Abstract
Barrett’s esophagus (BE) is a disease entity that is a sequela of chronic gastroesophageal reflux disease that may result in esophageal adenocarcinoma (EAC) due to columnar epithelial dysplasia. The histological degree of dysplasia is the sole biomarker frequently utilized by clinicians. However, the cost of endoscopy and the fact that the degree of dysplasia does not progress in many patients with BE diminish the effectiveness of histological grading as a perfect biomarker. Multiple or more quantitative biomarkers are required by clinicians since early diagnosis is crucial in esophageal adenocancers, which have a high mortality rate. The presence of epigenetic factors in the early stages of this neoplastic transformation holds promise as a predictive biomarker. In this review, current studies on DNA methylations, histone modifications, and noncoding RNAs (miRNAs) that have been discovered during the progression from BE dysplasia to EAC were collated.
1. Introduction
BE is a columnar cell dysplasia characterized as a phenotype of gastroesophageal reflux disease (GERD) with high-grade esophagitis, which can progress to esophageal adenocarcinoma (EAC) [1]. Dr. Norman Barrett first mentioned this type of dysplasia in his 1950 article titled “Chronic peptic ulcers of the esophagus and esophagitis” [2], and since then, thousands of articles have been published regarding this aberrance, particularly regarding its association with esophageal cancer. A meta-analysis reported a worldwide prevalence of 3–14% for histologically confirmed BE [3]. The incidence of BE is steadily increasing in Western societies, where it is more prevalent compared to Eastern societies [4,5]. BE can lead to the development of a more severe condition known as low-grade dysplasia (LGD) or high-grade dysplasia (HGD). These abnormal cells can progress to intramucosal carcinoma and eventually become invasive carcinoma without treatment [6]. Patients with GERD are 3.1 times more likely to develop EAC compared to those without GERD. However, the likelihood of developing EAC is significantly higher in patients with BE, which is 29.8 times greater compared to those without BE [7]. The prognosis for EAC is generally poor, as approximately >50% of cases are typically diagnosed at advanced stages (III–IV). This is a major contributing factor to the low 5-year survival rate of EAC patients, which has recently been reported to range between 20.1% and 23.4% [8,9].
The objective of regular endoscopic surveillance accompanied by histopathological examination in patients with BE and EAC is to identify dysplasia or neoplasia at an early stage [10]. Therefore, the identification of specific biomarkers in the detection of LGD or HGD and EAC is important due to its potential for early intervention and cancer stage determination, including its cost-effectiveness and applicability compared to upper gastrointestinal endoscopy [11]. It has become widely acknowledged that epigenetic factors, specifically those related to super-enhancers, DNA methylation, histone modifications, and non-noncoding RNAs, can be inherited somatically and can play a role in creating lasting but adaptable alterations in the development and advancement of HGD and EAC [12,13]. Early stages of cancer development involve the initiation of epigenetic modifications, which are indicative of the likelihood of progression. There are various epigenetic modifications that occur during carcinogenesis, including DNA methylation, changes to histone proteins after translation, certain types of miRNA, and alterations to nucleosome positioning [10,14].
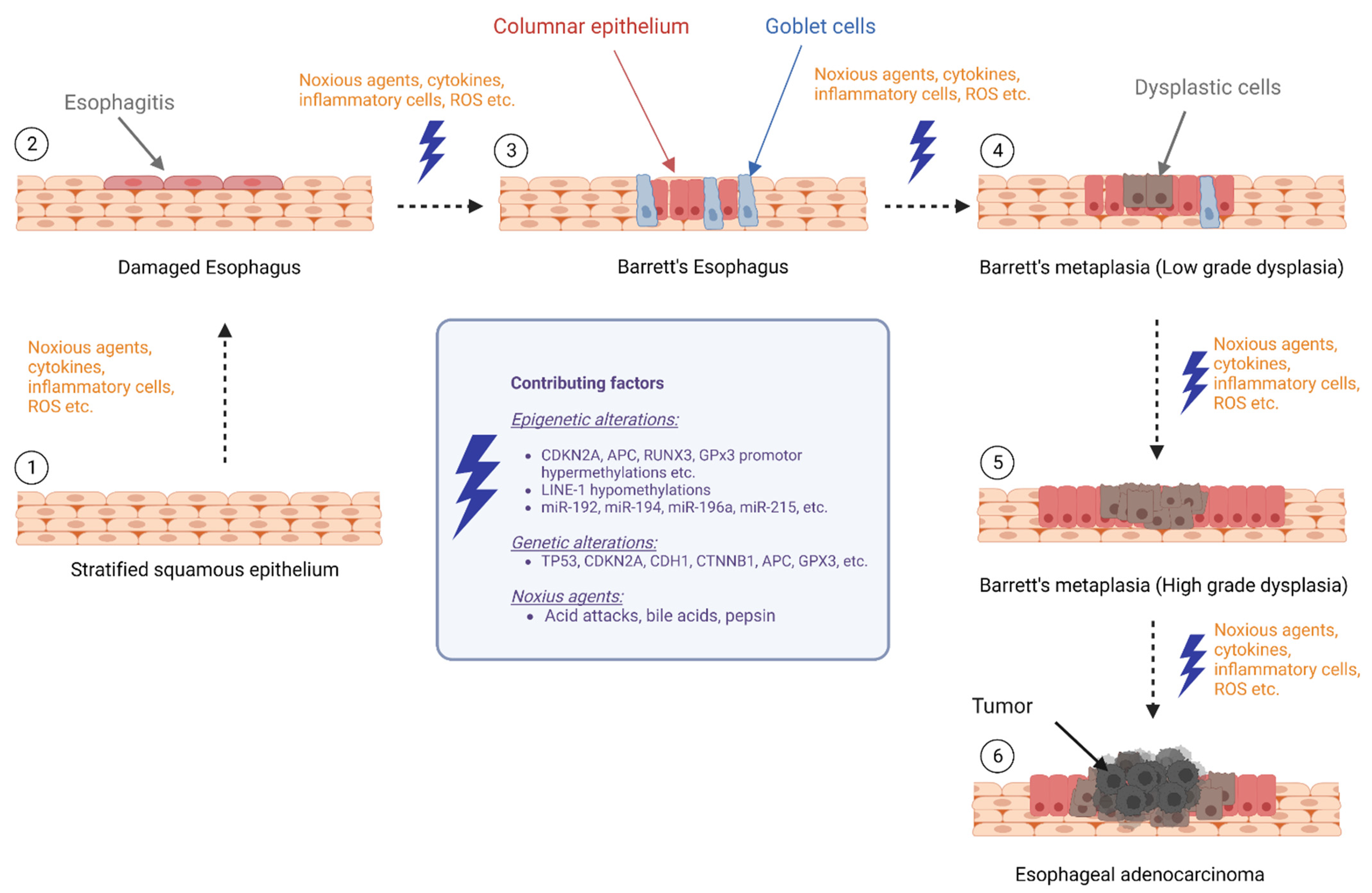
This review aimed to provide an overview of the current literature on biomarker research focusing on epigenetic changes and their potential role in the progression from BE to EAC, which summarizes DNA methylation, histone modifications, and noncoding RNAs that may contribute to the development of EAC in individuals with BE (Figure 1).
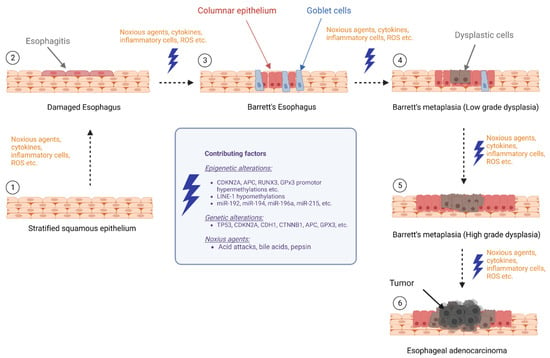
Figure 1.
