Submergence Stress Alters the Expression of Clock Genes and Configures New Zeniths and Expression of Outputs in Brachypodium distachyon
Abstract
1. Introduction
2. Results
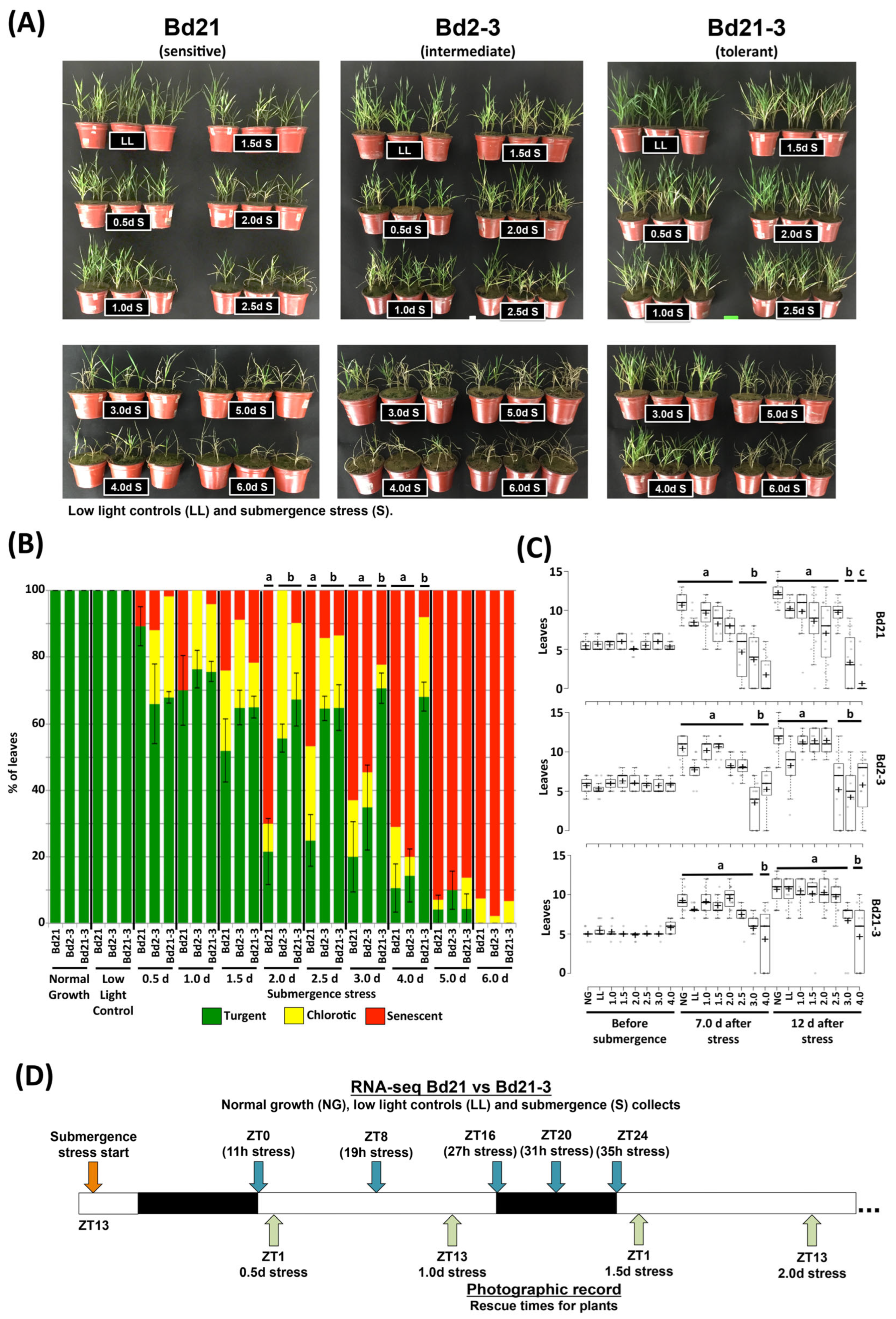
2.1. Brachypodium Ecotypes of Differential Submergence Stress Tolerance
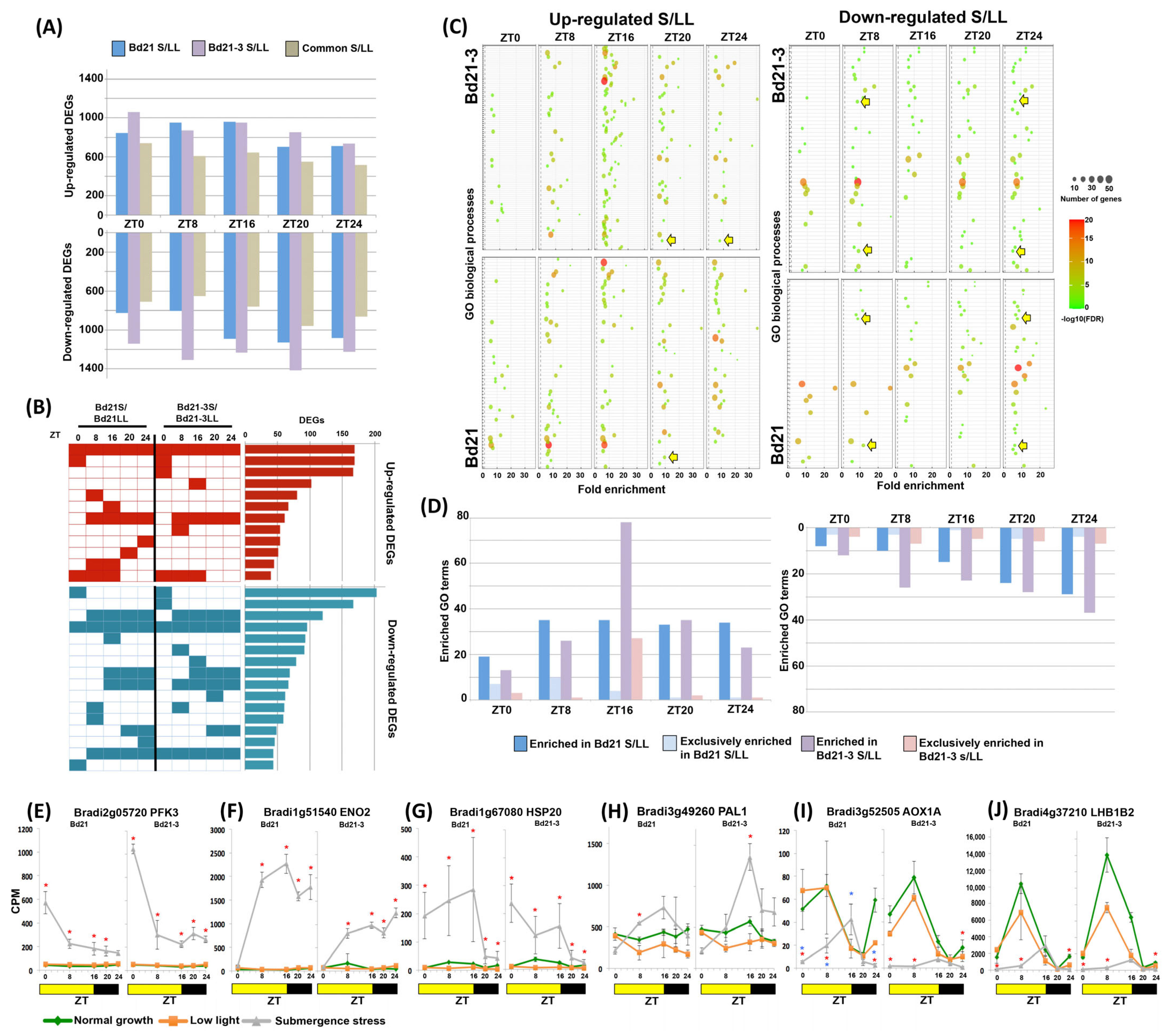
2.2. Brachypodium Transcriptome under Submergence Stress Responds to Time of Day
2.3. Submergence Stress Changed the Biological Processes Enriched at Each ZT
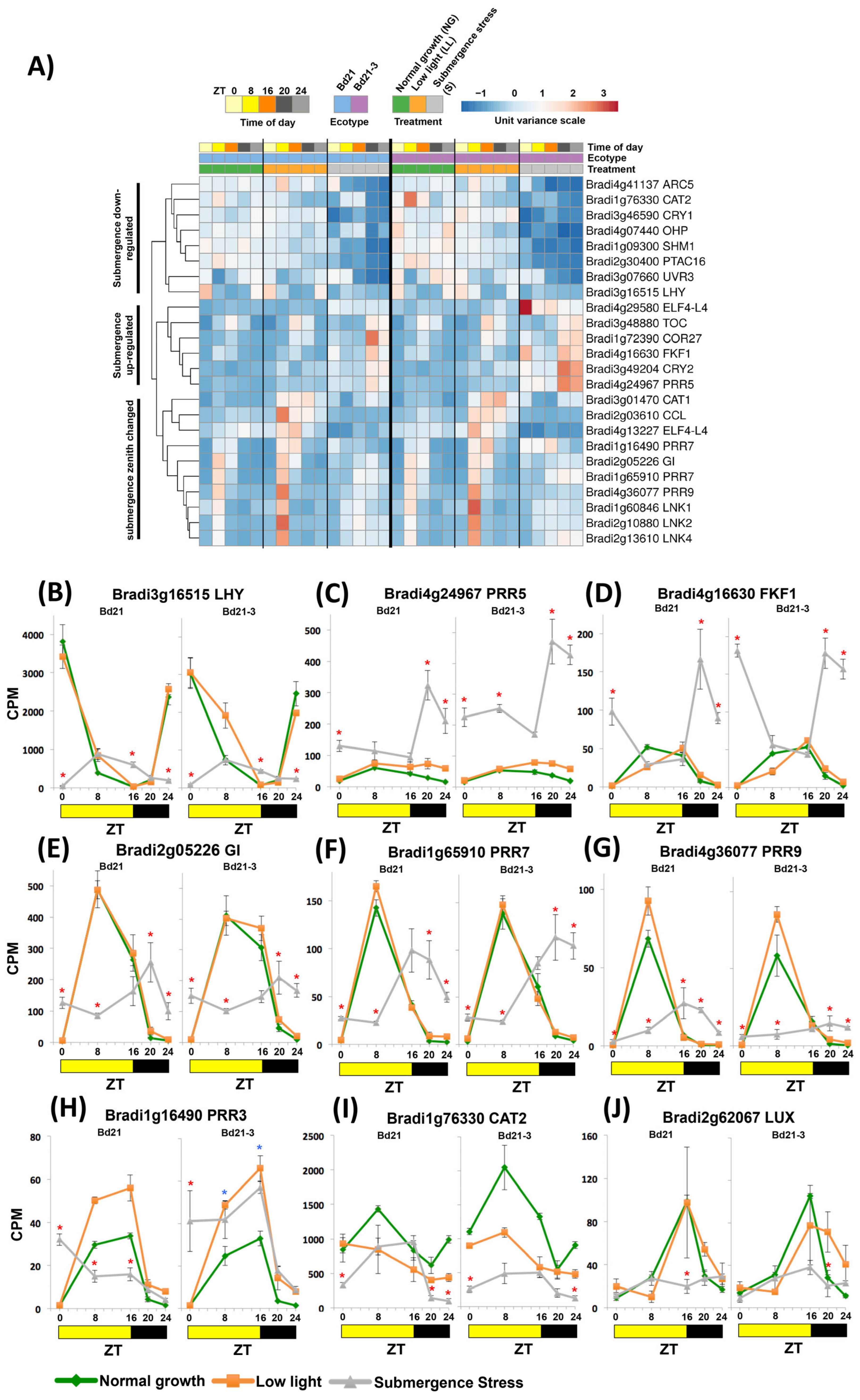
2.4. Submergence Stress Reshaped the Expression of Circadian Clock Genes
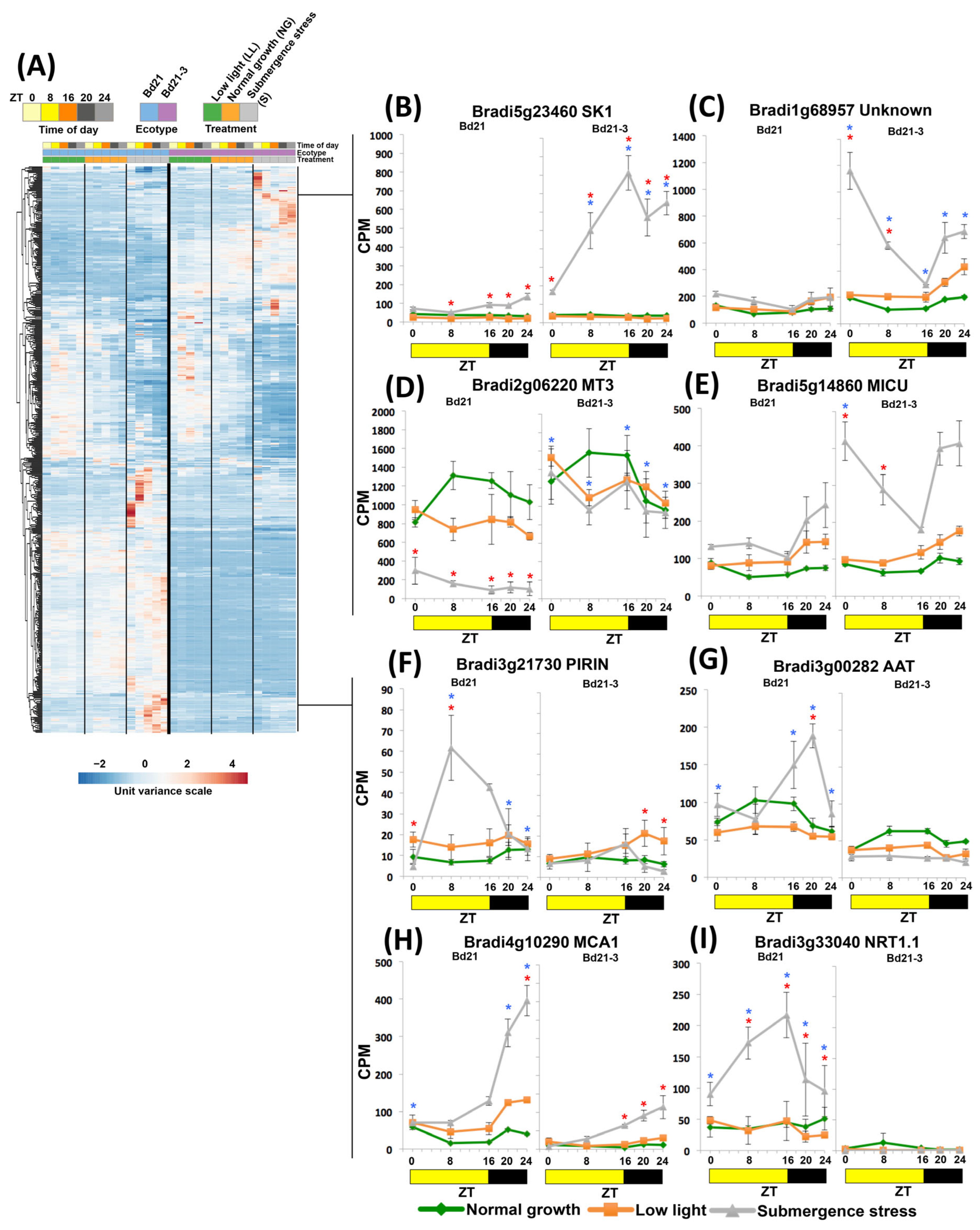
2.5. DEGS under Submergence Had ZT-Dependent Transcriptomic Responses
2.6. Ecotypes of Contrasting Tolerance Have Exclusive DEGs with Acquired ZT-Dependent Expression
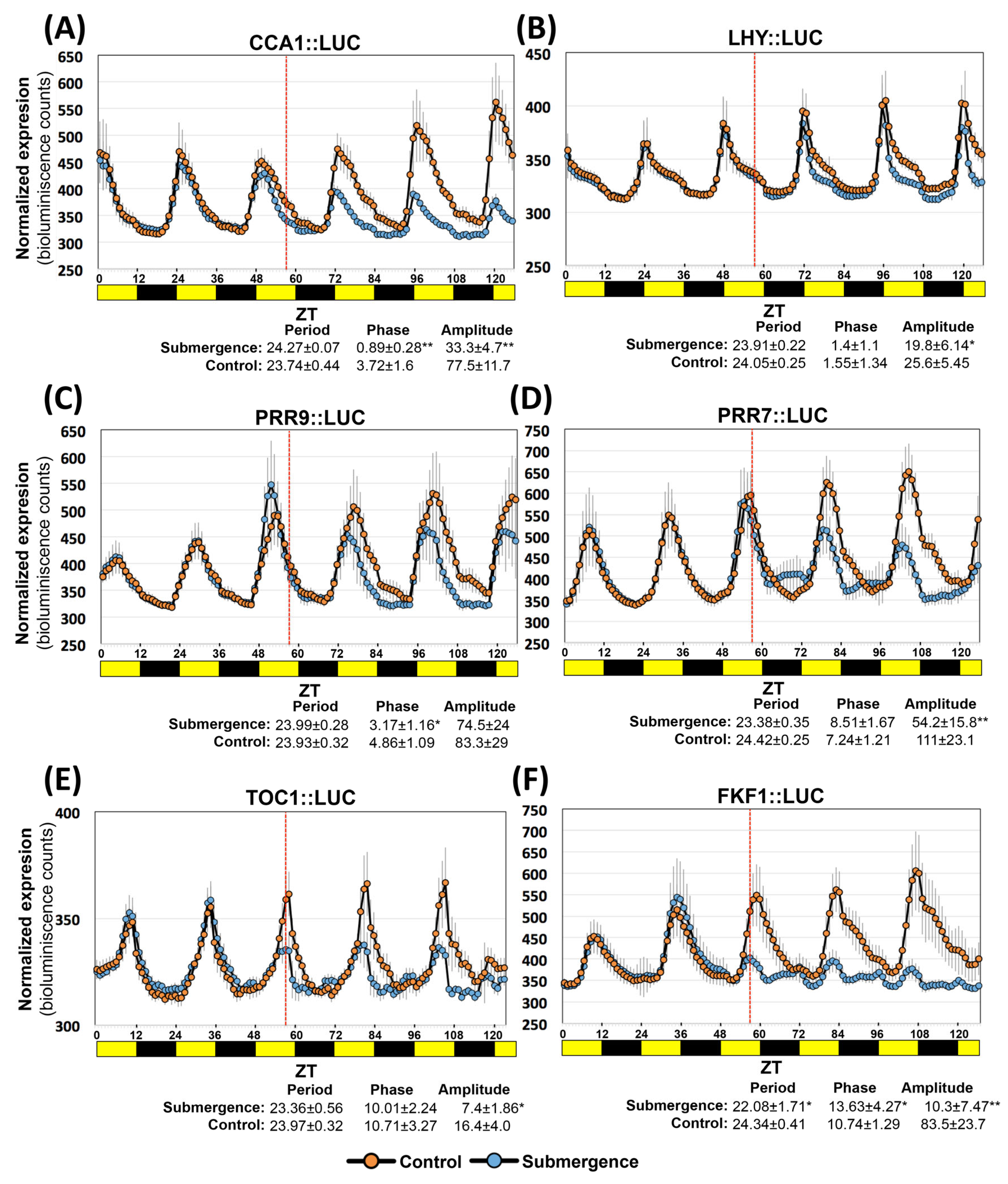
2.7. Arabidopsis Clock Genes Expression Is also Reshaped under Submergence Stress
2.8. The Hypoxia Core Genes in Brachypodium Are Early Responders
3. Discussion
3.1. The Transcriptomic Response of Brachypodium Is Dynamically Responding to Time of Day
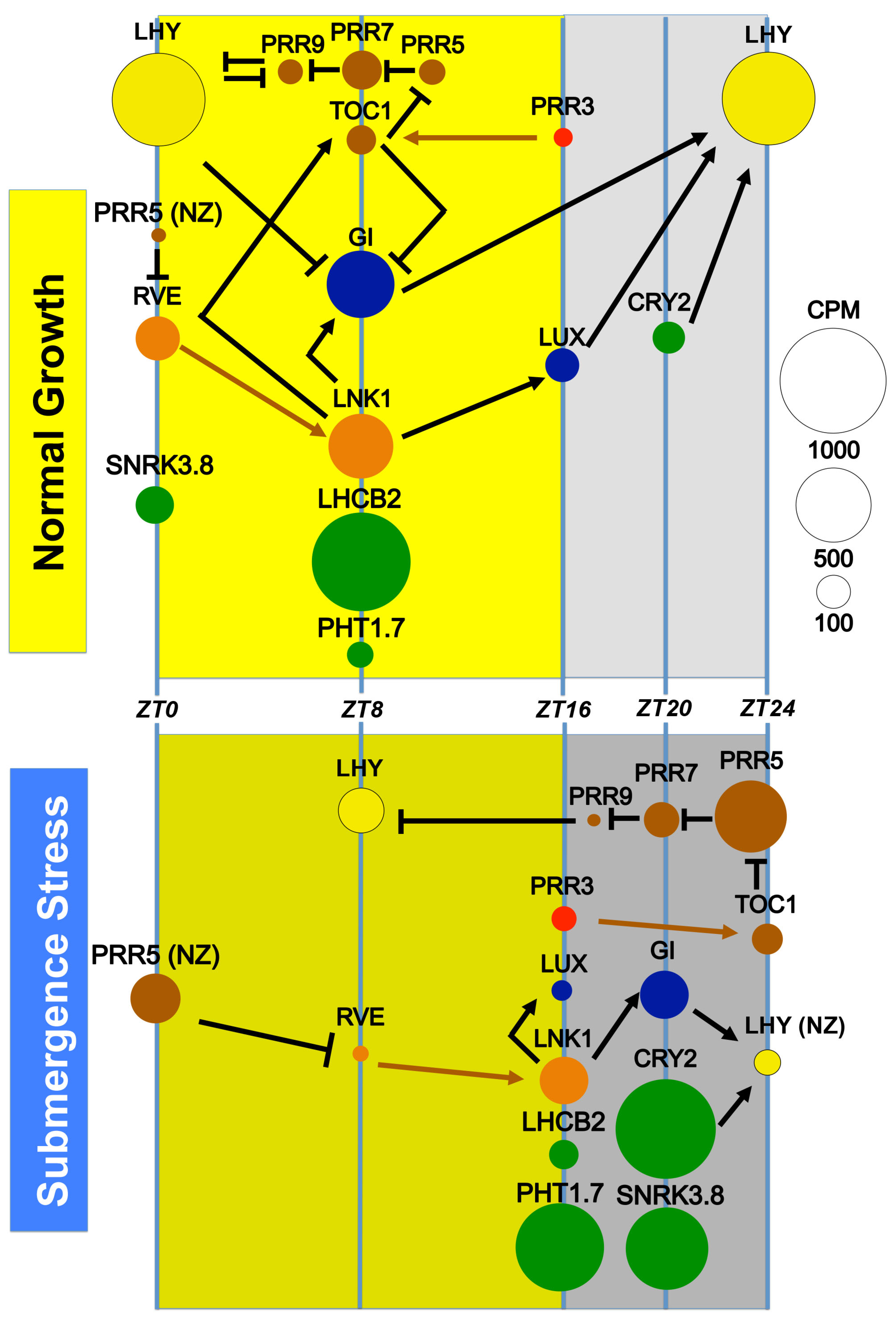
3.2. The Expression of Known Circadian Clock Genes and Outputs Is Affected by Submergence, a Multicomponent Stress
3.3. DEGs Present a Picture of Complex Interplay under Submergence between Metabolism, Signaling and Rhythmic Processes
3.4. Conclusions
4. Materials and Methods
4.1. Brachypodium Germplasm
4.2. Submergence Stress in Brachypodium
4.3. RNA Sequencing
4.4. Bioinformatic Analysis
4.5. Bioluminescence and Submergence Stress in Arabidopsis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bailey-Serres, J.; Lee, S.C.; Brinton, E. Waterproofing Crops: Effective Flooding Survival Strategies. Plant Physiol. 2012, 160, 1698–1709. [Google Scholar] [CrossRef] [PubMed]
- Science Breakthroughs to Advance Food and Agricultural Research by 2030; National Academies Press: Washington, DC, USA, 2019; ISBN 978-0-309-47392-7.
- Stevenson, S.; Coats, S.; Touma, D.; Cole, J.; Lehner, F.; Fasullo, J.; Otto-Bliesner, B. Twenty-first century hydroclimate: A continually changing baseline, with more frequent extremes. Proc. Natl. Acad. Sci. USA 2022, 119, e2108124119. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. The Impact of Disasters and Crises on Agriculture and Food Security; FAO: Rome, Italy, 2021. [Google Scholar]
