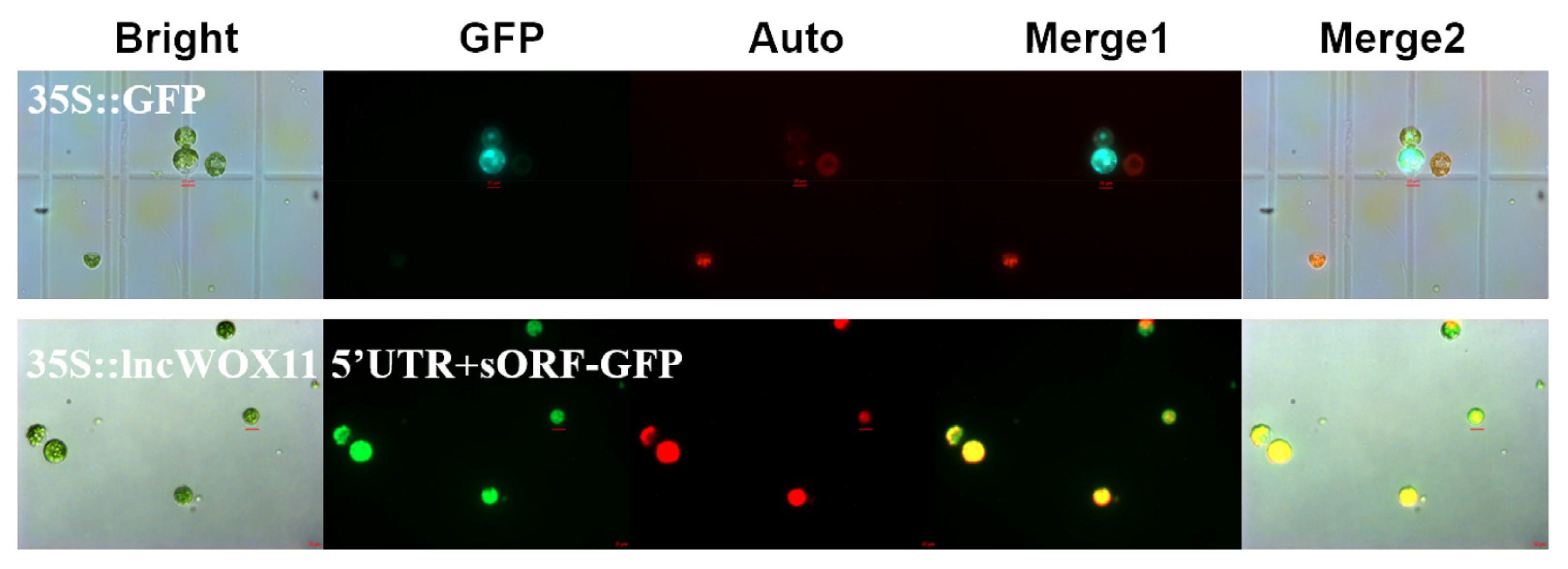
2.2. Verification of the Coding Ability of lncWOX11a
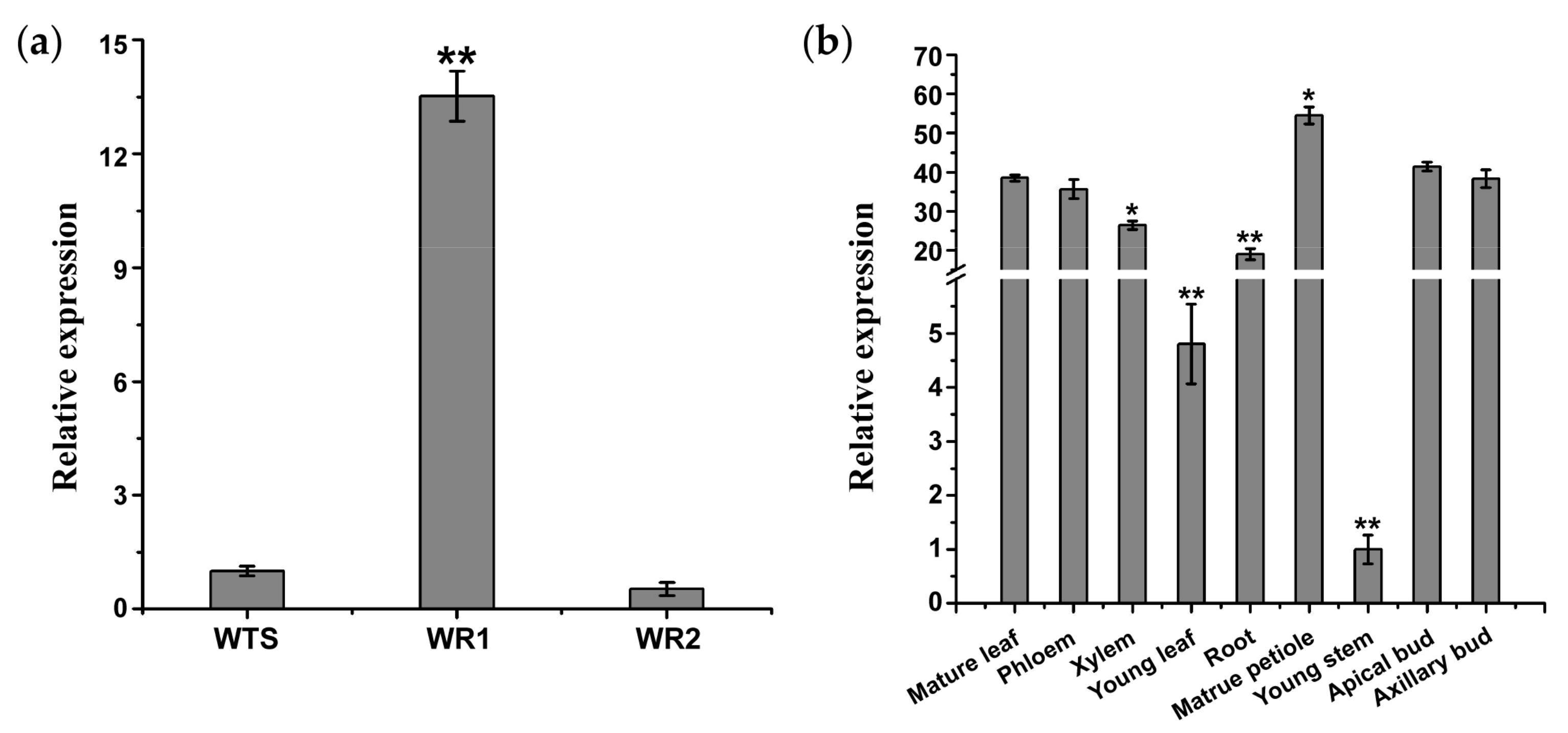
2.3. Organ-Specific Expression of lncWOX11a
2.4. The Overexpression of lncWOX11a Inhibits AR Development
2.5. The cis-Regulation Module for lncWOX11a
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cloning and Sequence Analysis
4.3. Transient Expression in Poplar Protoplasts
4.4. Quantitative Real-Time Quantification
4.5. Generation of Transgenic Poplar Lines
4.6. Target Gene Prediction
4.7. Construction of the pYLCRISPR/Cas9-lncWOX11a Vector
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Legué, V.; Rigal, A.; Bhalerao, R. Adventitious root formation in tree species: Involvement of transcription factors. Physiol. Plant. 2014, 151, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Dickmann, D.; Kuzovkina, J. Poplars and willows of the world, with emphasis on silviculturally important species. In Poplars and Willows: Trees for Society and the Environment; CABI: Wallingford, UK, 2014; pp. 8–91. [Google Scholar]
- Lin, Y.C.; Wang, J.; Delhomme, N.; Schiffthaler, B.; Sundstrom, G.; Zuccolo, A.; Nystedt, B.; Hvidsten, T.R.; de la Torre, A.; Cossu, R.M.; et al. Functional and evolutionary genomic inferences in Populus through genome and population sequencing of American and European aspen. Proc. Natl. Acad. Sci. USA 2018, 115, E10970–E10978. [Google Scholar] [CrossRef]
- Stettler, R.F.; Bradshaw, H.D.; Heilman, P.E.; Hinckley, T.M. Biology of Populus and Its Implications for Management and Conservation; NRC Research Press: Ottawa, ON, Canada, 1996. [Google Scholar]
- Zhai, S.; Cai, W.; Xiang, Z.-X.; Chen, C.-Y.; Lu, Y.-T.; Yuan, T.-T. PIN3-mediated auxin transport contributes to blue light-induced adventitious root formation in Arabidopsis. Plant Sci. 2021, 312, 111044. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Tayengwa, R.; Cheng, Z.M.; Peer, W.A.; Murphy, A.S.; Zhao, M. Auxin regulates adventitious root formation in tomato cuttings. BMC Plant Biol. 2019, 19, 435. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, D.; Meng, Y.; Li, K.; Wang, H.; Han, M. Inhibition of adventitious root development in apple rootstocks by cytokinin is based on its suppression of adventitious root primordia formation. Physiol. Plant. 2019, 166, 663–676. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, C.; Wang, N.; Wei, L.; Li, W.; Yao, Y.; Liao, W. Roles of Small-Molecule Compounds in Plant Adventitious Root Development. Biomolecules 2019, 9, 420. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, S.; Ruiz-Cano, H.; Fernandez, M.A.; Sanchez-Garcia, A.B.; Villanova, J.; Micol, J.L.; Perez-Perez, J.M. A Network-Guided Genetic Approach to Identify Novel Regulators of Adventitious Root Formation in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 461. [Google Scholar] [CrossRef]
- Lin, C.; Sauter, M. Polar Auxin Transport Determines Adventitious Root Emergence and Growth in Rice. Front. Plant Sci. 2019, 10, 444. [Google Scholar] [CrossRef]
- Singh, Z.; Singh, H.; Garg, T.; Mushahary, K.K.K.; Yadav, S.R. Genetic and Hormonal Blueprint of Shoot-Borne Adventitious Root Development in Rice and Maize. Plant Cell Physiol. 2022, 63, 1806–1813. [Google Scholar] [CrossRef]
- Steffens, B.; Rasmussen, A. The Physiology of Adventitious Roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef]
- Lorbiecke, R.; Sauter, M. Adventitious Root Growth and Cell-Cycle Induction in Deepwater Rice1. Plant Physiol. 1999, 119, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Druege, U.; Hilo, A.; Perez-Perez, J.M.; Klopotek, Y.; Acosta, M.; Shahinnia, F.; Zerche, S.; Franken, P.; Hajirezaei, M.R. Molecular and physiological control of adventitious rooting in cuttings: Phytohormone action meets resource allocation. Ann. Bot. 2019, 123, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Wan, Q.; Zhai, N.; Xie, D.; Liu, W.; Xu, L. WOX11: The founder of plant organ regeneration. Cell. Regen. 2023, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Baesso, B.; Chiatante, D.; Terzaghi, M.; Zenga, D.; Nieminen, K.; Mahonen, A.P.; Siligato, R.; Helariutta, Y.; Scippa, G.S.; Montagnoli, A. Transcription factors PRE3 and WOX11 are involved in the formation of new lateral roots from secondary growth taproot in A. thaliana. Plant Biol. 2018, 20, 426–432. [Google Scholar] [CrossRef]
- Zhang, T.; Li, R.; Xing, J.; Yan, L.; Wang, R.; Zhao, Y. The YUCCA-Auxin-WOX11 Module Controls Crown Root Development in Rice. Front. Plant Sci. 2018, 9, 523. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, Y.; Chen, Z.; Yue, Y.; Huang, H.; Wu, B.; Liu, Y.; Zhou, D.X.; Zhao, Y. ROS-stimulated Protein Lysine Acetylation Is Required for Crown Root Development in Rice. J. Adv. Res. 2022, in press. [Google Scholar] [CrossRef]