Epigenetic alterations during progression of Barrett’s esophagus to esophageal cancer (BE: Barrett’s esophagus, LGD: low-grade dysplasia, HGD: high-grade dysplasia, and EAC: esophageal adenocarcinoma) (The figure created with biorender.com, accessed on 23 March 2023).
1.1. DNA Methylation as Biomarkers BE and EAC
DNA methylation is an important epigenetic modification that involves the addition of a methyl group to cytosine residues in DNA. Normal methylation patterns are necessary for cell growth and metabolism, whereas abnormal methylation can lead to diseases such as tumors [15]. Other epigenetic modifications, such as histone modifications and noncoding RNAs, also play important roles in gene regulation and development. Environmental factors can also influence epigenetic patterns, highlighting the importance of gene–environment interactions. In vertebrates, DNA methylation occurs when a methyl group binds to the CpG sites by DNA methyltransferase; as a result, five methyl cytosines are formed. Methylated cytosines are found in approximately 75% of all CpG dinucleotides in the human genome [16]. The enzyme that adds methyl groups to the CpG islands in DNA is called DNA methyltransferase (DNMT). DNA methylations can inhibit gene expression either directly or indirectly through methyl-CpG binding domain proteins, thereby suppressing protein expression [17].
In all cancer types, hypermethylation and hypomethylation are observed in DNA. Wide areas of hypomethylation are observed globally, whereas hypermethylation is observed in specific regions such as CpG islands and localized areas of hypermethylation in gene promoter regions [14]. There are significant differences in the amount and distribution of DNA methylation between different vertebrate tissues because DNA methylation varies by species and tissue, highlighting the importance of tissue-specific epigenetic regulation for proper gene expression and differentiation [18].
1.2. DNA Hypermethylation Is a Frequent Event in BE and EAC
DNA methylation has also been studied extensively in BE. Hypermethylation of CpG islands and hypomethylation have distinctive hallmarks in BE progression [19]. Abnormal methylation of CpG islands has been examined in BE, dysplastic BE, and EAC. The hypermethylation of cyclin-dependent kinase inhibitor 2A (CDKN2A), a stabilizer of the tumor suppressor protein p53 and cell cycle G1 control inhibitor [20], has been shown in studies conducted nearly 20 years ago [21,22,23,24,25,26]. The CDKN2A gene is located on chromosome 9p21 and has two different upstream exons (1α and 1β) regulated by different promoters. The transcript that was initiated from the proximal promoter (1α) encodes CDKN2A–p16INK4A, while the latter (1β) encodes CDKN2A–p14ARF [20]. The majority of CDKN2A studies in BE, HGD, and EAC is on CDKN2A–p16INK4A promoter methylation. This chromosome loss and hypermethylation of the promoter of the CDKN2A–p16INK4A inhibit the activity of the CDKN2A gene. Inactivation of this gene has been reported in patients with BE, dysplasia, and EAC [14,21,26,27]. Although the frequency of CDKN2A hypermethylation in BE mucosa ranges from 3% to 77%, it varies between 11% and 75% in dysplastic tissues and between 16% and 85% in EAC (Table 1). Furthermore, CpG island hypermethylation of the CDKN2A promoter was either absent or very low in the normal squamous epithelium (Table 1). These findings suggest that CDKN2A methylation is an early change in BE formation.

Table 1.
Hypermethylated genes in Barrett’s Carcinoma.
However, hypermethylation of this gene is not specific to BE, suggesting the need for multiple methylation markers. A panel including TP53 loss of heterozygosity (LOH), CDKN2A LOH, and tetraploidy was screened in esophageal biopsies from 243 BE patients, and it was shown that the co-existence of these three abnormalities has an approximately 39-fold risk of cancer progression. However, it was stated that the methylation of CDKN2A by itself did not cause a significant statistical difference [55]. A retrospective cohort study of 50 progressive and 145 nonprogressive BE patients by Jin et al. reported that a panel of 8 biomarkers including CDKN2A and age factor had an area under the curve (AUC) of 0.732 at diagnosis [56]. Schulmann et al., in their retrospective cohort study with 77 EACs, 93 BEs, 20 dysplasias (n = 14 LGD, n = 6 HGD), and 64 NE patients, reported that CDKN2A, RUNX3, and HPP1 inactivation and progression risk increased 2 years before EAC diagnosis in patients with BE [24]. CDKN2A is also regulated independently by the p14ARF promotor [57]. However, Vieth et al. showed that p14ARF promoter hypermethylation can also be observed frequently in Barrett adenocarcinoma (20%) [25]. It has been reported that during the progression from normal epithelium to BE and EAC, there is a significant decrease in p14ARF expression [58].
Hypermethylation of the CDKN2A promoter may be induced by nicotinamide adenine dinucleotide phosphate hydrogen (NADPH) oxidase 5 (NOX5) overexpression in BE and EAC. NOX5 is a novel NADPH oxidase that produces superoxide. Reactive oxygen species (ROS) produced by NOX5 have been implicated in the immune system stimulation and signaling cascades during tumorigenesis [59]. Some cell culture studies in BE and EAC have shown that NOX5 messenger RNA (mRNA) levels are higher than in healthy tissues [60,61]. It has been reported that NOX5-mediated accumulation of hydrogen peroxide (H2O2) suppresses its own expression and causes CDKN2A promoter hypermethylation due to DNMT1 upregulation [61,62].
These studies show that CDKN2A promoter methylation is an important marker in the progression from BE to EAC. However, these studies are mostly qualitative, which means they cannot provide clear data regarding the degree of methylation that is crucial in the prognosis. In a recent study, lesion-containing sections of patients with LGD, HGD, and EAC were separated from histological sections by laser-capture microdissection, and p16 (encoded by CDKN2A) methylation analysis was performed with pyrosequencing. The authors reported that p16 methylation increased in parallel with the level of the carcinogenesis process [40].
In hypermethylation panel studies, such as the studies by Schulmann et al. [24] and Jin et al. [56], Runt-related transcription factor 3 (RUNX3) is also found to be hypermethylated in Barrett’s dysplasia (Table 1) [24,33,56]. This tumor suppressor gene, which has a role in the transforming growth factor β signaling pathway, has been frequently deleted or transcriptionally silenced in cancer [63]. In nine studies selected in a meta-analysis examining RUNX3 promoter methylation in the development of esophageal cancer, it was found that RUNX3 methylation of EAC was significantly higher than in healthy controls and patients with BE. RUNX3 promoter methylation has been reported to be an important independent risk factor for the progression of BE to HGD of the esophagus and EAC [64].
The downregulation of adenomatous polyposis coli (APC) hypermethylation is another topic of EAC progression. APC is a negative regulator of the Wnt/β-catenin pathway. The loss of APC expression causes the stabilization and nuclear accumulation of β-catenin, which can lead to the initiation of tumorigenesis [30]. Hypermethylation of the APC promoter has been studied in many cancers, including EAC (Table 1). In the combined hypermethylation panel study including the APC promoter, conducted by Clement et al. with 12 progressive BE and 16 nonprogressive BE patients, it was reported that the APC promoters of progressive patients and nonprogressive patients were 100% and 36% hypermethylated, respectively. However, no statistical analysis was performed in the study [29]. In a study examining the methylation status of APC, CDKN2A, mutL homolog 1 (hMLH), RUNX3, and Methylguanine methyltransferase (MGMT) genes, only APC gene hypermethylation was reported to be an independent predictor of EAC [65]. Wang et al. showed that p16 and APC promoter hypermethylation in 17 normal esophagus, 102 BE, and 42 adenocarcinoma patients is reported to be an important marker in dysplastic BE and EAC during a mean follow-up of 4.1 years [30]. In another study, it was reported that APC promoter hypermethylation was found in approximately 40% of BE patients and 92% of EAC patients in esophageal biopsies, whereas it was not detected in the esophagus of healthy controls. Plasma samples from the same patients were also taken, and methylated APC was found in 25% of the plasma of EAC patients [35].