- Sasidharan, R.; Bailey-Serres, J.; Ashikari, M.; Atwell, B.J.; Colmer, T.D.; Fagerstedt, K.; Fukao, T.; Geigenberger, P.; Hebelstrup, K.H.; Hill, R.D.; et al. Community recommendations on terminology and procedures used in flooding and low oxygen stress research. New Phytol. 2017, 214, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Barrera-Figueroa, B.E.; Juntawong, P.; Peña-Castro, J.M. Submergence and Waterlogging Stress in Plants: A Review Highlighting Research Opportunities and Understudied Aspects. Front. Plant Sci. 2019, 10, 340. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Hartman, S.; Liu, Z.; Martopawiro, S.; Sajeev, N.; van Veen, H.; Yeung, E.; Voesenek, L.A.C.J. Signal Dynamics and Interactions during Flooding Stress. Plant Physiol. 2018, 176, 1106–1117. [Google Scholar] [CrossRef]
- Jethva, J.; Schmidt, R.R.; Sauter, M.; Selinski, J. Try or Die: Dynamics of Plant Respiration and How to Survive Low Oxygen Conditions. Plants 2022, 11, 205. [Google Scholar] [CrossRef]
- Streb, S.; Zeeman, S.C. Starch Metabolism in Arabidopsis. Arab. Book 2012, 2012, e0160. [Google Scholar] [CrossRef]
- Loreti, E.; Valeri, M.C.; Novi, G.; Perata, P. Gene Regulation and Survival under Hypoxia Requires Starch Availability and Metabolism. Plant Physiol. 2018, 176, 1286–1298. [Google Scholar] [CrossRef]
- Peña-Castro, J.M.; van Zanten, M.; Lee, S.C.; Patel, M.R.; Voesenek, L.A.J.C.; Fukao, T.; Bailey-Serres, J. Expression of rice SUB1A and SUB1C transcription factors in Arabidopsis uncovers flowering inhibition as a submergence tolerance mechanism. Plant J. 2011, 67, 434–446. [Google Scholar] [CrossRef]
- Rivera-Contreras, I.K.; Zamora-Hernández, T.; Huerta-Heredia, A.A.; Capataz-Tafur, J.; Barrera-Figueroa, B.E.; Juntawong, P.; Peña-Castro, J.M. Transcriptomic analysis of submergence-tolerant and sensitive Brachypodium distachyon ecotypes reveals oxidative stress as a major tolerance factor. Sci. Rep. 2016, 6, 27686. [Google Scholar] [CrossRef]
- Goodspeed, D.; Chehab, E.W.; Min-Venditti, A.; Braam, J.; Covington, M.F. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 2012, 109, 4674–4677. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Park, J.-S.; Shin, S.Y.; Kim, S.-G.; Lee, G.; Kim, H.-S.; Jeon, J.H.; Cho, H.S. Submergence deactivates wound-induced plant defence against herbivores. Commun Biol 2020, 3, 651. [Google Scholar] [CrossRef]
