Downregulation of BUD31 Promotes Prostate Cancer Cell Proliferation and Migration via Activation of p-AKT and Vimentin In Vitro
Abstract
1. Introduction
2. Results
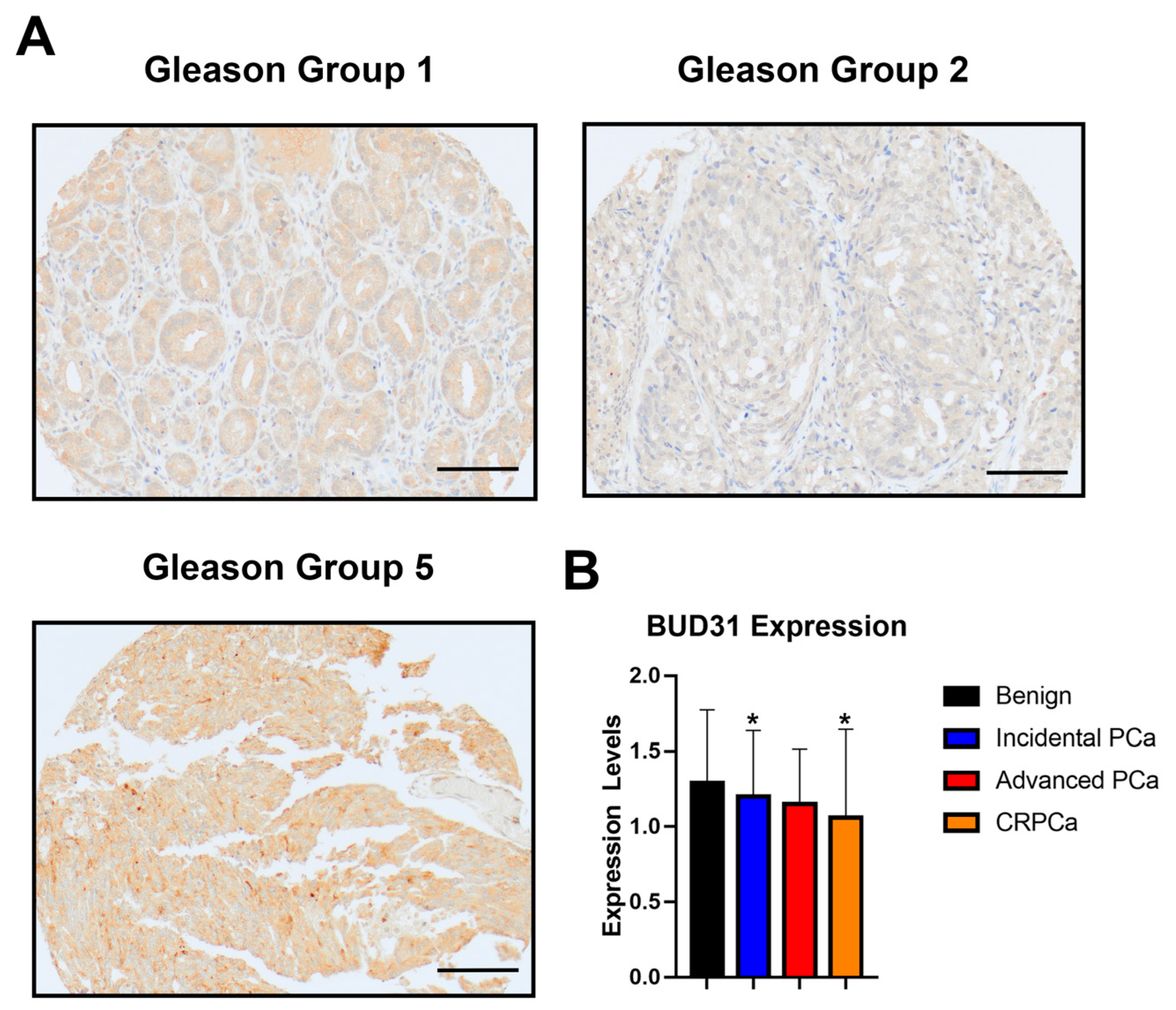
2.1. BUD31 Expression in Clinical Cohort with Prostate Cancer
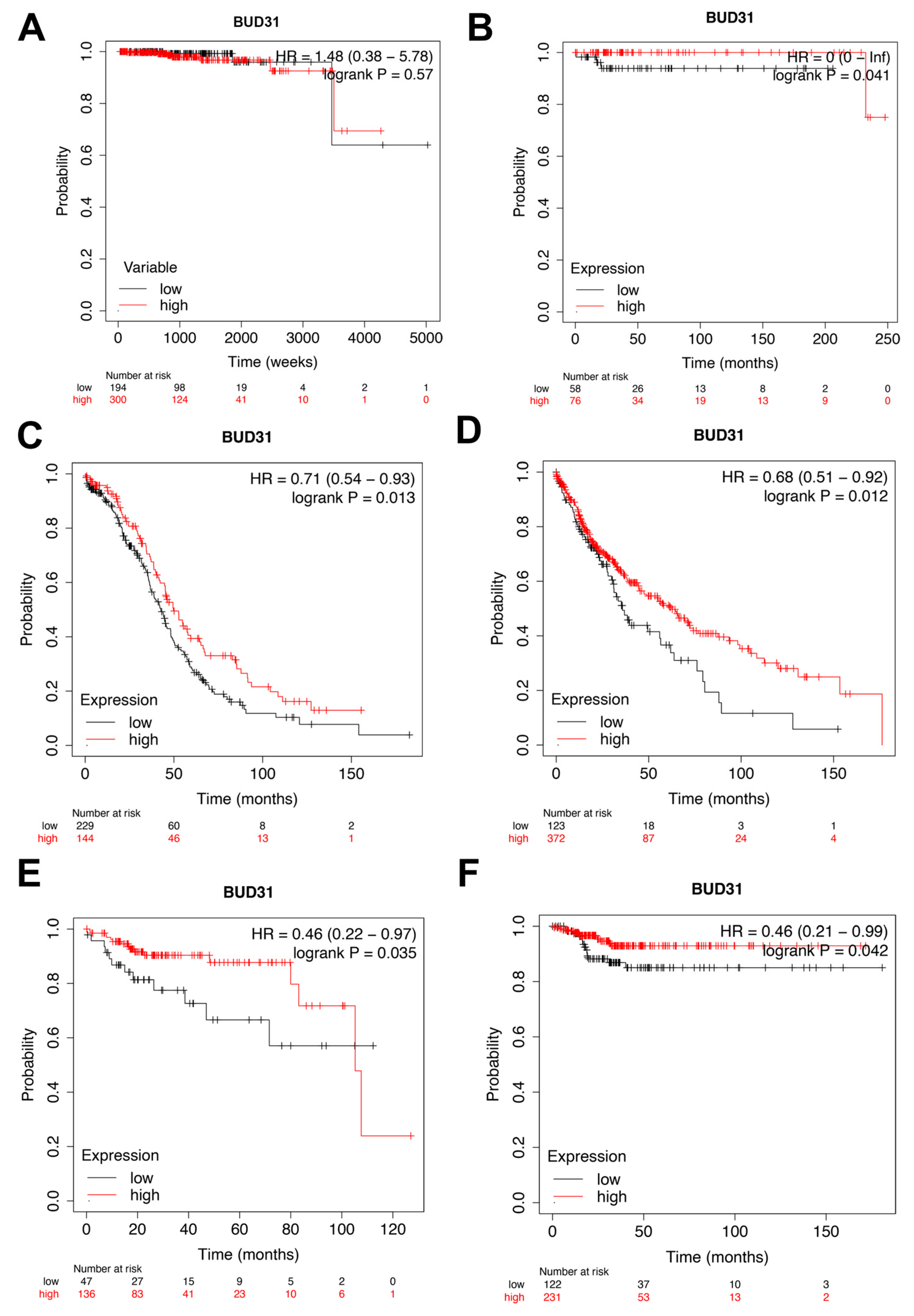
2.2. Low Expression of BUD31 Is Associated with Worse Overall Survival and Return Free Survival
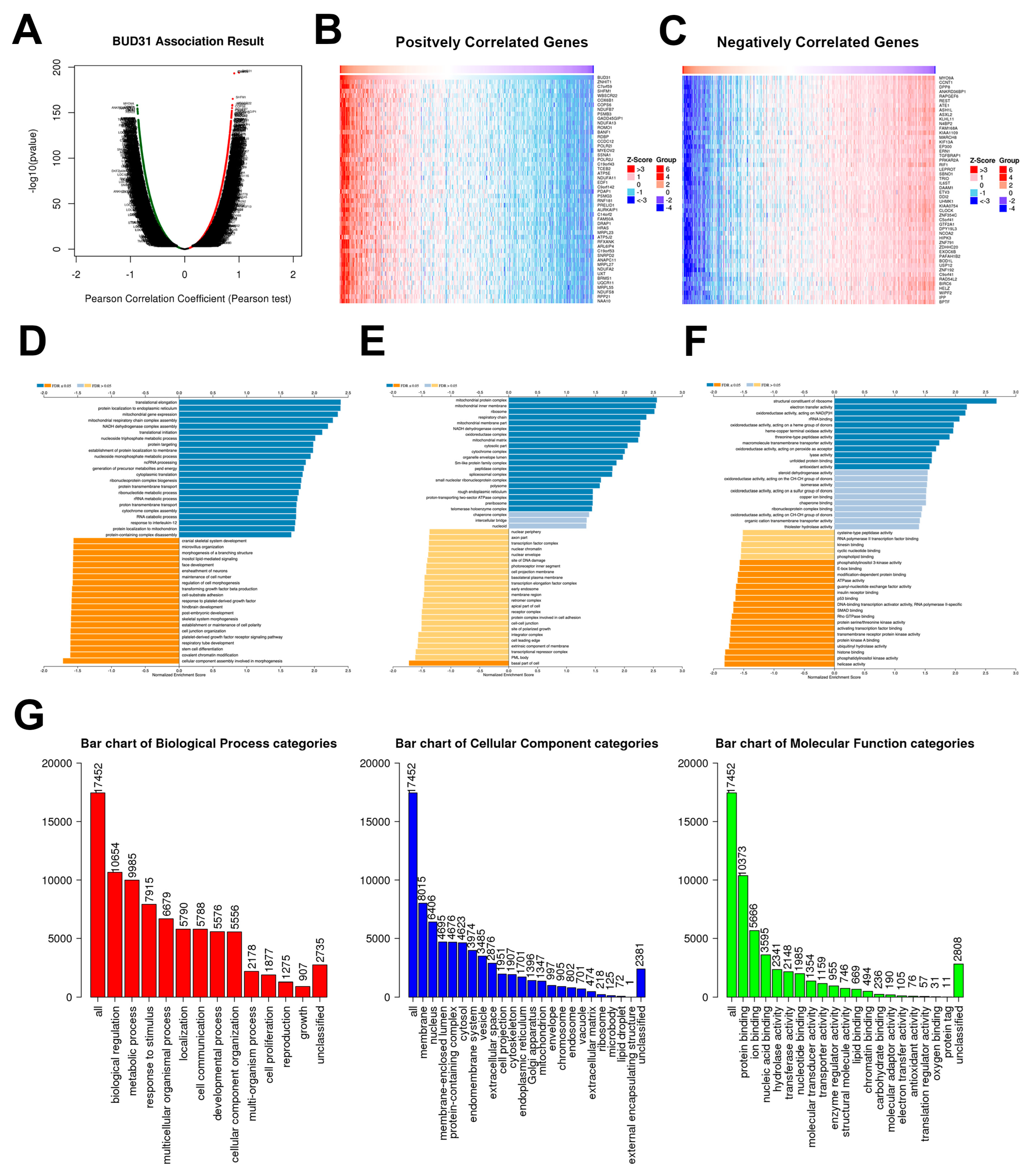
2.3. BUD31 Gene Set Enrichment Analysis in TCGA PRAD Database
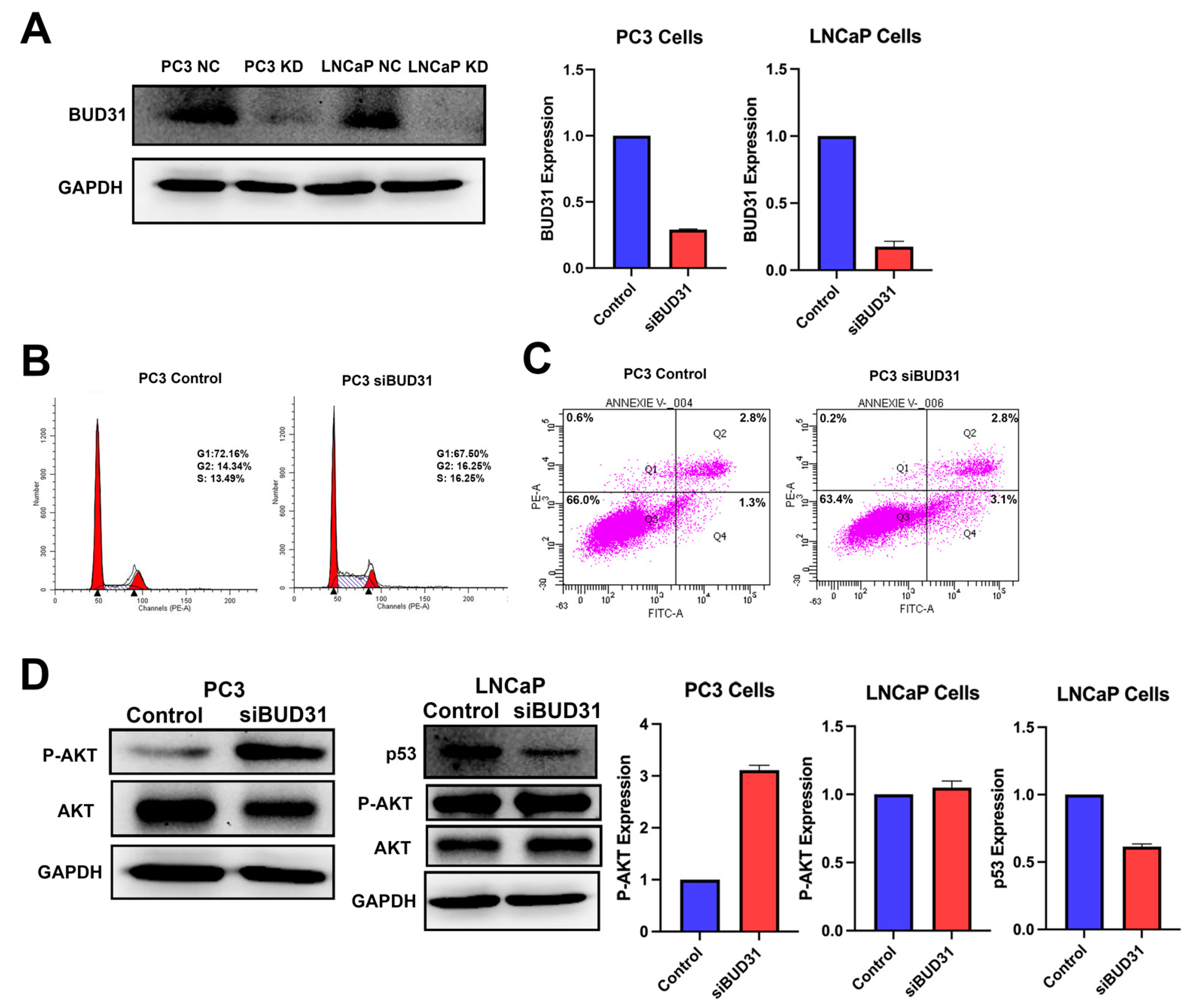
2.4. BUD31 Regulates Cell Cycle Progression of Prostate Cancer Cells In Vitro
2.5. Inhibition of BUD31 Enhances the Migration and Invasion of PCa
3. Discussion
4. Material and Methods
4.1. Study Population and Pathological Analysis
4.2. Immunohistochemistry (IHC)
4.3. TCGA PRAD Data Analysis
4.4. Cell Lines
4.5. Cell Line Transfection and RNA Silencing
4.6. Western Blot
4.7. Migration and Invasion Assay
4.8. Flow Cytometry
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PCa | Prostate Cancer |
| PRAD | Prostate Adenocarcinoma |
| CRPCa | Castrate Resistant PCa |
| BUD31 | Functional Spliceosome-Associated Protein 17 |
| ERG | Ets-Related Gene |
| PTEN | Phosphatase Tensin Homologue |
| goi | Gene of Interest |
| TCGA | The Cancer Genome Atlas |
| AR | Androgen Receptor |
| GSEA | Gene Set Enrichment Analysis |
| IHC | Immunohistochemistry |
| TMA | Tissue Micro-Array |
| FFPE | Formalin-Fixed Paraffin-Embedded |