These results suggest that aberrant methylation of this gene occurs in the early stages of the BE to EAC process. However, more precise data are warranted to determine the diagnostic power of APC methylation and its relationship with EAC progression. In a meta-analysis conducted by Wang et al. to investigate the early detection potential of APC hypermethylation in esophageal cancer, 18 studies showed that APC hypermethylation was higher in esophageal cancer (EC) and BE than controls in the data of 1008 ECs, 570 BEs, and 782 controls [66]. They also stated that APC methylation in ECs was similar to BE and not associated with tumor stage and survival. Researchers reported that the diagnostic performance of APC methylation shows an AUC of 0.94 in EC and 0.88 in BE.
β-catenin interacts with e-cadherin (CDH1), the intercellular synthesis molecule, and with APC, the tumor suppressor gene product. APC competes with e-cadherin for binding with beta-catenin [67]. E-cadherin is a tumor suppressor protein, and the downregulation of e-cadherin in tumor cells is frequently observed in metastasis [68]. In epithelial cancers, e-cadherin may be silenced by LOH in the 16q22 region, promoter hypermethylation [20], or inherited mutations such as germline large deletions [69] and transcriptional silencing [70] in gastric cancers. However, diagnostic inherited mutations in BE and EAC have not been demonstrated thus far in EC or BE. In a study with healthy controls and esophageal cancer patients, e-cadherin hypermethylation was not observed in healthy controls, whereas methylation was observed in 84% of esophageal tissues of cancer patients (Table 1) [37]. In a similar methylation panel study, 66% of patients with EAC were reported to have methylation [32]. Schildhaus et al. found that e-cadherin methylation was rare (1/10) in patients with Barrett carcinoma [38].
One of the main mechanisms triggering cancer is ROS-mediated DNA damage. ROS damages DNA, RNA, enzymes, and proteins, which are involved in the activation of oncogenes and inhibition of tumor suppressors [71]. The exposure of the esophageal epithelium to acid influences the pathogenesis of BE, and acid is considered a carcinogen [72]. Glutathione peroxidase 3 (GPx3) is the main scavenger of ROS in plasma and is responsible for H2O2 detoxification. GPx3 is expressed in many tissues of the human gastrointestinal tract, including the esophageal squamous epithelium [20]. One study showed that GPx3 mRNA expressions were reduced to approximately 90.5% in Barrett’s adenocarcinoma samples. In the same study, the authors concluded that 61.9% of BE, 81.8% of dysplastic samples, 88.2% of EAC samples, and 17% of normal squamous epithelium had GPx3 promoter hypermethylation (Table 1) [45]. However, they discussed that the methylation in normal samples may be attributed to abnormal contaminant cells. These results show that intestinal metaplasia cells lose their ability to ROS detoxification. In addition, GPx3 hypermethylation may be an important marker that develops in the early period of BE carcinogenesis.
As seen in Table 1, the frequency of hypermethylation in well-studied genes such as CDKN2A/p16 varies among studies. Although methylation-specific PCR is mostly used in these studies, the use of methods such as methylight [34] or melt curve analysis [33] may also contribute to this variability. Additionally, the difficulty in diagnosing and monitoring patient groups such as Barrett’s dysplasia, the lack of information on dysplasia grade in each study, and the small sample sizes may explain the differences in ranges.
1.3. DNA Hypomethylation in BE and EAC
In the prognosis of BE and EAC, global hypomethylation is an important distinguishing characteristic, along with hypermethylation of CpG islands. Hypomethylation levels in DNA have been linked to a higher likelihood of genome instability [73] and increased expression of oncogenes such as Cyclin D1 (CCND1), Cyclin E1 (CCNE1), KRAS, MYC, and cell division protein kinase 6 (CDK6) [74]. A study showed that global hypomethylation is present at the earliest stages of epithelial carcinogenesis in BE [75]. The study also found that epigenetic regulation during epithelial carcinogenesis may not be restricted to traditionally defined “CpG islands” but also occur through differential methylation outside these regions. Finally, they found that novel targets X-C motif chemokine ligand 1 (XCL1), matrix metallopeptidase 13 (XCL3), GATA binding protein 6 (GATA6), and deleted in malignant brain tumors 1 (DMBT1) were more highly expressed in NDBE and HGD/EAC tissues compared to normal squamous epithelium. This suggests that the observed hypomethylation may contribute to the development and progression of BE and EAC by activating genes that promote cell proliferation and survival. Another study showed that patients with BE and EAC have decreased DNA methylation levels outside the CpG islands and increased methylation in the CpG islands compared to the squamous epithelium [19].
Hypermethylation and hypomethylation lead to global changes in the transcriptome that are involved in the development of EAC and appear early in carcinogenesis. In a study examining differentially methylated CpG sites in patients with BE and EAC, there were 15 CpG sites that demonstrated differential methylation between the BE hypermethylated epigenotype and BE hypomethylated epigenotypes and 74 CpG sites in the EAC [14]. In a retrospective cohort study, methylation profiles of 150 BE and 285 EAC cases were examined and grouped into 4 subtypes: subtype 1 with aberrant DNA methylation, a high mutation rate, and multiple mutations in the cell cycle and receptor tyrosine signaling pathways; subtype 2 with a metabolic gene expression pattern and unmethylated transcription factor binding sites; subtype 3 with no changes in methylation; and subtype 4 with DNA hypomethylation, which is linked to structural changes and copy number variations, with increased amplification of CCNE1. Subtype 4 exhibited hypomethylation, large-scale genomic rearrangements, copy number alterations, and CCNE1 and erb-b2 receptor tyrosine kinase 2 (ERBB2) amplification. The findings of this study reveal the heterogeneity of DNA methylation patterns in BE and EAC and their impact on gene expression and genomic stability, providing evidence for the involvement of DNA methylation changes in EAC development [74]. Boldrin et al. stated that LINE-1 hypomethylation could be used as a biomarker not only to monitor EAC prognosis but also as an indicator in BE surveillance [76]. In a study examining the whole genome methylation of progressive and nonprogressive nondysplastic BE patients, 44 methylation profiles were found to be different between the two groups; particularly, hypomethylation at the OR3A4 position was identified as a differentiator between progressive and nonprogressive patients [13].
Studies have demonstrated that the epigenomes of BE and EAC are frequently hypomethylated in intragenic and noncoding regions [14]. Furthermore, hypomethylation may be more dominant than hypermethylation in BE progression [56,75]. A high-resolution methylome analysis study showed that in the early stages of BE progression, there is a greater tendency toward hypomethylation rather than hypermethylation as the primary epigenetic alteration, and early progression of BE is characterized by genome-wide hypomethylation affecting both coding and noncoding regions of the genome [77].
