Glutathione Contribution in Interactions between Turnip mosaic virus and Arabidopsis thaliana Mutants Lacking Respiratory Burst Oxidase Homologs D and F
Abstract
1. Introduction
2. Results
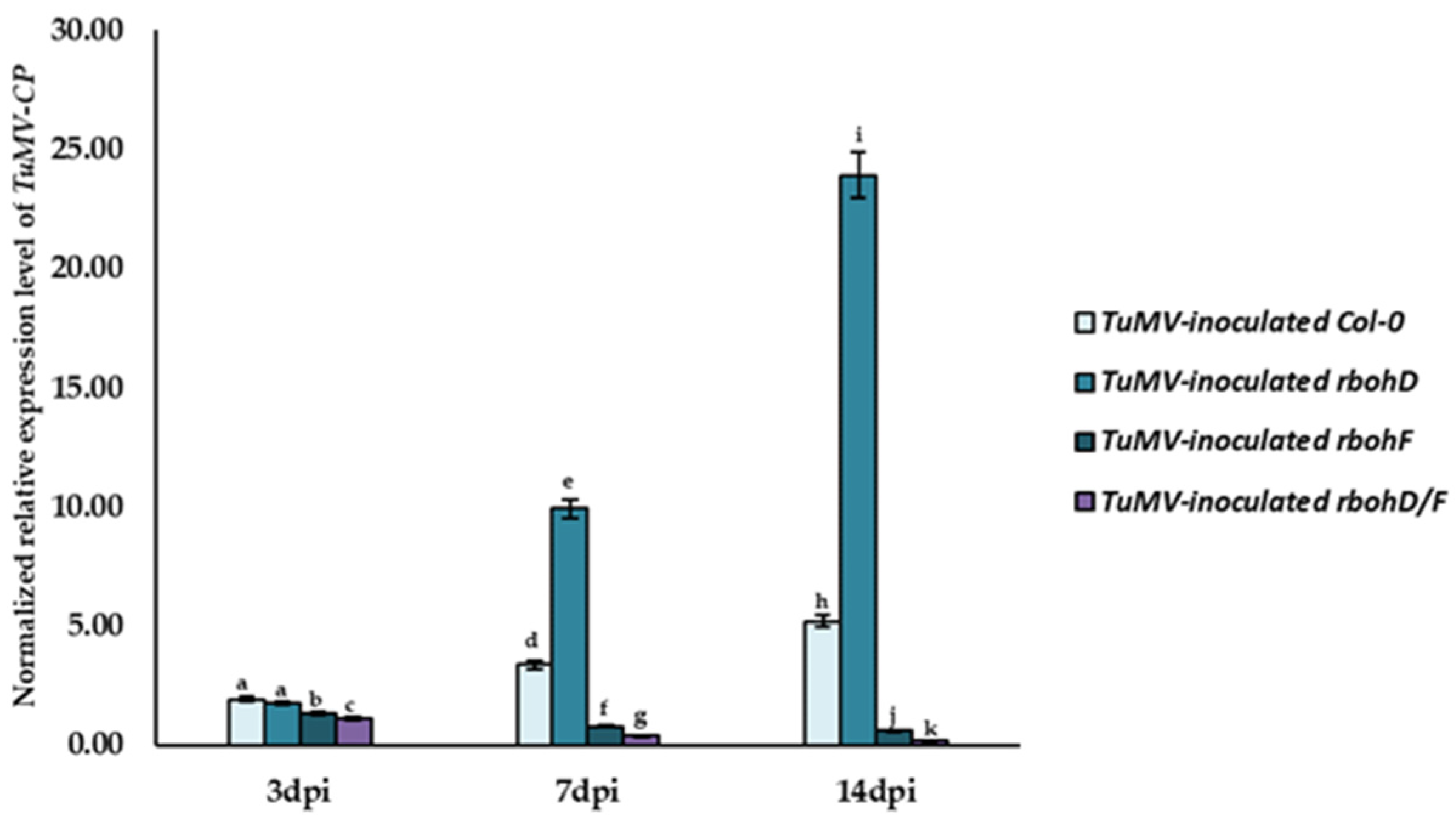
2.1. Significant Regulation of TuMV Concentration in Col-0–, rbohD–, rbohF–, and rbohD/F–TuMV Interactions
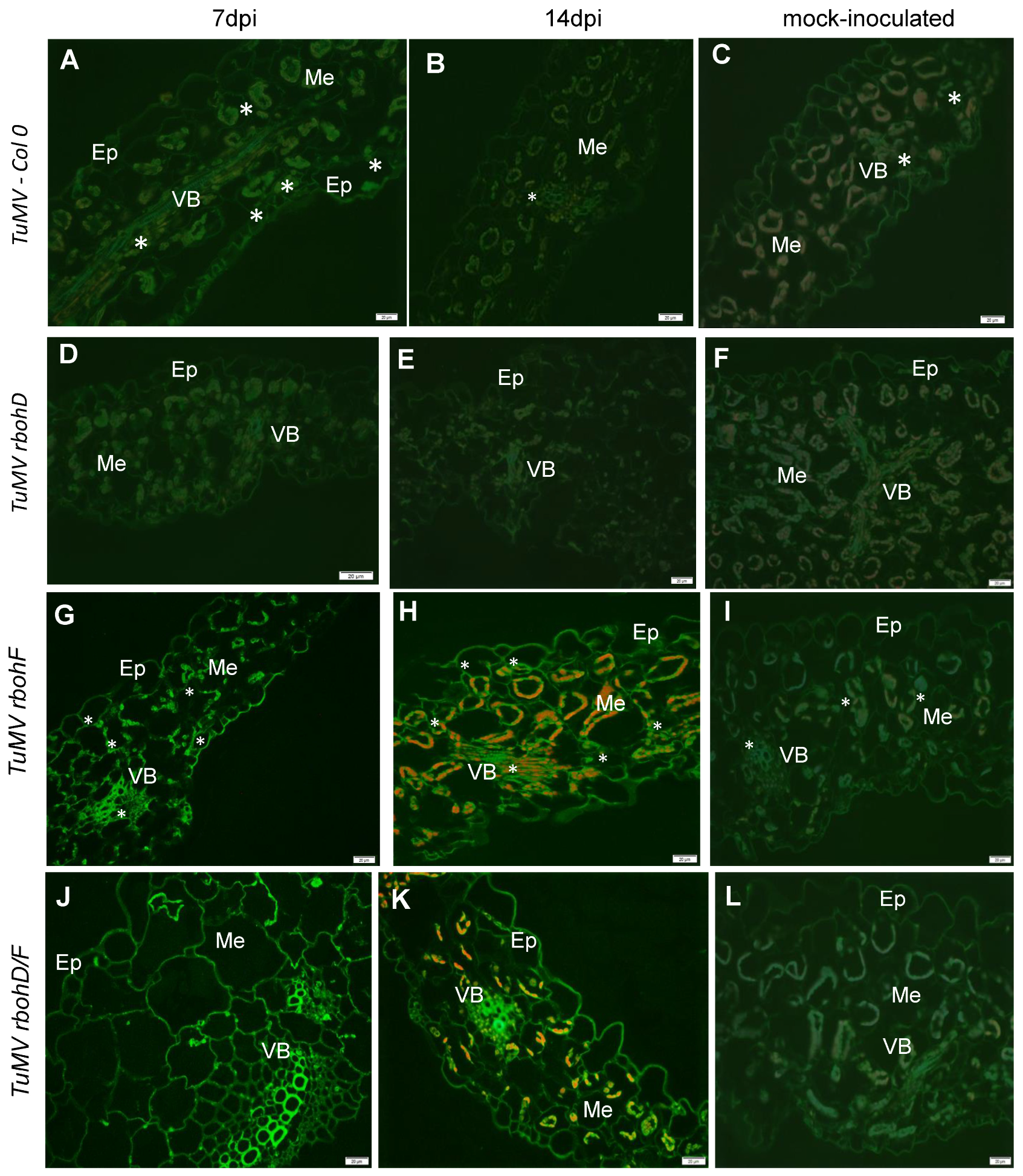
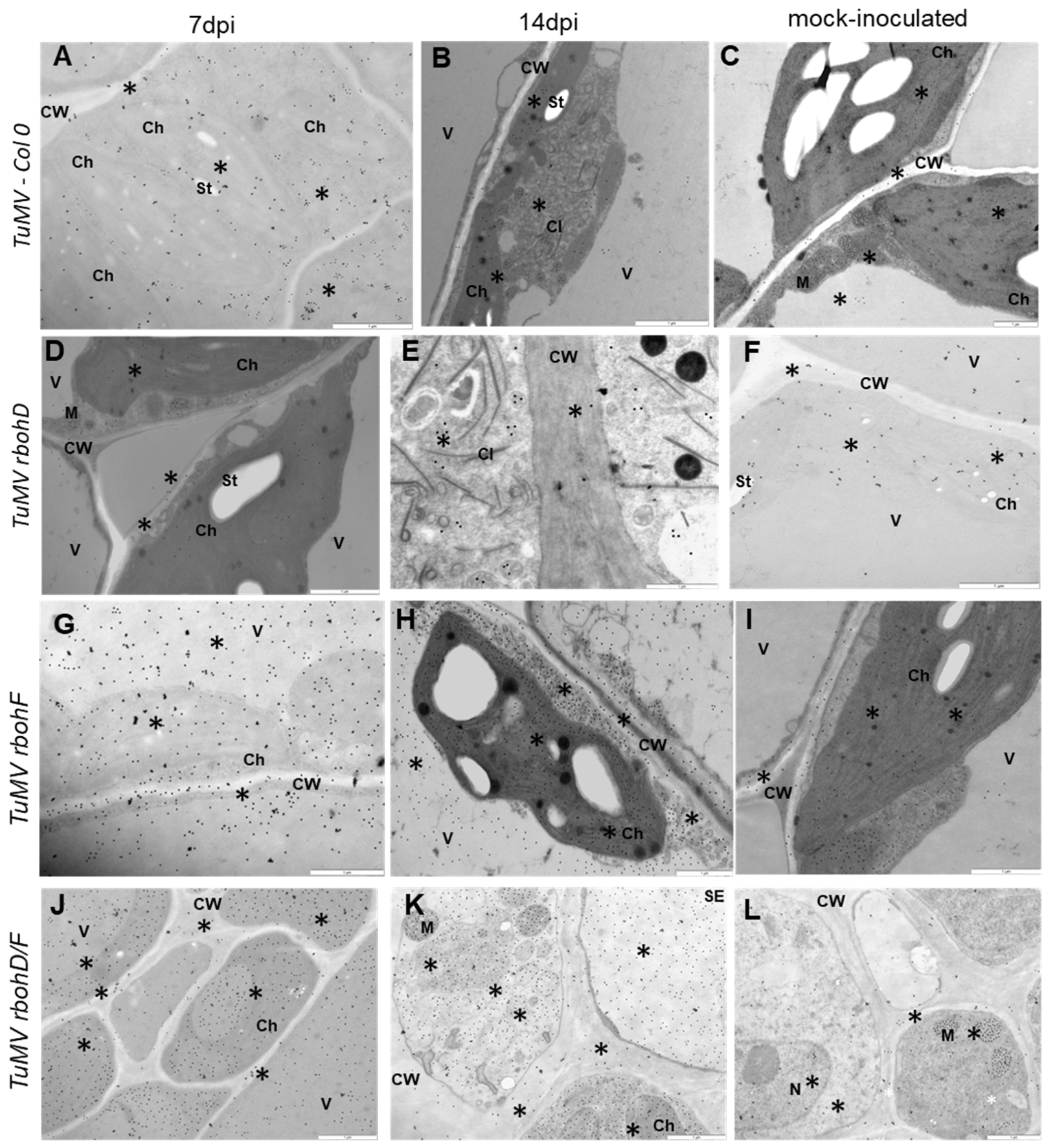
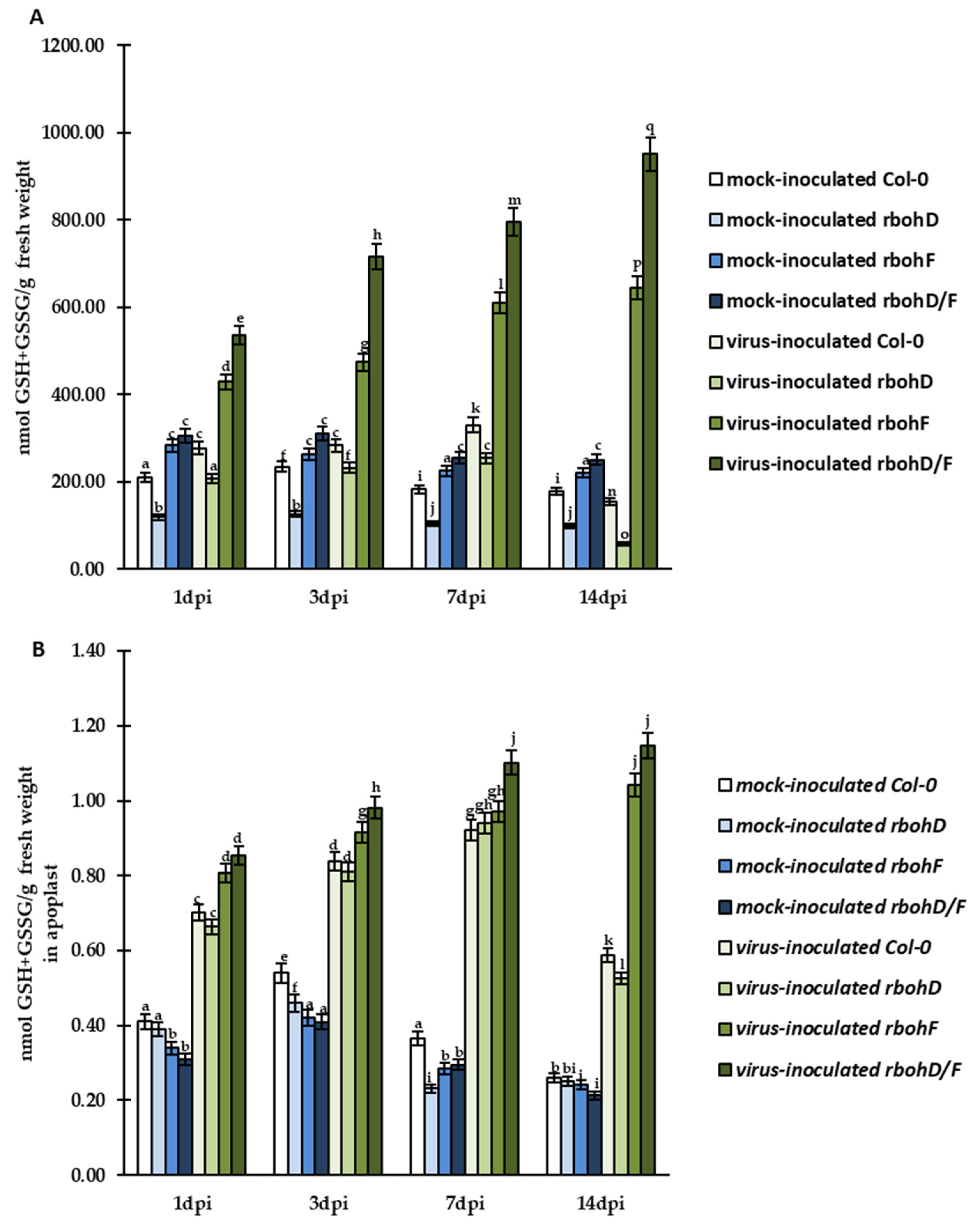
2.2. Crucial Cellular and Apoplastic Localization with Concentration of GSH (Reduced Glutathione) and GSSG (Oxidized Glutathione) Pools as an Element of Increased Susceptibility or Resistance Reaction in rbohD, rbohF, and rbohD/F Arabidopsis Mutants
2.3. Relative Expression of AtGGT1 and Selected AtGSTU Genes during Col-0–, rbohD–, rbohF–, and rbohD/F–TuMV Interactions Correlated with Increased Susceptibility or Resistance Reaction
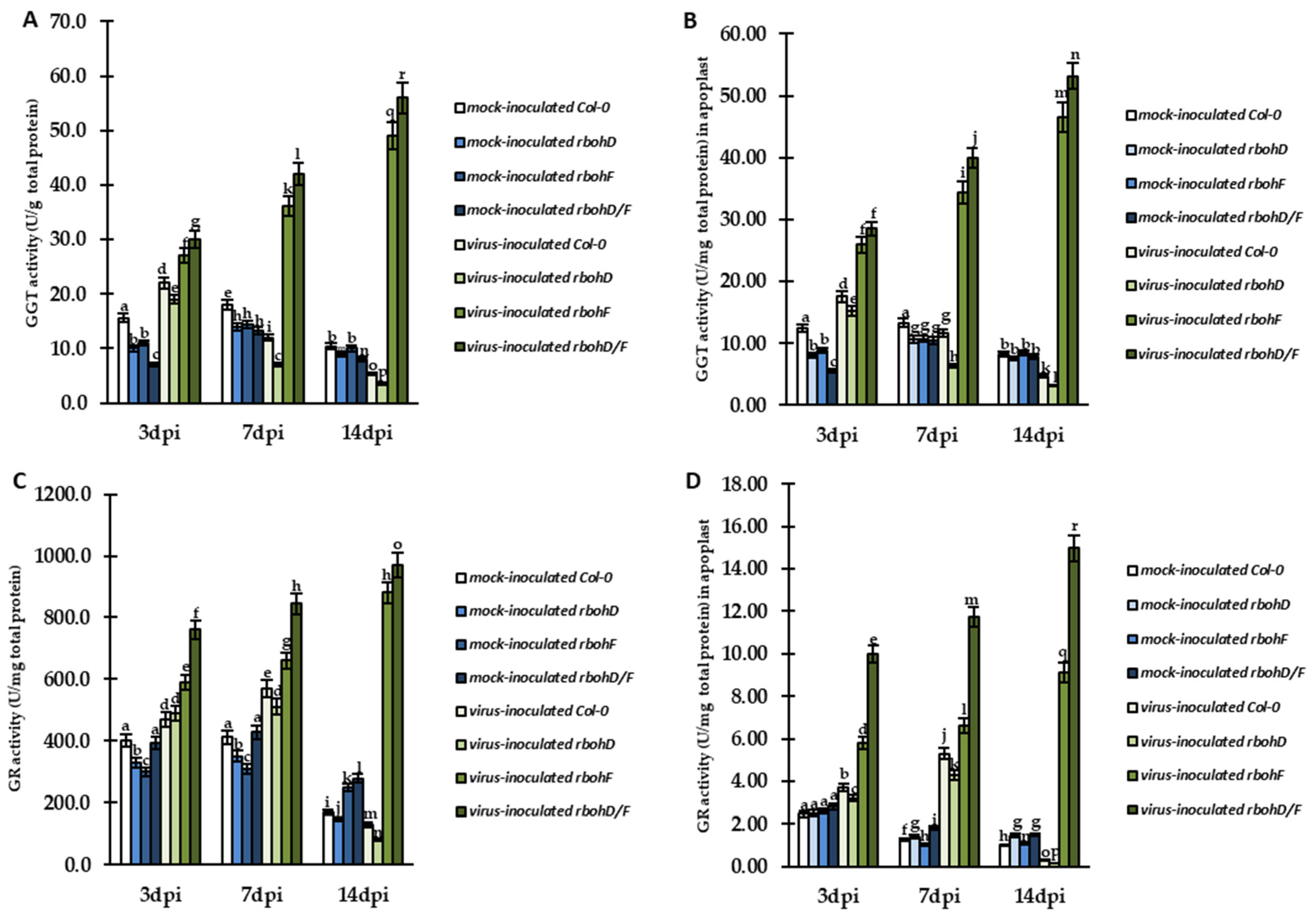
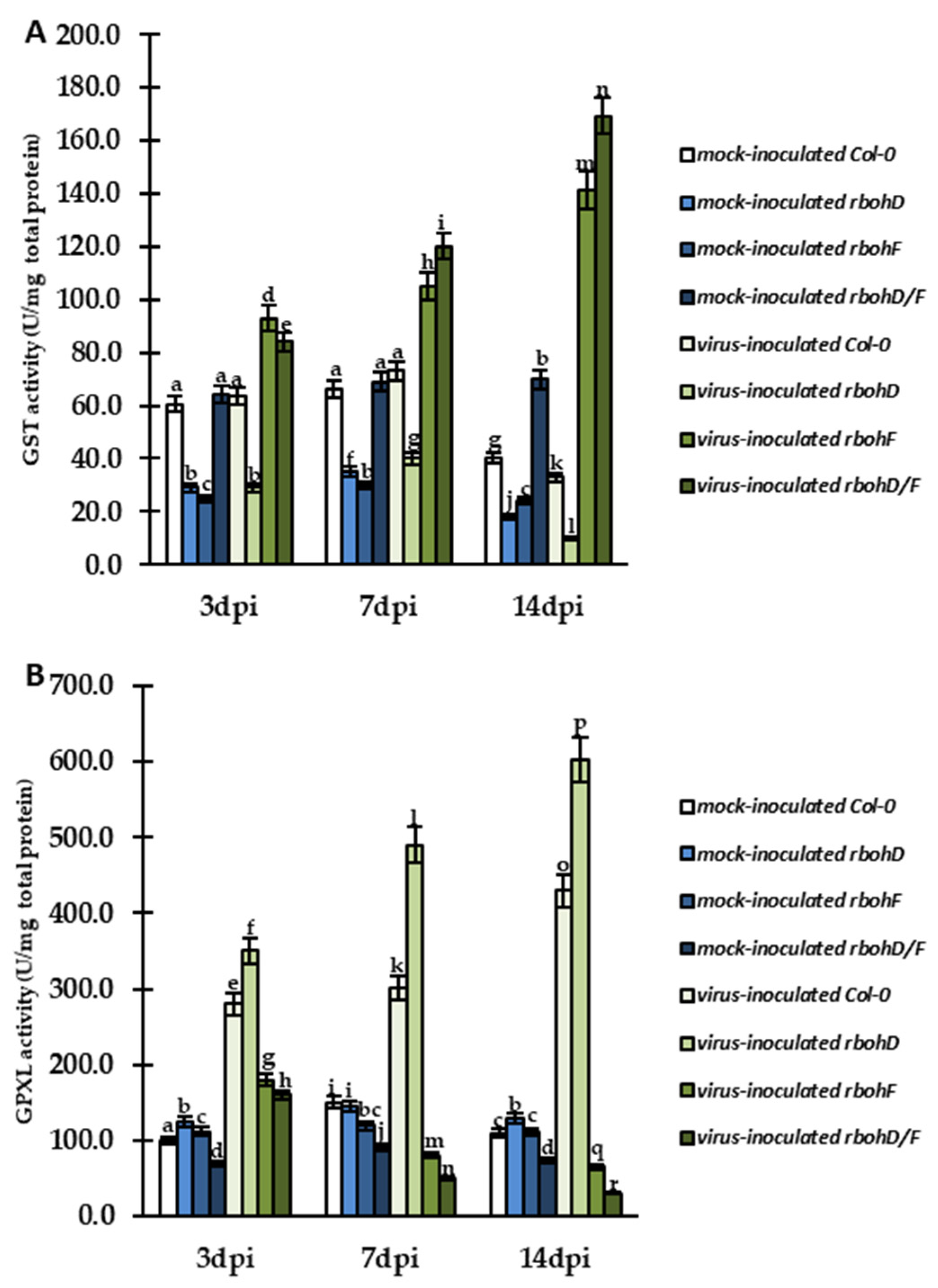
2.4. Changes in GGT and GST, with GR and GPXL Activity, as Modulators of Enhanced Susceptibility and Resistance during rbohD–, rbohF–, and rbohD/F–TuMV Interactions, along with Control of Lipid Peroxidation
3. Discussion
4. Materials and Methods
4.1. Plant Material, Virus Inoculation, and DAS-ELISA and Molecular Test for TuMV Content
4.2. Quantification of Immunofluorescence and Immunogold Localization of Total Glutathione in TuMV-Inoculated Col-0, rbohD, rbohF, and rbohD/F Arabidopsis Leaves
4.3. HPLC Analysis of Cellular and Apoplastic Content of GSH, GSSG, and Total Glutathione Accompanied by Estimation of Cytoplasmic Contamination
4.4. Isolation of RNA and Genomic DNA (gDNA) for AtGGT1 and Selected AtGSTU Genes in TuMV-Infected Col-0, rbohD, rbohF, and rbohD/F Plants
4.5. Analysis of Relative Expression of AtGGT1 and Selected AtGSTU Genes in TuMV-Infected Col-0, rbohD, rbohF, and rbohD/F Plants Using qPCR
4.6. Validation of GST, GR, GPXL, and GGT Activities and Lipid Peroxidation in Leaves of TuMV-Infected Col-0, rbohD, rbohF, and rbohD/F Plants
4.7. Validation of Apoplastic GR and GGT Activities in TuMV-Infected Col-0, rbohD, rbohF, and rbohD/F Leaves
4.8. Estimation of PCCs for Elements of Host Reaction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suzuki, N.; Miller, G.; Morales, J.; Shulaev, V.; Torres, M.A.; Mittler, R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 2011, 14, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Oda, T.; Hashimoto, H.; Kuwabara, N.; Akashi, S.; Hayashi, K.; Kojima, C.; Wong, H.L.; Kawasaki, T.; Shimamoto, K.; Sato, M.; et al. Structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications. J. Biol. Chem. 2010, 285, 1435–1445. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, D.; Zhang, X.; Liu, D.; Cheng, Y.; Shen, F. Role of plant respiratory burst oxidase homologs in stress responses. Free Radic. Res. 2018, 52, 826–839. [Google Scholar] [CrossRef]
- Sagi, M.; Fluhr, R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 2006, 141, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Groom, Q.J.; Torres, M.A.; Fordham-Skelton, A.P.; Hammond-Kosack, K.E.; Robinson, N.J.; Jones, J.D. rbohA, a rice homologue of the mammalian gp91phox respiratory burst oxidase gene. Plant J. 1996, 10, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Kakar, K.U.; Yang, Z.; Nawaz, Z.; Lin, S.; Guo, Y.; Ren, X.L.; Baloch, A.A.; Han, D. Systematic study of the stress-responsive Rboh gene family in Nicotiana tabacum: Genome-wide identification, evolution and role in disease resistance. Genomics 2020, 112, 1404–1418. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Ali, B.; Ahmed, W.; Tang, Y.; Li, H. Genome-wide Identification, Classification, Evolutionary Expansion and Expression of Rboh Family Genes in Pepper (Capsicum annuum L.). Trop. Plant Biol. 2021, 14, 251–266. [Google Scholar] [CrossRef]