- Xu, M.; Xie, W.; Huang, M. Two WUSCHEL-related HOMEOBOX genes, PeWOX11a and PeWOX11b, are involved in adventitious root formation of poplar. Physiol. Plant 2015, 155, 446–456. [Google Scholar] [CrossRef]
- Liu, R.; Wen, S.-S.; Sun, T.-T.; Wang, R.; Zuo, W.-T.; Yang, T.; Wang, C.; Hu, J.-J.; Lu, M.-Z.; Wang, L.-Q. PagWOX11/12a positively regulates the PagSAUR36 gene that enhances adventitious root development in poplar. J. Exp. Bot. 2022, 73, 7298–7311. [Google Scholar] [CrossRef]
- Baesso, B.; Terzaghi, M.; Chiatante, D.; Scippa, G.S.; Montagnoli, A. WOX genes expression during the formation of new lateral roots from secondary structures in Populus nigra (L.) taproot. Sci. Rep. 2020, 10, 18890. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, K.; Yu, R.; Zhou, B.; Huang, P.; Cao, Z.; Zhou, Y.; Wang, J. From “Dark Matter” to “Star”: Insight Into the Regulation Mechanisms of Plant Functional Long Non-Coding RNAs. Front. Plant Sci. 2021, 12, 650926. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding Transcription by Alternative RNA Polymerases Dynamically Regulates an Auxin-Driven Chromatin Loop. Mol. Cell 2014, 55, 383–396. [Google Scholar] [CrossRef]
- Herr, A.J.; Jensen, M.B.; Dalmay, T.; Baulcombe, D.C. RNA polymerase IV directs silencing of endogenous DNA. Science 2005, 308, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Zou, X.; Ali, F.; Jin, S.; Li, F.; Wang, Z. RNA-Seq with a novel glabrous-ZM24fl reveals some key lncRNAs and the associated targets in fiber initiation of cotton. BMC Plant Biol. 2022, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, J.; Wang, C.; Xu, Y.; Li, X.; Yang, J.; Chen, K.; Kang, Y.; Wang, Y.; Cao, P.; et al. Comprehensive Analysis of Long Non-coding RNA Modulates Axillary Bud Development in Tobacco (Nicotiana tabacum L.). Front. Plant Sci. 2022, 13, 809435. [Google Scholar] [CrossRef]
- Wang, H.; Chu, Z.; Chang, S.; Jia, S.; Pang, L.; Xi, C.; Liu, J.; Zhao, H.; Wang, Y.; Han, S. Transcriptomic identification of long noncoding RNAs and their hormone-associated nearby coding genes involved in the differential development of caryopses localized on different branches in rice. J. Plant Physiol. 2022, 271, 153663. [Google Scholar] [CrossRef]