1.4. Histone Modifications in BE and EAC Pathogenesis
Histones are a group of small proteins that are vital components of chromatin, the material that makes up chromosomes. They comprise four different types, known as H2A, H2B, H3, and H4. Each histone has a spherical section and a flexible charged tail, which extends from a protein complex called a nucleosome. A nucleosome is a cluster of 8 histone proteins that wrap around approximately 146 base pairs of DNA. These histone octamers are made up of two copies of each of the four core histones [78]. Chromatin is a nucleoprotein complex composed of DNA, histones, and nonhistone proteins that serves as the fundamental framework for storing eukaryotic genetic information. Regulation of gene expression is achieved through modifications to the histone tails, including acetylation, methylation, phosphorylation, ubiquitination, sumoylation, proline isomerization, and ADP ribosylation. These modifications are post-translational and allow fine-tuned control over gene expression [79]. The alterations to histone modifications can be undone and are regulated by enzymes that comprise histone acetyltransferases (HATs) and deacetylases (HDACs), as well as methyltransferases and demethylases [80]. For instance, histone acetylation is a chemical modification that influences DNA structure and gene transcription. When lysine residues are acetylated by HATs, the DNA structure relaxes, which facilitates gene transcription. Conversely, hypoacetylation of histones is a characteristic feature of inactive heterochromatin. In cancer cells, there is an impaired balance between HATs and HDACs, resulting in a significant alteration in the chromatin structure. This, in turn, leads to changes in gene expression, particularly those related to cell cycle regulation, differentiation, and apoptosis. Thus, cancer cells display aberrant gene expression patterns due to the disrupted balance between HATs and HDACs [10,81].
Despite the potential importance of histone modifications in BE and EAC, relatively few studies have been conducted in this area. Many of the studies that have been conducted have focused on esophageal squamous cell carcinoma (ESCC), a different type of esophageal cancer that is not closely related to BE or EAC [82]. The study investigated the expression of histone deacetylase 1 (HDAC1) and histone deacetylase 2 (HDAC2) genes in EAC and found that both genes were highly expressed in cancerous tissues. However, there was no correlation between HDAC1 expression and tumor stages. In contrast, the study revealed a significant correlation between overexpression of HDAC2 and increased lymphatic spread of the tumor, as well as aggressive tumor behavior [83].
To gain a comprehensive understanding of the role of histone modifications in the progression from BE to EAC, there is a need for further detailed and extensive investigations, similar to those that have been conducted for ESCC.
Moreover, the identification of additional histone modifications and the characterization of their roles in BE and EAC could provide further insights into the pathogenesis of these diseases and potentially lead to the development of novel therapeutic targets. In conclusion, the examination of histone modifications in BE and EAC is a promising area of research with significant implications for the diagnosis and treatment of these diseases, but further studies are needed to fully understand their roles and potential as biomarkers and therapeutic targets.
1.5. The Role of miRNAs as Biomarkers in BE and EAC
MicroRNAs (miRNAs) are small, noncoding RNA molecules of approximately 20–22 nucleotides that regulate gene expression by binding to target mRNAs leading to their post-transcriptional modifications. miRNAs play important roles in regulating important and disease-related cellular processes such as cell proliferation, differentiation, and apoptosis. In the context of cancer, certain microRNAs can play opposing roles in tumor development and progression. Oncomir microRNAs promote tumor growth and metastasis by downregulating tumor suppressor genes, whereas tumor suppressor microRNAs inhibit tumor growth by targeting oncogenes. These opposing actions highlight the complex role of microRNAs in cancer biology and their potential as therapeutic targets [84].
While the exact mechanisms underlying the transition from BE to EAC are not fully understood, research has shown that miRNAs might play a role in this process. Many studies have investigated miRNAs as potential biomarkers and therapeutic targets for the early detection and treatment of BE and EAC. These studies have identified specific miRNAs that are dysregulated in BE and EAC tissues and have demonstrated their potential as diagnostic and prognostic biomarkers. Despite these promising findings, miRNA-based biomarkers and therapeutics for BE and EAC have yet to be translated into clinical practice. Further research is needed to validate the diagnostic and prognostic utility of miRNAs and to develop effective miRNA-targeted therapies for these pathologies. Nevertheless, the identification of dysregulated miRNAs in BE and EAC tissues has provided valuable insights into the molecular mechanisms underlying these diseases and has opened up new avenues for their diagnosis and treatment.
miR-192 acts as a tumor suppressor by inhibiting cell proliferation and inducing apoptosis in different cancer types such as colon and prostate cancers [85,86]. Conversely, some studies have reported that miR-192 has oncogenic properties by promoting cell proliferation, migration, and invasion in different cancer types such as bladder and pancreatic cancers [87]. Studies examining the role of miR-192 in BE and EAC suggest that it generally acts as a tumor suppressor (Table 2). Hassan et al. found that miR-192 expression was significantly reduced in BE tissues compared with normal esophageal tissues. Reduced expression of miR-192 was associated with an increased risk of progression from BE to EAC. They also showed that overexpression of miR-192 in EAC cells inhibited cell proliferation and invasion, suggesting that miR-192 may function as a tumor suppressor in EAC [88]. miR-192 has the potential to be a noninvasive biomarker for the diagnosis of BE in general. Further studies are needed to confirm the findings in larger patient cohorts and to explore the clinical utility of microRNA-based diagnostic tests for BE [89,90].

Table 2.
Deregulated microRNAs in BE and EAC.
miR-194 is generally considered to be a tumor suppressor miRNA. Studies have shown that miR-194 expression is reduced in many types of cancer, including gastric, colorectal, and breast cancers. This reduction in miR-194 expression is often associated with increased tumor growth, invasion, and metastasis [101,102]. miR-194 has been identified as a predictive marker for the progression of BE and EAC. The research findings indicated that miR-194 expression levels were notably elevated in tissue samples obtained from patients who had developed EAC [90,92]. A thorough investigation conducted on serum samples collected from individuals with BE and EAC revealed that, similar to the findings from tissue samples, miR-194 expression levels were markedly elevated in the serum of these patients as compared with those with normal esophageal epithelium [103]. In addition, increased miR-194 gene expression has been associated with intestinal epithelial differentiation by Hino et al. [104]. Although strong evidence postulates that miR-194 is associated with metaplasia and neoplastic progression, reaching a definite conclusion regarding whether it has oncomir or tumor suppressor properties remains unattainable [105].
The miRNAs miR-192 and miR-194 have been extensively researched and found to exhibit elevated expression levels in BE and EAC. The high miR-192 and -194 expression in BE and EAC is due to hypomethylation in the promoter regions [89].
miR-215 can act as an oncomir or as a tumor suppressor, depending on the mRNA targets. For instance, this miRNA has tumor suppressor properties in many cancer types, while in gastric cancer, miR-215 has been shown to promote malignant progression by targeting the RUNX family transcription factor 1 (RUNX1) gene, which is involved in cell differentiation and apoptosis [106,107]. Several studies have investigated the expression of miR-215 in BE and EAC. The results of these studies have consistently shown that mir-215 expression is higher in both BE and EAC compared to normal esophageal tissue [98,108,109]. However, when the expression levels between BE and EAC were compared, two studies found that the expression of miR-215 was lower in EAC [90,92]. This finding is particularly significant for the potential use of miR-215 as a biomarker during the transition from BE to EAC. Additionally, the decreased expression of miR-215 in EAC suggests that it may have tumor suppressor properties.
miR-203 plays a crucial role as a tumor suppressor in various types of cancer [110]. The ability to bind the 3′ UTR of mRNA of jun proto-oncogene (c-Jun), which is a transcription factor involved in regulating cellular growth and differentiation, has potential in cancer treatment strategies. miR-203 can inhibit the c-Jun mRNA translation, which results in a reduction of c-Jun protein levels. Therefore, miR-203 is considered a promising therapeutic molecule in the treatment of a variety of cancers [111,112]. It also plays a role in suppressing the growth of tumors in EAC [109]. Research conducted by Hezova et al. has shown that EAC patients with low levels of miR-203 in their tumor tissue have a shorter period of time without disease recurrence [99]. Furthermore, studies have revealed that the expression of miR-203 tends to decrease during the progression from healthy esophageal tissue to EAC [92]. Another significant feature of miR-203 is its ability to target and downregulate tumor protein p63 mRNA. P63 is a protein that is expressed in differentiated suprabasal cells and promotes their division [113,114]. In cancers such as the EAC, the low expression of miR-203 is believed to be due to hypermethylation of the miR-203 gene, leading to the increased expression of p63 [115]. By inhibiting the translation of p63, miR-203 prevents the growth and proliferation of cancerous cells.