- Liu, J.; Lu, H.; Wan, Q.; Qi, W.; Shao, H. Genome-wide analysis and expression profiling of respiratory burst oxidase homologue gene family in Glycine max. Environ. Exp. Bot. 2018, 161, 344–356. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; He, Y.; Hu, W.; Zhang, Y.; Wang, X.; Tang, H. Identification of NADPH oxidase family members associated with cold stress in strawberry. FEBS Open Bio. 2018, 8, 593–605. [Google Scholar] [CrossRef]
- Li, D.; Wu, D.; Li, S.; Dai, Y.; Cao, Y. Evolutionary and functional analysis of the plant-specific NADPH oxidase gene family in Brassica rapa L. R. Soc. Open Sci. 2019, 6, 181727. [Google Scholar] [CrossRef]
- Huang, S.; Tang, Z.; Zhao, R.; Hong, Y.; Zhu, S.; Fan, R.; Ding, K.; Cao, M.; Luo, K.; Geng, M.; et al. Genome-wide identification of cassava MeRboh genes and functional analysis in Arabidopsis. Plant Physiol. Biochem. 2021, 167, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-H.; Wang, P.-Q.; Zhang, P.-P.; Nie, X.-M.; Li, B.-B.; Tai, L.; Liu, W.-T.; Li, W.-Q.; Chen, K.-M. NADPH Oxidases: The Vital Performers and Center Hubs during Plant Growth and Signaling. Cells 2020, 9, 437. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Fichman, Y.; Mittler, R. Vascular bundles mediate systemic reactive oxygen signaling during light stress. Plant Cell 2020, 32, 3425–3435. [Google Scholar] [CrossRef]
- Morales, J.; Kadota, Y.; Zipfel, C.; Molina, A.; Torres, M.A. The Arabidopsis NADPH Oxidases Rbohd and Rbohf Display Differential Expression Patterns and Contributions During Plant Immunity. J. Exp. Bot. 2016, 67, 1663–1676. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005, 37, 1130–1134. [Google Scholar] [CrossRef]
- Proels, R.K.; Oberhollenzer, K.; Pathuri, I.P.; Hensel, G.; Kumlehn, J.; Huckelhoven, R. Rbohf2 of Barley Is Required for Normal Development of Penetration Resistance to the Parasitic Fungus Blumeria graminis F. sp. Hordei. Mol. Plant-Microbe Interact. 2010, 23, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Chaouch, S.; Queval, G.; Noctor, G. Atrbohf Is a Crucial Modulator of Defence-Associated Metabolism and a Key Actor in the Interplay between Intracellular Oxidative Stress and Pathogenesis Responses in Arabidopsis. Plant J. 2012, 69, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, H.; Numata, N.; Nakajima, K.; Katou, S.; Kawakita, K.; Rowland, O.; Jones, J.D.G.; Doke, N. Nicotiana Benthamiana gp91(Phox) Homologs Nbrboha and Nbrbohb Participate in H2O2 Accumulation and Resistance to Phytophthora Infestans. Plant Cell 2003, 15, 706–718. [Google Scholar] [CrossRef]
- Zhang, Z.; van Esse, H.P.; van Damme, M.; Fradin, E.F.; Liu, C.M.; Thomma, B.P. Ve1-mediated resistance against Verticillium does not involve a hypersensitive response in Arabidopsis. Mol. Plant Pathol. 2013, 14, 719–727. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, Q.; Zhou, S.; Sun, Y.; Li, X.; Li, Y. H2Bub1 Regulates RbohD-Dependent Hydrogen Peroxide Signal Pathway in the Defense Responses to Verticillium ahlia Toxins. Plant Physiol. 2020, 182, 640–657. [Google Scholar] [CrossRef]
- Trujillo, M.; Altschmied, L..; Schweizer, P.; Kogel, K.H.; Huckelhoven, R. Respiratory burst oxidase homologue A of barley contributes to penetration by the powdery mildew fungus Blumeria graminis f. sp. Hordei. J. Exp. Bot. 2006, 57, 3781–3791. [Google Scholar] [CrossRef]
- Foley, R.C.; Gleason, C.A.; Anderson, J.P.; Hamann, T.; Singh, K.B. Genetic and Genomic Analysis of Rhizoctonia Solani Interactions with Arabidopsis; Evidence of Resistance Mediated through NADPH Oxidases. PLoS ONE 2013, 8, e56814. [Google Scholar] [CrossRef] [PubMed]