- Tian, J.; Zhang, G.; Zhang, F.; Ma, J.; Wen, C.; Li, H. Genome-Wide Identification of Powdery Mildew Responsive Long Non-Coding RNAs in Cucurbita pepo. Front. Genet. 2022, 13, 933022. [Google Scholar] [CrossRef]
- Liu, J.; Chen, S.; Liu, M.; Chen, Y.; Fan, W.; Lee, S.; Xiao, H.; Kudrna, D.; Li, Z.; Chen, X.; et al. Full-Length Transcriptome Sequencing Reveals Alternative Splicing and lncRNA Regulation during Nodule Development in Glycine max. Int. J. Mol. Sci. 2022, 23, 7371. [Google Scholar] [CrossRef]
- Xuhui, L.; Weiwei, C.; Siqi, L.; Junteng, F.; Hang, Z.; Xiangbo, Z.; Yongwen, Q. Full-length transcriptome analysis of maize root tips reveals the molecular mechanism of cold stress during the seedling stage. BMC Plant Biol. 2022, 22, 398. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Fang, M.; Li, Z.; Zhang, M.; Liu, X.; Peng, Y.; Wan, Y.; Chen, J. Third-Generation Sequencing Reveals LncRNA-Regulated HSP Genes in the Populus × canadensis Moench Heat Stress Response. Front. Genet. 2020, 11, 249. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chen, P.; Liu, P.; Bu, C.; Zhang, D. High-Temperature-Responsive Poplar lncRNAs Modulate Target Gene Expression via RNA Interference and Act as RNA Scaffolds to Enhance Heat Tolerance. Int. J. Mol. Sci. 2020, 21, 6808. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, L.Y.; Chen, X.; Shi, W.G.; Deng, S.R.; Luo, Z.B. Genome-Wide Identification and Characterization of Long Noncoding RNAs in Populus × canescens Roots Treated With Different Nitrogen Fertilizers. Front. Plant Sci. 2022, 13, 890453. [Google Scholar] [CrossRef]
- Lu, Y.; Deng, S.; Li, Z.; Wu, J.; Liu, Q.; Liu, W.; Yu, W.J.; Zhang, Y.; Shi, W.; Zhou, J.; et al. Competing Endogenous RNA Networks Underlying Anatomical and Physiological Characteristics of Poplar Wood in Acclimation to Low Nitrogen Availability. Plant Cell Physiol. 2019, 60, 2478–2495. [Google Scholar] [CrossRef]
- Xu, H.; Chen, B.; Zhao, Y.; Guo, Y.; Liu, G.; Li, R.; Zeisler-Diehl, V.V.; Chen, Y.; He, X.; Schreiber, L.; et al. Non-Coding RNA Analyses of Seasonal Cambium Activity in Populus tomentosa. Cells 2022, 11, 640. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, H.; Chen, B.; Grünhofer, P.; Schreiber, L.; Lin, J.; Zhao, Y. Genome-wide analysis of long non-coding RNAs in shoot apical meristem and vascular cambium in Populus tomentosa. J. Plant Physiol. 2022, 275, 153759. [Google Scholar] [CrossRef]