| GO | Gene Ontology |
| OS | Overall Survival |
| RFS | Return-Free Survival |
References
- Wang, L.; Lu, B.; He, M.; Wang, Y.; Wang, Z.; Du, L. Prostate Cancer Incidence and Mortality: Global Status and Temporal Trends in 89 Countries From 2000 to 2019. Front. Public Health 2022, 10, 811044. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Armstrong, A.J.; Dehm, S.M.; Luo, J. Androgen receptor variant-driven prostate cancer: Clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis. 2016, 19, 231–241. [Google Scholar] [CrossRef]
- Aparicio, A.M.; Harzstark, A.L.; Corn, P.G.; Wen, S.; Araujo, J.C.; Tu, S.M.; Pagliaro, L.C.; Kim, J.; Millikan, R.E.; Ryan, C.; et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin. Cancer Res. 2013, 19, 3621–3630. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, C.E.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.P.; White, T.A.; Stojanov, P.; Van Allen, E.; Stransky, N.; et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012, 44, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Wang, S.; Qiao, R.; Vivanco, I.; Watson, P.A.; Sawyers, C.L.; Wu, H. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer Res. 2007, 67, 6083–6091. [Google Scholar] [CrossRef]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Tran, S.; Boissier, R.; Perrin, J.; Karsenty, G.; Lechevallier, E. Review of the Different Treatments and Management for Prostate Cancer and Fertility. Urology 2015, 86, 936–941. [Google Scholar] [CrossRef]