miR-205 is a molecule known for its role as a tumor suppressor [96]. It has been found to be expressed at low levels in both BE and EAC tissues compared to normal epithelial tissue [88,97]. Interestingly, when comparing the expression levels, it was found that BE had relatively higher expression levels compared to EAC [100]. This finding is particularly significant for understanding the transition from BE to EAC. One important aspect of miR-205’s tumor suppressor function is its association with epithelial–mesenchymal transition (EMT). Low expression of miR-205 leads to the increased expression of its targets, zinc finger E-box binding homeobox 1 and 2 (ZEB-1 and ZEB-2, respectively), which in turn decrease the expression of e-cadherin, promoting EMT [116]. This relationship between miR-205 and EMT provides compelling evidence for miR-205’s tumor suppressor properties. However, Hezova et al. conducted a study on cell lines and found that miR-205 exhibits a tumor suppressor effect in EAC by regulating the EMT pathway, while in ESCC, it has an oncogenic effect through the regulation of matrix metalloproteinase-10 [117]. Additionally, in a different study in ESCC, overexpression of miR-205 led to the development of radiation-resistant properties, promoting cancer growth [118].
Profiling BE- and EAC-specific miRNAs is a crucial area of study to understand BE to EAC transition. Saller et al. endeavored to develop a prognostic method for the progression of BE to EAC. They conducted a comparative analysis of biopsy samples from patients with BE who did not experience dysplasia or carcinoma during a 7-year follow-up period (BE-nonprogressed, BEN) and those who developed carcinoma during a 3–4-year follow-up period (BE-progressed, BEP). To profile 24 biopsy samples of BE (comprising 13 BENs and 11 BEPs), the researchers utilized the NanoString nCounter miRNA assay. They determined the most significantly differentially expressed miRNAs between the 2 groups and selected the top 12 miRNAs (miR-1278, miR-1301, miR-1304-5p, miR-517b-3p, miR-584-5p, miR-599, miR-103a-3p, miR-1197, miR-1256, miR-509-3-5p, miR-544b, and miR-802) for principal component analysis. The 12-miRNA signature demonstrated high sensitivity and specificity in both the training and validation datasets for distinguishing between BEN and BEP. Overall, the researchers were successful in identifying a 12-miRNA signature that could reliably differentiate between BEN and BEP using miRNA profiling. This discovery has the potential to enhance the prediction of BE progression and facilitate the earlier detection of esophageal adenocarcinoma [119].
The process of identifying potential biomarkers for the transition from BE to EAC measured from esophageal samples should ideally be noninvasive. Li et al. endeavored to uncover a panel of miRNAs capable of accurately distinguishing between BE and normal esophageal tissue. To achieve this, they employed a noninvasive esophageal cell sampling device called cytosponge. The authors analyzed miRNA expression profiles using the Agilent microarray and Nanostring nCounter assays from two distinct sets of esophageal biopsy tissues obtained during endoscopy from 38 BE patients and 26 controls with a normal esophagus. Their analysis revealed 15 miRNAs that were significantly upregulated in BE tissues compared to controls, of which 11 were verified in Cytosponge samples. The most prominently upregulated miRNAs in BE tissues were miR-196a, miR-192, miR-194, and miR 215, each of which exhibited an area under the curve (AUC) value of 0.82 or more for distinguishing BE from control tissues. The researchers developed an optimized multivariable logistic regression model based on the expression levels of 6 miRNAs that could identify BE patients with an AUC value of 0.89, 86.2% sensitivity, and 91.6% specificity. Furthermore, the combination of miR-192, miR-196a, miR-199a, and trefoil factor 3 expression levels showed an AUC of 0.93, 93.1% sensitivity, and 93.7% specificity in identifying patients with BE. Finally, the researchers identified a miRNA expression pattern capable of identifying Cytosponge samples from BE patients with an AUC of 0.93. The overexpression of miR194 in BE samples through epigenetic mechanisms might be implicated in the pathogenesis of BE [89].
Circulating miRNAs, as well as tissue, have high potential as biomarkers for the early detection of EAC malignancy. In a systematic screening study conducted by Wang et al. using serum samples from patients with BE and EAC, they found that miR-130a was highly expressed in BE and EAC patients. Additionally, they found that high-grade BE patients highly expressed miR-130a than low-grade BE patients. When comparing early-stage and advanced-stage EAC patients, they also found that miR-130a expression increased as the stage advanced. These results provide significant evidence that miR-130a is associated with the development of BE and EAC [93]. In a recent study published by Hassan et al., they showed that seven miRNAs had ROC curves that could distinguish between BE, EAC, and HG neoplasia. Among the miRNAs with significant expression, miR-92a-3p’s data were compared with tissue samples, and it was determined that the main source of miR-92a-3p in circulation was epithelial cells supporting neoplastic cells [94].
miRNAs can not only be tools for diagnosis or demonstrate progression but also predictive biomarkers of response to anti-cancer therapies and potential therapeutic targets. Therefore, it is crucial to identify their target genes [120]. Yao and colleagues aimed to identify the miRNA–mRNA regulatory network for distinguishing BE from EAC, and they selected 16 molecules as hub genes based on enriched function and pathway analyses. They determined that CDH1, phosphoribosylglycinamide formyltransferase (GART), G2 and S-phase expressed 1 (GTSE1), NIMA-related kinase 2 (NEK2), miR-496, miR-214, and miR-15b were associated with survival [95].
The association of miRNA expressions with intricate molecular networks renders it challenging to determine miRNAs that can serve as both biomarkers and therapeutic targets. As a result, extensive research in this field is necessary to explore the potential for promising advancements.
2. Conclusions
BE results from chemical damage and is a significant risk factor for EAC. Regular endoscopic follow-ups and tissue biopsies are valuable tools for investigating the molecular changes associated with BE, including epigenetic modifications such as DNA methylation, histone modifications, and miRNA regulation. Notably, these epigenetic changes can occur in BE prior to the development of dysplasia or cancer. However, caution is necessary when utilizing these changes to predict disease progression since not all individuals with BE progress to dysplasia or cancer. Although the inactivation of tumor suppressor genes and activation of oncogenes is commonly observed in BE, more research is required to determine the potential of these epigenetic changes as markers for disease progression.
Author Contributions
Conceptualization, P.E., S.K. and S.B.; methodology, P.E. and S.K.; writing—original draft preparation, P.E. and S.K.; writing—review and editing, P.E., S.K. and S.B.; visualization, P.E. and S.K.; supervision, S.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to Ege University Planning and Monitoring Coordination of Organizational Development and the Directorate of Library and Documentation for their support in editing and proofreading this study.
Conflicts of Interest
The authors declare no conflict of interest.
Correction Statement
Due to an error in article production, incorrect references were previously listed in the table 1. This information has been updated and this change does not affect the scientific content of the article.