- Voesenek, L.A.C.J.; Harren, F.J.M.; Bögemann, G.M.; Blom, C.W.P.M.; Reuss, J. Ethylene Production and Petiole Growth in Rumex Plants Induced by Soil Waterlogging: The Application of a Continuous Flow System and a Laser Driven Intracavity Photoacoustic Detection System. Plant Physiol. 1990, 94, 1071–1077. [Google Scholar] [CrossRef]
- Lai, A.G.; Doherty, C.J.; Mueller-Roeber, B.; Kay, S.A.; Schippers, J.H.M.; Dijkwel, P.P. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 2012, 109, 17129–17134. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Li, L.; Narsai, R.; De Clercq, I.; Whelan, J.; Berkowitz, O. Mitochondrial signalling is critical for acclimation and adaptation to flooding in Arabidopsis thaliana. Plant J. 2020, 103, 227–247. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.H.; Prince, S.J.; Mutava, R.N.; Patil, G.; Li, S.; Chen, W.; Babu, V.; Joshi, T.; Khan, S.; Nguyen, H.T. Core clock, SUB1, and ABAR genes mediate flooding and drought responses via alternative splicing in soybean. J. Exp. Bot. 2015, 66, 7129–7149. [Google Scholar] [CrossRef]
- Lee, T.A.; Bailey-Serres, J. Integrative Analysis from the Epigenome to Translatome Uncovers Patterns of Dominant Nuclear Regulation during Transient Stress. Plant Cell 2019, 31, 2573–2595. [Google Scholar] [CrossRef]
- Bragg, J.N.; Wu, J.; Gordon, S.P.; Guttman, M.E.; Thilmony, R.; Lazo, G.R.; Gu, Y.Q.; Vogel, J.P. Generation and Characterization of the Western Regional Research Center Brachypodium T-DNA Insertional Mutant Collection. PLoS ONE 2012, 7, e41916. [Google Scholar] [CrossRef]
- Tsai, K.-J.; Lin, C.-Y.; Ting, C.-Y.; Shih, M.-C. Ethylene-Regulated Glutamate Dehydrogenase Fine-Tunes Metabolism during Anoxia-Reoxygenation. Plant Physiol. 2016, 172, 1548–1562. [Google Scholar] [CrossRef] [PubMed]
- Dutilleul, C.; Garmier, M.; Noctor, G.; Mathieu, C.; Chétrit, P.; Foyer, C.H.; de Paepe, R. Leaf Mitochondria Modulate Whole Cell Redox Homeostasis, Set Antioxidant Capacity, and Determine Stress Resistance through Altered Signaling and Diurnal Regulation. Plant Cell 2003, 15, 1212–1226. [Google Scholar] [CrossRef]