- Potter, S.R.; Epstein, J.I.; Partin, A.W. Seminal vesicle invasion by prostate cancer: Prognostic significance and therapeutic implications. Rev. Urol. 2000, 2, 190–195. [Google Scholar]
- Hyun, J.S. Prostate cancer and sexual function. World J. Mens. Health 2012, 30, 99–107. [Google Scholar] [CrossRef]
- Lee, S.C.; Abdel-Wahab, O. Therapeutic targeting of splicing in cancer. Nat. Med. 2016, 22, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Shilo, A.; Siegfried, Z.; Karni, R. The role of splicing factors in deregulation of alternative splicing during oncogenesis and tumor progression. Mol. Cell Oncol. 2015, 2, e970955. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Climente-Gonzalez, H.; Porta-Pardo, E.; Godzik, A.; Eyras, E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017, 20, 2215–2226. [Google Scholar] [CrossRef]
- Bertram, K.; Agafonov, D.E.; Liu, W.T.; Dybkov, O.; Will, C.L.; Hartmuth, K.; Urlaub, H.; Kastner, B.; Stark, H.; Luhrmann, R. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 2017, 542, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.L.; Liu, J.S.; Wu, P.L.; Guan, H.H.; Chen, Y.L.; Lin, A.C.; Ting, H.J.; Pang, S.T.; Yeh, S.D.; Ma, W.L.; et al. Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis. Mol. Oncol. 2014, 8, 1575–1587. [Google Scholar] [CrossRef] [PubMed]
- Alderton, G.K. MYC: Splicing up your survival. Nat. Rev. Cancer 2015, 15, 574–575. [Google Scholar] [CrossRef]