References
- Katzka, D.A.; Pandolfino, J.E.; Kahrilas, P.J. Phenotypes of Gastroesophageal Reflux Disease: Where Rome, Lyon, and Montreal Meet. Clin. Gastroenterol. Hepatol. 2020, 18, 767–776. [Google Scholar] [CrossRef]
- Barrett, N.R. Chronic Peptic Ulcerz of the Œophagus and ‘Œsophagitis’. Br. J. Surg. 2005, 38, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Eusebi, L.H.; Cirota, G.G.; Zagari, R.M.; Ford, A.C. Global Prevalence of Barrett’s Oesophagus and Oesophageal Cancer in Individuals with Gastro-Oesophageal Reflux: A Systematic Review and Meta-Analysis. Gut 2021, 70, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Bor, S.; Saritas Yuksel, E. How Is the Gastroesophageal Reflux Disease Prevalence, Incidence, and Frequency of Complications (Stricture/Esophagitis/Barrett’s Esophagus/Carcinoma) in Turkey Compared to Other Geographical Regions Globally? Turk. J. Gastroenterol. 2017, 28, 4–9. [Google Scholar] [CrossRef]
- Shimamura, Y.; Iwaya, Y.; Goda, K.; Teshima, C.W. Endoscopic Treatment of Barrett’s Esophagus: What Can We Learn from the Western Perspective? Dig. Endosc. 2018, 30, 182–191. [Google Scholar] [CrossRef]
- Snider, E.J.; Kaz, A.M.; Inadomi, J.M.; Grady, W.M. Chemoprevention of Esophageal Adenocarcinoma. Gastroenterol. Rep. 2020, 8, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Solaymani-Dodaran, M. Risk of Oesophageal Cancer in Barrett’s Oesophagus and Gastro-Oesophageal Reflux. Gut 2004, 53, 1070–1074. [Google Scholar] [CrossRef]
- Haiyu, Z.; Xiaofeng, P.; Xiangqiong, M.; Junlan, Q.; Xiaobin, Z.; Shuncong, W.; Huanhuan, S.; Haiqing, M. Incidence and Survival Changes in Patients with Esophageal Adenocarcinoma during 1984–2013. Biomed. Res. Int. 2019, 2019, 7431850. [Google Scholar] [CrossRef]
- Then, E.O.; Lopez, M.; Saleem, S.; Gayam, V.; Sunkara, T.; Culliford, A.; Gaduputi, V. Esophageal Cancer: An Updated Surveillance Epidemiology and End Results Database Analysis. World J. Oncol. 2020, 11, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Maslyonkina, K.S.; Konyukova, A.K.; Alexeeva, D.Y.; Sinelnikov, M.Y.; Mikhaleva, L.M. Barrett’s Esophagus: The Pathomorphological and Molecular Genetic Keystones of Neoplastic Progression. Cancer Med. 2022, 11, 447–478. [Google Scholar] [CrossRef]
- Yu, M.; Moinova, H.R.; Willbanks, A.; Cannon, V.K.; Wang, T.; Carter, K.; Kaz, A.; Reddi, D.; Inadomi, J.; Luebeck, G.; et al. Novel DNA Methylation Biomarker Panel for Detection of Esophageal Adenocarcinoma and High-Grade Dysplasia. Clin. Cancer Res. 2022, 28, 3761–3769. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Yao, Z.; Fang, W.; He, Q.; Xu, W.W.; Li, B. Epigenetics in Esophageal Cancer: From Mechanisms to Therapeutics. Small Methods 2020, 4, 2000391. [Google Scholar] [CrossRef]
- Dilworth, M.P.; Nieto, T.; Stockton, J.D.; Whalley, C.M.; Tee, L.; James, J.D.; Noble, F.; Underwood, T.J.; Hallissey, M.T.; Hejmadi, R.; et al. Whole Genome Methylation Analysis of Nondysplastic Barrett Esophagus That Progresses to Invasive Cancer. Ann. Surg. 2019, 269, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Kaz, A.M.; Grady, W.M.; Stachler, M.D.; Bass, A.J. Genetic and Epigenetic Alterations in Barrett’s Esophagus and Esophageal Adenocarcinoma. Gastroenterol. Clin. N. Am. 2015, 44, 473–489. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA Methylation Landscape of Cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Kim, J.K.; Samaranayake, M.; Pradhan, S. Epigenetic Mechanisms in Mammals. Cell. Mol. Life Sci. 2009, 66, 596. [Google Scholar] [CrossRef]
- Leighton, G.; Williams, D.C. The Methyl-CpG–Binding Domain 2 and 3 Proteins and Formation of the Nucleosome Remodeling and Deacetylase Complex. J. Mol. Biol. 2020, 432, 1624–1639. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA Methylation in Cancer: Too Much, but Also Too Little. Oncogene 2002, 21, 5400–5413. [Google Scholar] [CrossRef]
- Xu, E.; Gu, J.; Hawk, E.T.; Wang, K.K.; Lai, M.; Huang, M.; Ajani, J.; Wu, X. Genome-Wide Methylation Analysis Shows Similar Patterns in Barrett’s Esophagus and Esophageal Adenocarcinoma. Carcinogenesis 2013, 34, 2750–2756. [Google Scholar] [CrossRef]
- Kalatskaya, I. Overview of Major Molecular Alterations during Progression from Barrett’s Esophagus to Esophageal Adenocarcinoma. Ann. N. Y. Acad. Sci. 2016, 1381, 74–91. [Google Scholar] [CrossRef]
- Bian, Y.; Osterheld, M.; Fontolliet, C.; Bosman, F.T.; Benhattar, J. P16 Inactivation by Methylation of the CDKN2A Promoter Occurs Early during Neoplastic Progression in Barrett’s Esophagus. Gastroenterology 2002, 122, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Klump, B.; Hsieh, C.-J.; Holzmann, K.; Gregor, M.; Porschen, R. Hypermethylation of the CDKN2/P16 Promoter during Neoplastic Progression in Barrett’s Esophagus. Gastroenterology 1998, 115, 1381–1386. [Google Scholar] [CrossRef] [PubMed]
- Maley, C.C.; Galipeau, P.C.; Li, X.; Sanchez, C.A.; Paulson, T.G.; Reid, B.J. Selectively Advantageous Mutations and Hitchhikers in Neoplasms. Cancer Res. 2004, 64, 3414–3427. [Google Scholar] [CrossRef] [PubMed]
- Schulmann, K.; Sterian, A.; Berki, A.; Yin, J.; Sato, F.; Xu, Y.; Olaru, A.; Wang, S.; Mori, Y.; Deacu, E.; et al. Inactivation of P16, RUNX3, and HPP1 Occurs Early in Barrett’s-Associated Neoplastic Progression and Predicts Progression Risk. Oncogene 2005, 24, 4138–4148. [Google Scholar] [CrossRef]
- Vieth, M.; Schneider-Stock, R.; Röhrich, K.; May, A.; Ell, C.; Markwarth, A.; Roessner, A.; Stolte, M.; Tannapfel, A. INK4a-ARF Alterations in Barrett?S Epithelium, Intraepithelial Neoplasia and Barrett?S Adenocarcinoma. Virchows Arch. 2004, 445, 135–141. [Google Scholar] [CrossRef]