- Cervela-Cardona, L.; Yoshida, T.; Zhang, Y.; Okada, M.; Fernie, A.; Mas, P. Circadian Control of Metabolism by the Clock Component TOC1. Front. Plant Sci. 2021, 12, 683516. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.-S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated Transcription of Key Pathways in Arabidopsis by the Circadian Clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef] [PubMed]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef]
- Higgins, J.A.; Bailey, P.C.; Laurie, D.A. Comparative Genomics of Flowering Time Pathways Using Brachypodium distachyon as a Model for the Temperate Grasses. PLoS ONE 2010, 5, e10065. [Google Scholar] [CrossRef]
- Para, A.; Farré, E.M.; Imaizumi, T.; Pruneda-Paz, J.L.; Harmon, F.G.; Kay, S.A. PRR3 Is a Vascular Regulator of TOC1 Stability in the Arabidopsis Circadian Clock. Plant Cell 2007, 19, 3462–3473. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.; Matiolli, C.C.; Viana, A.J.C.; Hearn, T.J.; Kusakina, J.; Belbin, F.E.; Newman, D.W.; Yochikawa, A.; Cano-Ramirez, D.L.; Chembath, A.; et al. Circadian Entrainment in Arabidopsis by the Sugar-Responsive Transcription Factor bZIP63. Curr. Biol. 2018, 28, 2597–2606.e6. [Google Scholar] [CrossRef] [PubMed]
- Mockler, T.C.; Michael, T.P.; Priest, H.D.; Shen, R.; Sullivan, C.M.; Givan, S.A.; McEntee, C.; Kay, S.A.; Chory, J. The Diurnal Project: Diurnal and Circadian Expression Profiling, Model-based Pattern Matching, and Promoter Analysis. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 353–363. [Google Scholar] [CrossRef]
- Chrost, B.; Kolukisaoglu, U.; Schulz, B.; Krupinska, K. An α-galactosidase with an essential function during leaf development. Planta 2007, 225, 311–320. [Google Scholar] [CrossRef]
- Lee, S.C.; Mustroph, A.; Sasidharan, R.; Vashisht, D.; Pedersen, O.; Oosumi, T.; Voesenek, L.A.C.J.; Bailey-Serres, J. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 2011, 190, 457–471. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic. Acids. Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, Y.; Liu, Y.; Li, N.; Yan, J.; Luo, L. Wound-induced polypeptides improve resistance against Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 504, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.J. The Intracellular Dynamics of Circadian Clocks Reach for the Light of Ecology and Evolution. Annu. Rev. Plant Biol. 2016, 67, 595–618. [Google Scholar] [CrossRef]