- Chen, P.; Song, Y.; Liu, X.; Xiao, L.; Bu, C.; Liu, P.; Zhao, L.; Ingvarsson, P.K.; Wu, H.X.; El-Kassaby, Y.A.; et al. LncRNA PMAT-PtoMYB46 module represses PtoMATE and PtoARF2 promoting Pb(2+) uptake and plant growth in poplar. J. Hazard Mater. 2022, 433, 128769. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Z.; Xu, M. Identification and characterization of long non-coding RNAs involved in the formation and development of poplar adventitious roots. Ind. Crops Prod. 2018, 118, 334–346. [Google Scholar] [CrossRef]
- Heo Jae, B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Wang, L.Q.; Wen, S.S.; Wang, R.; Wang, C.; Gao, B.; Lu, M.Z. PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant Biotechnol. J. 2021, 19, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Wang, Y.; Liu, Y.; Wang, G.; She, X. Reactive oxygen species regulate auxin levels to mediate adventitious root induction in Arabidopsis hypocotyl cuttings. J. Integr. Plant Biol. 2020, 62, 912–926. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, B.K.; Singh, S.; Tripathi, D.K.; Sharma, S.; Prasad, S.M.; Chauhan, D.K.; Kumar, V.; Singh, V.P. New adventitious root formation and primary root biomass accumulation are regulated by nitric oxide and reactive oxygen species in rice seedlings under arsenate stress. J. Hazard. Mater. 2019, 361, 134–140. [Google Scholar] [CrossRef]
- Chekanova, J.A.; Gregory, B.D.; Reverdatto, S.V.; Chen, H.; Kumar, R.; Hooker, T.; Yazaki, J.; Li, P.; Skiba, N.; Peng, Q.; et al. Genome-Wide High-Resolution Mapping of Exosome Substrates Reveals Hidden Features in the Arabidopsis Transcriptome. Cell 2007, 131, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Froberg, J.E.; Lee, J.T. Long noncoding RNAs: Fresh perspectives into the RNA world. Trends Biochem. Sci. 2014, 39, 35–43. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Chua, N.H. Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 2015, 13, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Dilokpimol, A.; Geshi, N. Arabidopsis thaliana glucuronosyltransferase in family GT14. Plant Signal Behav. 2014, 9, e28891. [Google Scholar] [CrossRef]