- Wong, D.J.; Barrett, M.T.; Stöger, R.; Emond, M.J.; Reid, B.J. P16INK4a Promoter Is Hypermethylated at a High Frequency in Esophageal Adenocarcinomas. Cancer Res. 1997, 57, 2619–2622. [Google Scholar]
- Sarbia, M.; Geddert, H.; Klump, B.; Kiel, S.; Iskender, E.; Gabbert, H.E. Hypermethylation of Tumor Suppressor Genes (P16INK4A,P14ARF AndAPC) in Adenocarcinomas of the Upper Gastrointestinal Tract. Int. J. Cancer 2004, 111, 224–228. [Google Scholar] [CrossRef]
- Jin, Z.; Hamilton, J.P.; Yang, J.; Mori, Y.; Olaru, A.; Sato, F.; Ito, T.; Kan, T.; Cheng, Y.; Paun, B.; et al. Hypermethylation of the AKAP12 Promoter Is a Biomarker of Barrett’s-Associated Esophageal Neoplastic Progression. Cancer Epidemiol. Biomark. Prev. 2008, 17, 111–117. [Google Scholar] [CrossRef]
- Clément, G.; Braunschweig, R.; Pasquier, N.; Bosman, F.T.; Benhattar, J. Methylation OfAPC, TIMP3, AndTERT: A New Predictive Marker to Distinguish Barrett’s Oesophagus Patients at Risk for Malignant Transformation. J. Pathol. 2006, 208, 100–107. [Google Scholar] [CrossRef]
- Wang, J.S.; Guo, M.; Montgomery, E.A.; Thompson, R.E.; Cosby, H.; Hicks, L.; Wang, S.; Herman, J.G.; Canto, M.I. DNA Promoter Hypermethylation of P16 and APC Predicts Neoplastic Progression in Barrett’s Esophagus. Am. J. Gastroenterol. 2009, 104, 2153–2160. [Google Scholar] [CrossRef]
- Eads, C.A.; Lord, R.V.; Kurumboor, S.K.; Wickramasinghe, K.; Skinner, M.L.; Long, T.I.; Peters, J.H.; DeMeester, T.R.; Danenberg, K.D.; Danenberg, P.V.; et al. Fields of Aberrant CpG Island Hypermethylation in Barrett’s Esophagus and Associated Adenocarcinoma. Cancer Res. 2000, 60, 5021–5026. [Google Scholar] [PubMed]
- Brock, M.V.; Gou, M.; Akiyama, Y.; Muller, A.; Wu, T.-T.; Montgomery, E.; Deasel, M.; Germonpré, P.; Rubinson, L.; Heitmiller, R.F.; et al. Prognostic Importance of Promoter Hypermethylation of Multiple Genes in Esophageal Adenocarcinoma. Clin. Cancer Res. 2003, 9, 2912–2919. [Google Scholar] [PubMed]
- Smith, E.; De Young, N.J.; Pavey, S.J.; Hayward, N.K.; Nancarrow, D.J.; Whiteman, D.C.; Smithers, B.M.; Ruszkiewicz, A.R.; Clouston, A.D.; Gotley, D.C.; et al. Similarity of Aberrant DNA Methylation in Barrett’s Esophagus and Esophageal Adenocarcinoma. Mol. Cancer 2008, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Eads, C.A.; Lord, R.V.; Wickramasinghe, K.; Long, T.I.; Kurumboor, S.K.; Bernstein, L.; Peters, J.H.; DeMeester, S.R.; DeMeester, T.R.; Skinner, K.A.; et al. Epigenetic Patterns in the Progression of Esophageal Adenocarcinoma. Cancer Res. 2001, 61, 3410–3418. [Google Scholar]
- Kawakami, K.; Brabender, J.; Lord, R.V.; Groshen, S.; Greenwald, B.D.; Krasna, M.J. Hypermethylated APC DNA in Plasma and Prognosis of Patients With Esophageal Adenocarcinoma. J. Natl. Cancer Inst. 2000, 92, 1805–1811. [Google Scholar] [CrossRef]
- Moinova, H.R.; LaFramboise, T.; Lutterbaugh, J.D.; Chandar, A.K.; Dumot, J.; Faulx, A.; Brock, W.; De la Cruz Cabrera, O.; Guda, K.; Barnholtz-Sloan, J.S.; et al. Identifying DNA Methylation Biomarkers for Non-Endoscopic Detection of Barrett’s Esophagus. Sci. Transl. Med. 2018, 10, eaao5848. [Google Scholar] [CrossRef]
- Corn, P.G.; Heath, E.I.; Heitmiller, R.; Fogt, F.; Forastiere, A.A.; Herman, J.G.; Wu, T.T. Frequent Hypermethylation of the 5′ CpG Island of E-Cadherin in Esophageal Adenocarcinoma. Clin. Cancer Res. 2001, 7, 2765–2769. [Google Scholar]
- Schildhaus, H.-U.; Kröckel, I.; Lippert, H.; Malfertheiner, P.; Roessner, A.; Schneider-Stock, R. Promoter Hypermethylation of P16INK4a, E-Cadherin, O6-MGMT, DAPK and FHIT in Adenocarcinomas of the Esophagus, Esophagogastric Junction and Proximal Stomach. Int. J. Oncol. 2005, 26, 1493–1500. [Google Scholar] [CrossRef]
- Jin, Z.; Cheng, Y.; Olaru, A.; Kan, T.; Yang, J.; Paun, B.; Ito, T.; Hamilton, J.P.; David, S.; Agarwal, R.; et al. Promoter Hypermethylation of CDH13 Is a Common, Early Event in Human Esophageal Adenocarcinogenesis and Correlates with Clinical Risk Factors. Int. J. Cancer 2008, 123, 2331–2336. [Google Scholar] [CrossRef]
- Chueca, E.; Valero, A.; Hördnler, C.; Puertas, A.; Carrera, P.; García-González, M.A.; Strunk, M.; Lanas, A.; Piazuelo, E. Quantitative Analysis of P16 Methylation in Barrett’s Carcinogenesis. Ann. Diagn. Pathol. 2020, 47, 151554. [Google Scholar] [CrossRef]
- Hardie, L.J.; Darnton, S.J.; Wallis, Y.L.; Chauhan, A.; Hainaut, P.; Wild, C.P.; Casson, A.G. P16 Expression in Barrett’s Esophagus and Esophageal Adenocarcinoma: Association with Genetic and Epigenetic Alterations. Cancer Lett. 2005, 217, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.; Hauge, T.; Jeanmougin, M.; Pharo, H.D.; Kresse, S.H.; Honne, H.; Winge, S.B.; Five, M.-B.; Kumar, T.; Mala, T.; et al. Targeted Genetic and Epigenetic Profiling of Esophageal Adenocarcinomas and Non-Dysplastic Barrett’s Esophagus. Clin. Epigenet. 2022, 14, 77. [Google Scholar] [CrossRef] [PubMed]
- Kuester, D.; Dar, A.A.; Moskaluk, C.C.; Krueger, S.; Meyer, F.; Hartig, R.; Stolte, M.; Malfertheiner, P.; Lippert, H.; Roessner, A.; et al. Early Involvement of Death-Associated Protein Kinase Promoter Hypermethylation in the Carcinogenesis of Barrett’s Esophageal Adenocarcinoma and Its Association with Clinical Progression. Neoplasia 2007, 9, 236–245. [Google Scholar] [CrossRef]
- Zou, H.; Osborn, N.K.; Harrington, J.J.; Klatt, K.K.; Molina, J.R.; Burgart, L.J.; Ahlquist, D.A. Frequent Methylation of Eyes Absent 4 Gene in Barrett’s Esophagus and Esophageal Adenocarcinoma. Cancer Epidemiol. Biomark. Prev. 2005, 14, 830–834. [Google Scholar] [CrossRef]