- Doke, N.; Ohashi, Y. Involvement of an O2− generating system in the induction of necrotic lesions on tobacco leaves infected with tobacco mosaic virus. Physiol. Mol. Plant Pathol. 1988, 32, 163–175. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Valverde, R.A. The respiratory burst oxidase homolog d (rbohd) cell and tissue distribution in potato–Potato virus Y (PVYntn) hypersensitive and susceptible reactions. Int. J. Mol. Sci. 2019, 20, 2741. [Google Scholar] [CrossRef] [PubMed]
- Otulak-Kozieł, K.; Kozieł, E.; Bujarski, J.J.; Frankowska-Łukawska, J.; Torres, M.A. Respiratory Burst Oxidase Homologs RBOHD and RBOHF as Key Modulating Components of Response in Turnip Mosaic Virus—Arabidopsis thaliana (L.) Heyhn System. Int. J. Mol. Sci. 2020, 21, 8510. [Google Scholar] [CrossRef]
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; KluÈsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium Channels Activated by Hydrogen Peroxide Mediate Abscisic Acid Signalling in Guard Cells. Nature 2000, 406, 732–734. [Google Scholar] [CrossRef]
- Boudsocq, M.; Willmann, M.R.; McCormack, M.; Lee, H.; Shan, L.; He, P.; Bush, J.; Cheng, S.H.; Sheen, J. Differential Innate Immune Signalling via Ca2+ Sensor Protein Kinases. Nature 2010, 464, 418–422. [Google Scholar] [CrossRef]
- Lin, F.; Ding, H.; Wang, J.; Zhang, H.; Zhang, A.; Zhang, Y.; Tan, M.; Dong, W.; Jiang, M. Positive Feedback Regulation of Maize NADPH Oxidase by Mitogen-Activated Protein Kinase Cascade in Abscisic Acid Signalling. J. Exp. Bot. 2009, 60, 3221–3238. [Google Scholar] [CrossRef]
- Yun, B.W.; Feechan, A.; Yin, M.; Saidi, N.B.; Bihan, T.L.; Yu, M.; Moore, J.W.; Kang, J.G.; Kwon, E.; Spoel, S.H.; et al. S-Nitrosylation of NADPH Oxidase Regulates Cell Death in Plant Immunity. Nature 2011, 478, 264–268. [Google Scholar] [CrossRef]
- Cairns, N.G.; Pasternak, M.; Wachter, A.; Cobbett, C.S.; Meyer, A.J. Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 2006, 141, 446–455. [Google Scholar] [CrossRef]
- Noctor, G.; Queval, G.; Mhamdi, A.; Chaouch, S.; Foyer, C.H. Glutathione. Arab. Book 2011, 9, e0142. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Foyer, C.H.; Theodoulou, F.L.; Delrot, S. The functions of inter- and intracellular glutathione transport systems in plants. Trends Plant Sci. 2001, 6, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Sabetta, W.; Paradiso, A.; Paciolla, C.; de Pinto, M.C. Chemistry, biosynthesis, and antioxidative function of glutathione in plants. In Glutathione in Plant Growth, Development, and Stress Tolerance, 1st ed.; Hossain, M.A., Mostofa, M.G., Diaz-Vivancos, P., Burritt, D.J., Fujita, M., Tran, S.L.P., Eds.; Springer Nature: Cham, Switzerland, 2017; Volume 1, pp. 1–27. [Google Scholar]
- Diaz-Vivancos, P.; Wolff, T.; Markovic, J.; Pallardó, F.V.; Foyer, C.H. A nuclear glutathione cycle within the cell cycle. Biochem. J. 2010, 431, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Zechmann, B.; Zellnig, G.; Urbanek-Krajnc, A.; Müller, M. Artificial elevation of glutathione affects symptom development in ZYMV-infected Cucurbita pepo L. plants. Arch. Virol. 2007, 152, 747–762. [Google Scholar] [CrossRef]
- Gullner, G.; Tobia, I.; Fodor, J.; Kömives, T. Elevation of glutathione level and activation of glutathione-related enzymes affect virus infection in tobacco. Free Radic. Res. 1999, 31, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Király, L.; Albert, R.; Zsemberi, O.; Schwarczinger, I.; Hafez, Y.M.; Künstler, A. Reactive Oxygen Species Contribute to Symptomless, Extreme Resistance to Potato virus X in Tobacco. Phytopathology 2021, 111, 1870–1884. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Przewodowski, W.; Ciacka, K.; Przewodowska, A. Glutathione Modulation in PVYNTN susceptible and resistant potato plant interactions. Int. J. Mol. Sci. 2022, 23, 3797. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Horváth, E.; Csiszár, J. AtGSTU19 and AtGSTU24 as Moderators of the Response of Arabidopsis thaliana to Turnip mosaic virus. Int. J. Mol. Sci. 2022, 23, 11531. [Google Scholar] [CrossRef]