- Kreft, H.; Jetz, W. Global patterns and determinants of vascular plant diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 5925–5930. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Miguez-Macho, G.; Jobbágy, E.G.; Jackson, R.B.; Otero-Casal, C. Hydrologic regulation of plant rooting depth. Proc. Natl. Acad. Sci. USA 2017, 114, 10572–10577. [Google Scholar] [CrossRef] [PubMed]
- Pucciariello, C.; Perata, P. Quiescence in rice submergence tolerance: An evolutionary hypothesis. Trends Plant Sci. 2013, 18, 377–381. [Google Scholar] [CrossRef]
- Vashisht, D.; Hesselink, A.; Pierik, R.; Ammerlaan, J.M.H.; Bailey-Serres, J.; Visser, E.J.W.; Pedersen, O.; van Zanten, M.; Vreugdenhil, D.; Jamar, D.C.L.; et al. Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol. 2011, 190, 299–310. [Google Scholar] [CrossRef]
- Campbell, M.T.; Proctor, C.A.; Dou, Y.; Schmitz, A.J.; Phansak, P.; Kruger, G.R.; Zhang, C.; Walia, H. Genetic and Molecular Characterization of Submergence Response Identifies Subtol6 as a Major Submergence Tolerance Locus in Maize. PLoS ONE 2015, 10, e0120385. [Google Scholar] [CrossRef]
- van Veen, H.; Mustroph, A.; Barding, G.A.; Vergeer-van Eijk, M.; Welschen-Evertman, R.A.M.; Pedersen, O.; Visser, E.J.W.; Larive, C.K.; Pierik, R.; Bailey-Serres, J.; et al. Two Rumex Species from Contrasting Hydrological Niches Regulate Flooding Tolerance through Distinct Mechanisms. Plant Cell 2013, 25, 4691–4707. [Google Scholar] [CrossRef]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet 2015, 16, 237–251. [Google Scholar] [CrossRef]
- Yeung, E.; van Veen, H.; Vashisht, D.; Sobral Paiva, A.L.; Hummel, M.; Rankenberg, T.; Steffens, B.; Steffen-Heins, A.; Sauter, M.; de Vries, M.; et al. A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, E6085–E6094. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Thalor, S.K.; Berberich, T.; Lee, S.S.; Yang, S.H.; Zhu, X.; Imai, R.; Takahashi, Y.; Kusano, T. Deregulation of Sucrose-Controlled Translation of a bZIP-Type Transcription Factor Results in Sucrose Accumulation in Leaves. PLoS ONE 2012, 7, e33111. [Google Scholar] [CrossRef] [PubMed]
- Haydon, M.J.; Mielczarek, O.; Robertson, F.C.; Hubbard, K.E.; Webb, A.A.R. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 2013, 502, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.J.; Kay, S.A. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA 1996, 93, 15491–15496. [Google Scholar] [CrossRef] [PubMed]