- Masciadri, B.; Areces, L.B.; Carpinelli, P.; Foiani, M.; Draetta, G.; Fiore, F. Characterization of the BUD31 gene of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2004, 320, 1342–1350. [Google Scholar] [CrossRef]
- Yang, H.; Beutler, B.; Zhang, D. Emerging roles of spliceosome in cancer and immunity. Protein Cell 2022, 13, 559–579. [Google Scholar] [CrossRef]
- Yan, C.; Wan, R.; Shi, Y. Molecular Mechanisms of pre-mRNA Splicing through Structural Biology of the Spliceosome. Cold Spring Harb. Perspect. Biol. 2019, 11, a032409. [Google Scholar] [CrossRef]
- Saha, D.; Khandelia, P.; O’Keefe, R.T.; Vijayraghavan, U. Saccharomyces cerevisiae NineTeen complex (NTC)-associated factor Bud31/Ycr063w assembles on precatalytic spliceosomes and improves first and second step pre-mRNA splicing efficiency. J. Biol. Chem. 2012, 287, 5390–5399. [Google Scholar] [CrossRef]
- Saha, D.; Banerjee, S.; Bashir, S.; Vijayraghavan, U. Context dependent splicing functions of Bud31/Ycr063w define its role in budding and cell cycle progression. Biochem. Biophys. Res. Commun. 2012, 424, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Kilpinen, S.; Ruusulehto, A.; Lothe, R.A.; Skotheim, R.I. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016, 35, 2413–2427. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.Y.; Simon, L.M.; Neill, N.J.; Marcotte, R.; Sayad, A.; Bland, C.S.; Echeverria, G.V.; Sun, T.; Kurley, S.J.; Tyagi, S.; et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 2015, 525, 384–388. [Google Scholar] [CrossRef]