- Ohkama-Ohtsu, N.; Radwan, S.; Peterson, A.; Zhao, P.; Badr, A.F.; Xiang, C.; Oliver, D.J. Characterization of the extracellular gamma-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. Plant J. 2007, 49, 865–877. [Google Scholar] [CrossRef]
- Hodges, D.M.; DeLong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Stone, S.; Yang, X.; Antico, J.; Callis, J.; Ramonell, K.M.; Somerville, S. ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses. PLoS ONE 2010, 5, e14426. [Google Scholar] [CrossRef] [PubMed]
- Lukan, T.; Pompe-Novak, M.; Baebler, Š.; Tušek-Žnidarič, M.; Kladnik, A.; Križnik, M.; Blejec, A.; Zagorščak, M.; Stare, K.; Dušak, B.; et al. Precision transcriptomics of viral foci reveals the spatial regulation of immune-signaling genes and identifies RBOHD as an important player in the incompatible interaction between potato virus Y and potato. Plant J. 2020, 104, 645–661. [Google Scholar] [CrossRef]
- Pogany, M.; von Rad, U.; Grun, S.; Dongo, A.; Pintye, A.; Simoneau, P.; Bahnweg, G.; Kiss, L.; Barna, B.; Durner, J. Dual Roles of Reactive Oxygen Species and NADPH Oxidase Rbohd in an Arabidopsis-Alternaria Pathosystem. Plant Physiol. 2009, 151, 1459–1475. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Dangl, J.L.; Jones, J.D.G. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A Burst of Plant NADPH Oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef]
- Hakmaoui, A.; Pérez-Bueno, M.L.; García-Fontana, B.; Camejo, D.; Jiménez, A.; Sevilla, S.; Barón, M. Analysis of the antioxidant response of Nicotiana benthamiana to infection with two strains of Pepper mild mottle virus. J. Exp. Bot. 2012, 63, 5487–5496. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.A.; Barba, E.; Diaz-Vivancos, P. Glutathione-Mediated biotic stress tolerance. In Glutathione in Plant Growth, Development, and Stress Tolerance, 1st ed.; Hossain, M.A., Mostofa, M.G., Diaz-Vivancos, P., Burritt, D.J., Fujita, M., Tran, S.L.P., Eds.; Springer Nature: Cham, Switzerland, 2017; Volume 1, pp. 309–329. [Google Scholar]
- Glazebrook, J.; Rogers, E.E.; Ausubel, F.M. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 1996, 143, 973–982. [Google Scholar] [CrossRef]
- Parisy, V.; Poinssot, B.; Owsianowski, L.; Buchala, A.; Glazebrook, J.; Mauch, F. Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 2007, 49, 159–172. [Google Scholar] [CrossRef]
- Ball, L.; Accotto, G.-P.; Bechtold, U.; Creissen, G.; Funck, D.; Jimenez, A.; Kular, B.; Leyland, N.; Mejia-Carranza, J.; Reynolds, H.; et al. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 2004, 16, 2448–2462. [Google Scholar] [CrossRef]
- Kuźniak, E.; SkŁodowska, M. Differential Implication of Glutathione, Glutathione-Metabolizing Enzymes and Ascorbate in Tomato Resistance to Pseudomonas syringae. J. Phytopathol. 2004, 152, 529–536. [Google Scholar] [CrossRef]
- Vanacker, H.; Carver, T.L.; Foyer, C.H. Early H2O2 accumulation in mesophyll cells leads to induction of glutathione during the hyper-sensitive response in the Barley-Powdery Mildew interaction. Plant Physiol. 2000, 123, 1289–1300. [Google Scholar] [CrossRef]
- Tolin, S.; Arrigoni, G.; Trentin, A.R.; Veljovic-Jovanovic, S.; Pivato, M.; Zechman, B.; Masi, A. Biochemical and quantitative proteomics investigations in Arabidopsis ggt1 mutant leaves reveal a role for the gamma-glutamyl cycle in plant’s adaptation to environment. Proteomics 2013, 13, 2031–2045. [Google Scholar] [PubMed]
- Singh, Y.J.; Grewal, S.K.; Gill, R.K. Role of glutathione in methylglyoxal detoxification pathway during yellow mosaic virus (YMV) infection in black gram (Vigna mungo (L.) Hepper). Physiol. Mol. Plant Pathol. 2020, 111, 101513. [Google Scholar] [CrossRef]