- de Montaigu, A.; Giakountis, A.; Rubin, M.; Tóth, R.; Cremer, F.; Sokolova, V.; Porri, A.; Reymond, M.; Weinig, C.; Coupland, G. Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc. Natl. Acad. Sci. USA 2015, 112, 905–910. [Google Scholar] [CrossRef]
- Blair, E.J.; Bonnot, T.; Hummel, M.; Hay, E.; Marzolino, J.M.; Quijada, I.A.; Nagel, D.H. Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Sci Rep 2019, 9, 4814. [Google Scholar] [CrossRef]
- Bonnot, T.; Nagel, D.H. Time of the day prioritizes the pool of translating mRNAs in response to heat stress. Plant Cell 2021, 33, 2164–2182. [Google Scholar] [CrossRef]
- Wilkins, O.; Waldron, L.; Nahal, H.; Provart, N.J.; Campbell, M.M. Genotype and time of day shape the Populus drought response. Plant J. 2009, 60, 703–715. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, W.; Karapetyan, S.; Mwimba, M.; Marqués, J.; Buchler, N.E.; Dong, X. Redox rhythm reinforces the circadian clock to gate immune response. Nature 2015, 523, 472–476. [Google Scholar] [CrossRef]
- Adams, S.; Grundy, J.; Veflingstad, S.R.; Dyer, N.P.; Hannah, M.A.; Ott, S.; Carré, I.A. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018, 220, 893–907. [Google Scholar] [CrossRef] [PubMed]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Balasubramanian, V.K.; Chopra, R.; Mendu, V. The Photoperiodic Flowering Time Regulator FKF1 Negatively Regulates Cellulose Biosynthesis. Plant Physiol. 2019, 180, 2240–2253. [Google Scholar] [CrossRef]
- Scandola, S.; Mehta, D.; Li, Q.; Rodriguez Gallo, M.C.; Castillo, B.; Uhrig, R.G. Multi-omic analysis shows REVEILLE clock genes are involved in carbohydrate metabolism and proteasome function. Plant Physiol. 2022, 190, 1005–1023. [Google Scholar] [CrossRef]
- Hernández-Verdeja, T.; Strand, Å. Retrograde Signals Navigate the Path to Chloroplast Development. Plant Physiol. 2018, 176, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Gong, Q.; Ma, Y.; Li, P.; Li, J.; Yang, S.; Yuan, L.; Yu, Y.; Pan, D.; Xu, F.; et al. cpSecA, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J. Exp. Bot. 2010, 61, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Behera, S.; De Bortoli, S.; Logan, D.C.; Fuchs, P.; Carraretto, L.; Teardo, E.; Cendron, L.; Nietzel, T.; Füßl, M.; et al. The EF-Hand Ca2+ Binding Protein MICU Choreographs Mitochondrial Ca2+ Dynamics in Arabidopsis. Plant Cell 2015, 27, 3190–3212. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Nakagawa, Y.; Mori, K.; Nakano, M.; Imamura, T.; Kataoka, H.; Terashima, A.; Iida, K.; Kojima, I.; Katagiri, T.; et al. MCA1 and MCA2 That Mediate Ca2+ Uptake Have Distinct and Overlapping Roles in Arabidopsis. Plant Physiol. 2010, 152, 1284–1296. [Google Scholar] [CrossRef]
- Shingaki-Wells, R.N.; Huang, S.; Taylor, N.L.; Carroll, A.J.; Zhou, W.; Millar, A.H. Differential Molecular Responses of Rice and Wheat Coleoptiles to Anoxia Reveal Novel Metabolic Adaptations in Amino Acid Metabolism for Tissue Tolerance. Plant Physiol. 2011, 156, 1706–1724. [Google Scholar] [CrossRef]
- García-Aguilar, A.; Cuezva, J.M. A Review of the Inhibition of the Mitochondrial ATP Synthase by IF1 in vivo: Reprogramming Energy Metabolism and Inducing Mitohormesis. Front. Physiol. 2018, 9, 1322. [Google Scholar] [CrossRef]
- Chen, C.; Meng, Y.; Shopan, J.; Whelan, J.; Hu, Z.; Yang, J.; Zhang, M. Identification and characterization of Arabidopsis thaliana mitochondrial F1F0-ATPase inhibitor factor 1. J. Plant Physiol. 2020, 254, 153264. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Meng, Y.; Hu, Z.; Yang, J.; Zhang, M. Identification of New Proteins and Potential Mitochondrial F1F0-ATPase Inhibitor Factor 1-Associated Mechanisms in Arabidopsis thaliana Using iTRAQ-Based Quantitative Proteomic Analysis. Plants 2021, 10, 2385. [Google Scholar] [CrossRef] [PubMed]
- Khalid, M.; Nan, H.U.I.; Kayani, S.I.; Kexuan, T.A.N.G. Diversity and versatile functions of metallothioneins produced by plants: A review. Pedosphere 2020, 30, 577–588. [Google Scholar] [CrossRef]
- Spitzer, M.; Wildenhain, J.; Rappsilber, J.; Tyers, M. BoxPlotR: A web tool for generation of box plots. Nat. Methods 2014, 11, 121–122. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup the Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Bonnot, T.; Gillard, M.B.; Nagel, D.H. A Simple Protocol for Informative Visualization of Enriched Gene Ontology Terms. Bio-Protocol 2019, e3429. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Salomé, P.A.; McClung, C.R. PSEUDO-RESPONSE REGULATOR 7 and 9 Are Partially Redundant Genes Essential for the Temperature Responsiveness of the Arabidopsis Circadian Clock. Plant Cell 2005, 17, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.-H.; Tobin, E.M.; et al. F-Box Proteins FKF1 and LKP2 Act in Concert with ZEITLUPE to Control Arabidopsis Clock Progression. Plant Cell 2010, 22, 606–622. [Google Scholar] [CrossRef]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal Regulation Between TOC1 and LHY/CCA1 Within the Arabidopsis Circadian Clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef]
- Zielinski, T.; Moore, A.M.; Troup, E.; Halliday, K.J.; Millar, A.J. Strengths and Limitations of Period Estimation Methods for Circadian Data. PLoS ONE 2014, 9, e96462. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medina-Chávez, L.; Camacho, C.; Martínez-Rodríguez, J.A.; Barrera-Figueroa, B.E.; Nagel, D.H.; Juntawong, P.; Peña-Castro, J.M. Submergence Stress Alters the Expression of Clock Genes and Configures New Zeniths and Expression of Outputs in Brachypodium distachyon. Int. J. Mol. Sci. 2023, 24, 8555. https://doi.org/10.3390/ijms24108555
Medina-Chávez L, Camacho C, Martínez-Rodríguez JA, Barrera-Figueroa BE, Nagel DH, Juntawong P, Peña-Castro JM. Submergence Stress Alters the Expression of Clock Genes and Configures New Zeniths and Expression of Outputs in Brachypodium distachyon. International Journal of Molecular Sciences. 2023; 24(10):8555. https://doi.org/10.3390/ijms24108555
Chicago/Turabian StyleMedina-Chávez, Lucisabel, Christian Camacho, Jorge Arturo Martínez-Rodríguez, Blanca Estela Barrera-Figueroa, Dawn H. Nagel, Piyada Juntawong, and Julián Mario Peña-Castro. 2023. "Submergence Stress Alters the Expression of Clock Genes and Configures New Zeniths and Expression of Outputs in Brachypodium distachyon" International Journal of Molecular Sciences 24, no. 10: 8555. https://doi.org/10.3390/ijms24108555
APA StyleMedina-Chávez, L., Camacho, C., Martínez-Rodríguez, J. A., Barrera-Figueroa, B. E., Nagel, D. H., Juntawong, P., & Peña-Castro, J. M. (2023). Submergence Stress Alters the Expression of Clock Genes and Configures New Zeniths and Expression of Outputs in Brachypodium distachyon. International Journal of Molecular Sciences, 24(10), 8555. https://doi.org/10.3390/ijms24108555