- Ye, C.-Y.; Li, T.; Tuskan, G.A.; Tschaplinski, T.J.; Yang, X. Comparative analysis of GT14/GT14-like gene family in Arabidopsis, Oryza, Populus, Sorghum and Vitis. Plant Sci. 2011, 181, 688–695. [Google Scholar] [CrossRef]
- Tan, B.; Xu, M.; Chen, Y.; Huang, M. Transient expression for functional gene analysis using Populus protoplasts. Plant Cell Tissue Organ Cult. 2013, 114, 11–18. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bruegmann, T.; Polak, O.; Deecke, K.; Nietsch, J.; Fladung, M. Poplar Transformation. In Transgenic Plants: Methods and Protocols; Kumar, S., Barone, P., Smith, M., Eds.; Springer: New York, NY, USA, 2019; pp. 165–177. [Google Scholar]
- Eck, J.V.; Kirk, D.D.; Walmsley, A.M. Tomato (Lycopersicum esculentum). In Agrobacterium Protocols; Wang, K., Ed.; Humana Press: Totowa, NJ, USA, 2006; pp. 459–474. [Google Scholar]
- Zhu, Q.; Liu, Y.-G.; Ma, X.; Zeng, D.; Xie, X. A protocol for CRISPR/Cas9-based multi-gene editing and sequence decoding of mutant sites in plants. Sci. Sin. Vitae 2018, 48, 783–794. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, Y.-G. CRISPR/Cas9-Based Multiplex Genome Editing in Monocot and Dicot Plants. Curr. Protoc. Mol. Biol. 2016, 115, 31.36.1–31.36.21. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ran, N.; Liu, S.; Qi, H.; Wang, J.; Shen, T.; Xu, W.; Xu, M. Long Non-Coding RNA lncWOX11a Suppresses Adventitious Root Formation of Poplar by Regulating the Expression of PeWOX11a. Int. J. Mol. Sci. 2023, 24, 5766. https://doi.org/10.3390/ijms24065766
Ran N, Liu S, Qi H, Wang J, Shen T, Xu W, Xu M. Long Non-Coding RNA lncWOX11a Suppresses Adventitious Root Formation of Poplar by Regulating the Expression of PeWOX11a. International Journal of Molecular Sciences. 2023; 24(6):5766. https://doi.org/10.3390/ijms24065766
Chicago/Turabian StyleRan, Na, Sian Liu, Haoran Qi, Jiali Wang, Tengfei Shen, Wenlin Xu, and Meng Xu. 2023. "Long Non-Coding RNA lncWOX11a Suppresses Adventitious Root Formation of Poplar by Regulating the Expression of PeWOX11a" International Journal of Molecular Sciences 24, no. 6: 5766. https://doi.org/10.3390/ijms24065766
APA StyleRan, N., Liu, S., Qi, H., Wang, J., Shen, T., Xu, W., & Xu, M. (2023). Long Non-Coding RNA lncWOX11a Suppresses Adventitious Root Formation of Poplar by Regulating the Expression of PeWOX11a. International Journal of Molecular Sciences, 24(6), 5766. https://doi.org/10.3390/ijms24065766