- Künstler, A.; Király, L.; Kátay, G.; Enyedi, A.J.; Gullner, G. Glutathione can compensate for salicylic acid deficiency in tobacco to maintain resistance to tobacco mosaic virus. Front. Plant Sci. 2019, 10, 1115. [Google Scholar] [CrossRef]
- Király, Z.; Barna, B.; Kecskés, A.; Fodor, J. Down-regulation of antioxidative capacity in a transgenic tobacco which fails to develop acquired resistance to necrotization caused by tobacco mosaic virus. Free Radic. Res. 2002, 36, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chaouch, S.; Mhamdi, A.; Queval, G.; Zechmann, B.; Noctor, G. Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signalling. Antioxid. Redox Signal. 2013, 18, 2106–2121. [Google Scholar] [CrossRef]
- Dorion, S.; Ouellet, J.C.; Rivoal, J. Glutathione Metabolism in Plants under Stress: Beyond Reactive Oxygen Species Detoxification. Metabolites 2021, 11, 641. [Google Scholar] [CrossRef]
- Martin, M.N.; Slovin, J.P. Purified gamma-glutamyl transpeptidases from tomato exhibit high affinity for glutathione and glutathione S-conjugates. Plant Physiol. 2000, 122, 1417–1426. [Google Scholar] [CrossRef][Green Version]
- Storozhenko, S.; Belles-Boix, E.; Babiychuk, E.; Herouart, D.; Davey, M.W.; Slooten, L.; Van Montagu, M.; Inze, D.; Kushnir, S. γ-glutamyl transpeptidase in transgenic tobacco plants. Cellular localization, processing, and biochemical properties. Plant Physiol. 2002, 128, 1109–1119. [Google Scholar] [CrossRef]
- Destro, T.; Prasad, D.; Martignago, D.; Bernet, I.L.; Trentin, A.R.; Renu, I.K.; Ferretti, M.; Masi, A. Compensatory expression and substrate inducibility of gamma-glutamyl transferase GGT2 isoform in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.N.; Saladores, P.H.; Lambert, E.; Hudson, A.O.; Leustek, T. Localization of members of the gamma-glutamyl transpeptidase family identifies sites of glutathione and glutathione S-conjugate hydrolysis. Plant Physiol. 2007, 144, 1715–1732. [Google Scholar] [CrossRef]
- Masi, A.; Trentin, A.R.; Agrawal, G.K.; Rakwal, R. Gamma-glutamyl cycle in plants: A bridge connecting the environment to the plant cell? Front. Plant Sci. 2015, 6, 252. [Google Scholar] [CrossRef] [PubMed]
- Ohkama-Ohtsu, N.; Zhao, P.; Xiang, C.B.; Oliver, D.J. Glutathione conjugates in the vacuole are degraded by gamma-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J. 2007, 49, 878–888. [Google Scholar] [CrossRef]
- Zhang, K.; Shen, Y.; Wang, T.; Wang, Y.; Xue, S.; Luan, H.; Wang, L.; Li, K.; Guo, D.; Zhi, H. GmGSTU13 is related to the development of mosaic symptoms in soybean plants infected with Soybean mosaic virus. Phytopathology 2022, 112, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Piślewska-Bednarek, M.; Nakano, R.T.; Hiruma, K.; Pastorczyk, M.; Sanchez-Vallet, A.; Singkaravanit-Ogawa, S.; Ciesiołka, D.; Takano, Y.; Molina, A.; Schulze-Lefert, P.; et al. Glutathione transferase U13 functions in pathogen-triggered glucosinolate metabolism. Plant Physiol. 2018, 176, 538–551. [Google Scholar] [CrossRef] [PubMed]
- Pavan Kumar, B.K.; Kanakala, S.; Malathi, V.G.; Gopal, M.; Usha, R. Transcriptomic and proteomic analysis of yellow mosaic diseased soybean. J. Plant Biochem. Biotechnol. 2017, 26, 224–234. [Google Scholar] [CrossRef]
- Chen, I.H.; Chiu, M.H.; Cheng, S.F.; Hsu, Y.H.; Tsai, C.H. The glutathione transferase of Nicotiana benthamiana NbGSTU4 plays a role in regulating the early replication of Bamboo mosaic virus. New Phytol. 2013, 199, 749–757. [Google Scholar] [CrossRef]
- Méndez-López, E.; Donaire, L.; Gosálvez, B.; Díaz-Vivancos, P.; Sánchez-Pina, M.A.; Tilsner, J.; Aranda, M.A. Tomato SlGSTU38 interacts with the PepMV coat protein and promotes viral infection. New Phytol. 2023, 1, 1–17. [Google Scholar] [CrossRef]