- Ozaki, T.; Nakagawara, A. Role of p53 in Cell Death and Human Cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef]
- Chang, F.; Lee, J.T.; Navolanic, P.M.; Steelman, L.S.; Shelton, J.G.; Blalock, W.L.; Franklin, R.A.; McCubrey, J.A. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: A target for cancer chemotherapy. Leukemia 2003, 17, 590–603. [Google Scholar] [CrossRef] [PubMed]
- Caspari, T. How to activate p53. Curr. Biol. 2000, 10, R315–R317. [Google Scholar] [CrossRef]
- Flatt, P.M.; Tang, L.J.; Scatena, C.D.; Szak, S.T.; Pietenpol, J.A. p53 regulation of G(2) checkpoint is retinoblastoma protein dependent. Mol. Cell Biol. 2000, 20, 4210–4223. [Google Scholar] [CrossRef]
- Wu, S.; Du, Y.; Beckford, J.; Alachkar, H. Upregulation of the EMT marker vimentin is associated with poor clinical outcome in acute myeloid leukemia. J. Transl. Med. 2018, 16, 170. [Google Scholar] [CrossRef]
- Wei, J.; Xu, G.; Wu, M.; Zhang, Y.; Li, Q.; Liu, P.; Zhu, T.; Song, A.; Zhao, L.; Han, Z.; et al. Overexpression of vimentin contributes to prostate cancer invasion and metastasis via src regulation. Anticancer Res. 2008, 28, 327–334. [Google Scholar]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. Linkedomics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Brenner, J.C.; Sabolch, A.; Jackson, W.; Speers, C.; Wilder-Romans, K.; Knudsen, K.E.; Lawrence, T.S.; Chinnaiyan, A.M.; Feng, F.Y. Targeted radiosensitization of ETS fusion-positive prostate cancer through PARP1 inhibition. Neoplasia 2013, 15, 1207–1217. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choudhry, M.; Gamallat, Y.; Khosh Kish, E.; Seyedi, S.; Gotto, G.; Ghosh, S.; Bismar, T.A. Downregulation of BUD31 Promotes Prostate Cancer Cell Proliferation and Migration via Activation of p-AKT and Vimentin In Vitro. Int. J. Mol. Sci. 2023, 24, 6055. https://doi.org/10.3390/ijms24076055
Choudhry M, Gamallat Y, Khosh Kish E, Seyedi S, Gotto G, Ghosh S, Bismar TA. Downregulation of BUD31 Promotes Prostate Cancer Cell Proliferation and Migration via Activation of p-AKT and Vimentin In Vitro. International Journal of Molecular Sciences. 2023; 24(7):6055. https://doi.org/10.3390/ijms24076055
Chicago/Turabian StyleChoudhry, Muhammad, Yaser Gamallat, Ealia Khosh Kish, Sima Seyedi, Geoffrey Gotto, Sunita Ghosh, and Tarek A. Bismar. 2023. "Downregulation of BUD31 Promotes Prostate Cancer Cell Proliferation and Migration via Activation of p-AKT and Vimentin In Vitro" International Journal of Molecular Sciences 24, no. 7: 6055. https://doi.org/10.3390/ijms24076055
APA StyleChoudhry, M., Gamallat, Y., Khosh Kish, E., Seyedi, S., Gotto, G., Ghosh, S., & Bismar, T. A. (2023). Downregulation of BUD31 Promotes Prostate Cancer Cell Proliferation and Migration via Activation of p-AKT and Vimentin In Vitro. International Journal of Molecular Sciences, 24(7), 6055. https://doi.org/10.3390/ijms24076055