- Király, L.; Künstler, A.; Fattinger, M.; Höller, K.; Juhász, C.; Müller, M.; Gullner, G.; Zechmann, B. Sulfate supply influences compartment specific glutathione metabolism and confers enhanced resistance to tobacco mosaic virus during a hypersensitive response. Plant Physiol. Biochem. 2012, 59, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Kondoh, H.; De Leon, T.B.; Macalalad, R.J.A.; Cabunagan, R.C.; Cabauatan, P.Q.; Mauleon, R.; Kichuchi, S.; Choi, I.R. Gene expression responses to Rice tungro spherical virus in susceptible and resistant near-isogenic rice plants. Virus Res. 2013, 171, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, J.; Jin, P.; Guo, M.; Guo, J.; Cheng, P.; Li, Q.; Wang, B. Glutathione S-transferase interactions enhance wheat resistance to powdery mildew but not wheat stripe rust. Plant Physiol. 2022, 190, 1418–1439. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Han, Z.; Wang, S.; Wang, X.; Sun, A.; Zu, X.; Chen, Y. Comparative proteomic analysis of the plant-virus interaction in resistant and susceptible ecotypes of maize infected with sugarcane mosaic virus. J. Proteom. 2013, 89, 124–140. [Google Scholar] [CrossRef]
- Diaz-Vivancos, P.; Clemente-Moreno, M.J.; Rubio, M.; Olmos, E.; Garcia, J.A.; Martinez-Gomez, P.; Hernandez, J.A. Alteration in the chloroplastic metabolism leads to ROS accumulation in pea plants in response to plum pox virus. J. Exp. Bot. 2008, 59, 2147–2160. [Google Scholar] [CrossRef]
- Amari, K.; Díaz-Vivancos, P.; Pallás, V.; Sánchez-Pina, M.A.; Hernández, J.A. Oxidative stress induction by Prunus necrotic ringspot virus infection in apricot seeds. Physiol. Plant. 2007, 131, 302–310. [Google Scholar] [CrossRef]
- Clarke, S.F.; Guy, P.L.; Burritt, D.J.; Jameson, P.E. Changes in the activities of antioxidant enzymes in response to virus infection and hormone treatment. Physiol. Plant. 2002, 114, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Attacha, S.; Solbach, D.; Bela, K.; Moseler, A.; Wagner, S.; Schwarzländer, M.; Aller, I.; Müller, S.J.; Meyer, A.J. Glutathione peroxidase-like enzymes cover five distinct cell compartments and membrane surfaces in Arabidopsis thaliana. Plant Cell Environ. 2017, 40, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Bela, K.; Bangash, S.A.K.; Csiszár, J. Plant Glutathione Peroxidases: Antioxidant Enzymes in Plant Stress Responses and Tolerance. In Glutathione in Plant Growth, Development, and Stress Tolerance; Springer: Cham, Switzerland, 2017; pp. 113–126. [Google Scholar]
- Bela, K.; Riyazuddin, R.; Csiszár, J. Plant Glutathione Peroxidases: Non-Heme Peroxidases with Large Functional Flexibility as a Core Component of ROS-Processing Mechanisms and Signalling. Antioxidants 2022, 11, 1624. [Google Scholar] [CrossRef]
- Passaia, G.; Fonini, L.S.; Caverzan, A.; Jardim-Messeder, D.; Christoff, A.P.; Gaeta, M.L.; de Araujo Mariath, J.E.; Margis, R.; Margis-Pinheiro, M. The Mitochondrial Glutathione Peroxidase GPX3 Is Essential for H2O2 Homeostasis and Root and Shoot Development in Rice. Plant Sci. 2013, 208, 93–101. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Bela, K.; Horváth, E.; Rigó, G.; Gallé, Á.; Szabados, L.; Fehér, A.; Csiszár, J. Overexpression of the Arabidopsis Glutathione Peroxidase-like 5 Gene (AtGPXL5) Resulted in Altered Plant Development and Redox Status. Environ. Exp. Bot. 2019, 167, 103849. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Bela, K.; Poór, P.; Szepesi, Á.; Horváth, E.; Rigó, G.; Szabados, L.; Fehér, A.; Csiszár, J. Crosstalk between the Arabidopsis Glutathione Peroxidase-Like 5 Isoenzyme (AtGPXL5) and Ethylene. Int. J. Mol. Sci. 2022, 23, 5749. [Google Scholar] [CrossRef] [PubMed]
- Milla, M.A.R.; Maurer, A.; Huete, A.R.; Gustafson, J.P. Glutathione Peroxidase Genes in Arabidopsis Are Ubiquitous and Regulated by Abiotic Stresses through Diverse Signaling Pathways. Plant J. 2003, 36, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Herbette, S.; de Labrouhe, D.T.; Drevet, J.R.; Roeckel-Drevet, P. Transgenic tomatoes showing higher glutathione peroxidase antioxidant activity are more resistant to an abiotic stress but more susceptible to biotic stresses. Plant Sci. 2011, 180, 548–553. [Google Scholar] [CrossRef]
- Iqbal, A.; Yabuta, Y.; Takeda, T.; Nakano, Y.; Shigeoka, S. Hydroperoxide reduction by thioredoxin-specific glutathione peroxidase isoenzymes of Arabidopsis thaliana. FEBS J. 2006, 273, 5589–5597. [Google Scholar] [CrossRef]
- Navrot, N.; Collin, V.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.P.; Rouhier, N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef]
- Kalapos, B.; Juhász, C.; Balogh, E.; Kocsy, G.; Tóbiás, I.; Gullner, G. Transcriptome profiling of pepper leaves by RNA-Seq during an incompatible and a compatible pepper-tobamovirus interaction. Sci. Rep. 2021, 11, 20680. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Jahan, B.; AlAjmi, M.F.; Rehman, M.T.; Khan, N.A. Ethephon Mitigates Nickel Stress by Modulating Antioxidant System, Glyoxalase System and Proline Metabolism in Indian Mustard. Physiol. Mol. Biol. Plants 2020, 26, 1201–1213. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, J.A. Turnip mosaic virus. In MI/AAB Descriptions of Plant Viruses; CMI/AAB: Surrey, UK, 1970. [Google Scholar]
- Walsh, J.A.; Jenner, C.E. Turnip mosaic virus and the quest for durable resistance. Mol. Plant Pathol. 2002, 3, 289–300. [Google Scholar] [CrossRef]
- Jenner, C.E.; Keane, G.J.; Jones, J.E.; Walsh, J.A. Serotypic variation in Turnip mosaic virus. Plant Pathol. 1999, 48, 101–108. [Google Scholar] [CrossRef]
- Arous, S.; Harmon, C.L.; Capobianco, H.M.; Polston, J.E. Comparison of genus-specific primers in RT-PCR for the broad-spectrum detection of viruses in the genus Potyvirus by plant diagnostic laboratories. J. Virol. Methods 2018, 258, 29–34. [Google Scholar] [CrossRef]
- Kozieł, E.; Otulak-Kozieł, K.; Bujarski, J.J. Ultrastructural analysis of Prune dwarf virus intercellular transport and pathogenesis. Int. J. Mol. Sci. 2018, 19, 2570. [Google Scholar] [CrossRef]
- Kozieł, E.; Otulak, K.; Lockhart, B.E.L.; Garbaczewska, G. Subcelullar localization of proteins associated with Prune dwarf virus replication. Eur. J. Plant Pathol. 2017, 149, 653–668. [Google Scholar] [CrossRef]
- Kozieł, E.; Otulak-Kozieł, K.; Bujarski, J.J. Modifications in Tissue and Cell Ultrastructure as Elements of Immunity-Like Reaction in Chenopodium quinoa against Prune Dwarf Virus (PDV). Cells 2020, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Otulak-Kozieł, K.; Kozieł, E.; Lockhart, B.E.L.; Bujarski, J.J. The Expression of Potato Expansin A3 (StEXPA3) and Extensin4 (StEXT4) Genes with Distribution of StEXPAs and HRGPs-Extensin Changes as an Effect of Cell Wall Rebuilding in Two Types of PVYNTN–Solanum tuberosum Interactions. Viruses 2020, 12, 66. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Lockhart, B.E.L. Plant cell wall dynamics in compatible and incompatible potato response to infection caused by Potato virus Y (PVYNTN). Int. J. Mol. Sci. 2018, 19, 862. [Google Scholar] [CrossRef] [PubMed]
- Luschin-Ebengreuth, N.; Zechmann, B. Compartment-specific investigations of antioxidants and hydrogen peroxide in leaves of Arabidopsis thaliana during dark-induced senescence. Acta Physiol. Plant. 2016, 38, 133. [Google Scholar] [CrossRef]
- Kranner, I. Determination of glutathione, glutathione disulphide and two related enzymes, glutathione reductase and glucose-6-phosphate dehydrogenase, in fungal and plant cells. In Mycorrhiza Manual, 1st ed.; Varma, A., Ed.; Springer Nature: Heidelberg, Germany, 2012; pp. 227–241. [Google Scholar]
- Borniego, M.L.; Molina, M.C.; Guiamét, J.J.; Martinez, D.E. Physiological and Proteomic Changes in the Apoplast Accompany Leaf Senescence in Arabidopsis. Front Plant Sci. 2020, 10, 1635. [Google Scholar] [CrossRef]
- Vanacker, H.; Carver, T.L.M.; Foyer, C.H. Pathogen induced changes in the antioxidant status of the apoplast in barley leaves. Plant Phyiol. 1998, 117, 1103–1114. [Google Scholar] [CrossRef]
- Vanacker, H.; Foyer, C.H.; Carver, T.L.M. Changes in apoplastic antioxidants induced by powdery mildew attack in oat genotypes with race non-specific resistance. Planta 1998, 208, 444–452. [Google Scholar] [CrossRef]
- Weimar, M.; Rothe, G. Preparation of extracts from mature spruce needles for enzymatic analysis. Physiol. Plant. 1987, 69, 692–698. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef]
- TAIR Database. Available online: https://www.arabidopsis.org/ (accessed on 21 February 2023).
- Islam, S.; Das Sajib, S.; Sultana Jui, Z.; Arabia, S.; Islam, T.; Ghosh, A. Genome-Wide identification of glutathione S-transferase gene family in pepper, its classification, and expression profiling under different anatomical and environmental conditions. Sci. Rep. 2019, 9, 9101. [Google Scholar] [CrossRef]
- Islam, S.; Rahman, I.A.; Islam, T.; Ghosh, A. Genome-wide identification and expression analysis of GST gene family in tomato: Gaining an insight to their physiological and stress-specific roles. PLoS ONE 2017, 12, e0187504. [Google Scholar] [CrossRef]
- Bela, K.; Riyazuddin, R.; Horváth, E.; Hurton, Á.; Gallé, Á.; Takács, Z.; Zsigmond, L.; Szabados, L.; Tari, I.; Csiszár, J. Comprehensive analysis of antioxidant mechanisms in Arabidopsis glutathione peroxidase-likemutants under salt-and osmotic stress reveals organ-specific significance of the AtGPXL’s activities. Environ. Exp. Bot. 2018, 150, 127–140. [Google Scholar] [CrossRef]
- Tate, S.S.; Meister, A. γ-Glutamyl transpeptidase from kidney. Meth. Enzymol. 1985, 113, 400–419. [Google Scholar]
- Burian, M.; Podgórska, A.; Ostaszewska-Bugajska, M.; Szal, B. Respiratory Burst Oxidase Homolog D as a Modulating Component of Oxidative Response under Ammonium Toxicity. Antioxidants 2022, 11, 703. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Eghbali, M.; Ou, J.; Lu, R.; Toro, L.; Stefani, E. Quantitative determination of spatial protein-protein correlations in fluorescence confocal microscopy. Biophys. J. 2010, 98, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Manders, E.M.; Stap, J.; Aten, J.A. Dynamics of three-dimensional replication patterns during the S-phase, analyzed by double labeling of DNA and confocal microscopy. J. Cell Sci. 1992, 103, 857–862. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otulak-Kozieł, K.; Kozieł, E.; Treder, K.; Király, L. Glutathione Contribution in Interactions between Turnip mosaic virus and Arabidopsis thaliana Mutants Lacking Respiratory Burst Oxidase Homologs D and F. Int. J. Mol. Sci. 2023, 24, 7128. https://doi.org/10.3390/ijms24087128
Otulak-Kozieł K, Kozieł E, Treder K, Király L. Glutathione Contribution in Interactions between Turnip mosaic virus and Arabidopsis thaliana Mutants Lacking Respiratory Burst Oxidase Homologs D and F. International Journal of Molecular Sciences. 2023; 24(8):7128. https://doi.org/10.3390/ijms24087128
Chicago/Turabian StyleOtulak-Kozieł, Katarzyna, Edmund Kozieł, Krzysztof Treder, and Lóránt Király. 2023. "Glutathione Contribution in Interactions between Turnip mosaic virus and Arabidopsis thaliana Mutants Lacking Respiratory Burst Oxidase Homologs D and F" International Journal of Molecular Sciences 24, no. 8: 7128. https://doi.org/10.3390/ijms24087128
APA StyleOtulak-Kozieł, K., Kozieł, E., Treder, K., & Király, L. (2023). Glutathione Contribution in Interactions between Turnip mosaic virus and Arabidopsis thaliana Mutants Lacking Respiratory Burst Oxidase Homologs D and F. International Journal of Molecular Sciences, 24(8), 7128. https://doi.org/10.3390/ijms24087128