- Lee, O.-J.; Schneider-Stock, R.; McChesney, P.A.; Kuester, D.; Roessner, A.; Vieth, M.; Moskaluk, C.A.; El-Rifai, W. Hypermethylation, Loss of Expression of Glutathione Peroxidase-3 in Barrett’s Tumorigenesis. Neoplasia 2005, 7, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.F.; Razvi, M.; Chen, H.; Washington, K.; Roessner, A.; Schneider-Stock, R.; El-Rifai, W. DNA Hypermethylation Regulates the Expression of Members of the Mu-Class Glutathione S-Transferases and Glutathione Peroxidases in Barrett’s Adenocarcinoma. Gut 2009, 58, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Kuester, D.; El-Rifai, W.; Peng, D.; Ruemmele, P.; Kroeckel, I.; Peters, B.; Moskaluk, C.A.; Stolte, M.; Mönkemüller, K.; Meyer, F.; et al. Silencing of MGMT Expression by Promoter Hypermethylation in the Metaplasia–Dysplasia–Carcinoma Sequence of Barrett’s Esophagus. Cancer Lett. 2009, 275, 117–126. [Google Scholar] [CrossRef]
- Baumann, S.; Keller, G.; Pühringer, F.; Napieralski, R.; Feith, M.; Langer, R.; Höfler, H.; Stein, H.J.; Sarbia, M. The Prognostic Impact OfO6-Methylguanine-DNA Methyltransferase (MGMT) Promotor Hypermethylation in Esophageal Adenocarcinoma. Int. J. Cancer 2006, 119, 264–268. [Google Scholar] [CrossRef]
- Jin, Z.; Mori, Y.; Yang, J.; Sato, F.; Ito, T.; Cheng, Y.; Paun, B.; Hamilton, J.P.; Kan, T.; Olaru, A.; et al. Hypermethylation of the Nel-like 1 Gene Is a Common and Early Event and Is Associated with Poor Prognosis in Early-Stage Esophageal Adenocarcinoma. Oncogene 2007, 26, 6332–6340. [Google Scholar] [CrossRef]
- Zou, H.; Molina, J.R.; Harrington, J.J.; Osborn, N.K.; Klatt, K.K.; Romero, Y.; Burgart, L.J.; Ahlquist, D.A. Aberrant Methylation of Secreted Frizzled-Related Protein Genes in Esophageal Adenocarcinoma and Barrett’s Esophagus. Int. J. Cancer 2005, 116, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Tischoff, I.; Hengge, U.R.; Vieth, M.; Ell, C.; Stolte, M.; Weber, A.; Schmidt, W.E.; Tannapfel, A. Methylation of SOCS-3 and SOCS-1 in the Carcinogenesis of Barrett’s Adenocarcinoma. Gut 2007, 56, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Olaru, A.; Yang, J.; Sato, F.; Cheng, Y.; Kan, T.; Mori, Y.; Mantzur, C.; Paun, B.; Hamilton, J.P.; et al. Hypermethylation of Tachykinin-1 Is a Potential Biomarker in Human Esophageal Cancer. Clin. Cancer Res. 2007, 13, 6293–6300. [Google Scholar] [CrossRef] [PubMed]
- Darnton, S.J.; Hardie, L.J.; Muc, R.S.; Wild, C.P.; Casson, A.G. Tissue Inhibitor of Metalloproteinase-3 (TIMP-3) Gene Is Methylated in the Development of Esophageal Adenocarcinoma: Loss of Expression Correlates with Poor Prognosis. Int. J. Cancer 2005, 115, 351–358. [Google Scholar] [CrossRef]
- Moinova, H.; Leidner, R.S.; Ravi, L.; Lutterbaugh, J.; Barnholtz-Sloan, J.S.; Chen, Y.; Chak, A.; Markowitz, S.D.; Willis, J.E. Aberrant Vimentin Methylation Is Characteristic of Upper Gastrointestinal Pathologies. Cancer Epidemiol. Biomark. Prev. 2012, 21, 594–600. [Google Scholar] [CrossRef]
- Galipeau, P.C.; Li, X.; Blount, P.L.; Maley, C.C.; Sanchez, C.A.; Odze, R.D.; Ayub, K.; Rabinovitch, P.S.; Vaughan, T.L.; Reid, B.J. NSAIDs Modulate CDKN2A, TP53, and DNA Content Risk for Progression to Esophageal Adenocarcinoma. PLoS Med. 2007, 4, e67. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Cheng, Y.; Gu, W.; Zheng, Y.; Sato, F.; Mori, Y.; Olaru, A.V.; Paun, B.C.; Yang, J.; Kan, T.; et al. A Multicenter, Double-Blinded Validation Study of Methylation Biomarkers for Progression Prediction in Barrett’s Esophagus. Cancer Res. 2009, 69, 4112–4115. [Google Scholar] [CrossRef] [PubMed]
- Barradas, M.; Anderton, E.; Acosta, J.C.; Li, S.; Banito, A.; Rodriguez-Niedenführ, M.; Maertens, G.; Banck, M.; Zhou, M.-M.; Walsh, M.J.; et al. Histone Demethylase JMJD3 Contributes to Epigenetic Control of INK4a/ARF by Oncogenic RAS. Genes Dev. 2009, 23, 1177–1182. [Google Scholar] [CrossRef]
- Huang, Y.; Peters, C.J.; Fitzgerald, R.C.; Gjerset, R.A. Progressive Silencing of P14ARF in Oesophageal Adenocarcinoma. J. Cell. Mol. Med. 2009, 13, 398–409. [Google Scholar] [CrossRef]
- Antony, S.; Jiang, G.; Wu, Y.; Meitzler, J.L.; Makhlouf, H.R.; Haines, D.C.; Butcher, D.; Hoon, D.S.; Ji, J.; Zhang, Y.; et al. NADPH Oxidase 5 (NOX5)-Induced Reactive Oxygen Signaling Modulates Normoxic HIF-1α and P27 Kip1 Expression in Malignant Melanoma and Other Human Tumors. Mol. Carcinog. 2017, 56, 2643–2662. [Google Scholar] [CrossRef]
- Fu, X.; Beer, D.G.; Behar, J.; Wands, J.; Lambeth, D.; Cao, W. CAMP-Response Element-Binding Protein Mediates Acid-Induced NADPH Oxidase NOX5-S Expression in Barrett Esophageal Adenocarcinoma Cells. J. Biol. Chem. 2006, 281, 20368–20382. [Google Scholar] [CrossRef]
- Hong, J.; Resnick, M.; Behar, J.; Wang, L.J.; Wands, J.; DeLellis, R.A.; Souza, R.F.; Spechler, S.J.; Cao, W. Acid-Induced P16 Hypermethylation Contributes to Development of Esophageal Adenocarcinoma via Activation of NADPH Oxidase NOX5-S. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G697–G706. [Google Scholar] [CrossRef]
- Hong, J.; Li, D.; Cao, W. Rho Kinase ROCK2 Mediates Acid-Induced NADPH Oxidase NOX5-S Expression in Human Esophageal Adenocarcinoma Cells. PLoS ONE 2016, 11, e0149735. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.H.; Matsuo, J.; Douchi, D.; Bte Mawan, N.A.; Ito, Y. RUNX3 in Stem Cell and Cancer Biology. Cells 2023, 12, 408. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qin, X.; Wu, J.; Qi, B.; Tao, Y.; Wang, W.; Liu, F.; Li, H.; Zhao, B. Association of Promoter Methylation of RUNX3 Gene with the Development of Esophageal Cancer: A Meta Analysis. PLoS ONE 2014, 9, e107598. [Google Scholar] [CrossRef]
- Fukui, S.; Watari, J.; Tomita, T.; Yamasaki, T.; Okugawa, T.; Kondo, T.; Kono, T.; Tozawa, K.; Ikehara, H.; Ohda, Y.; et al. Localization of Specialized Intestinal Metaplasia and the Molecular Alterations in Barrett Esophagus in a Japanese Population: An Analysis of Biopsy Samples Based on the “Seattle” Biopsy Protocol. Hum. Pathol. 2016, 51, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Song, H.; Jiang, H.; Fu, Y.; Ding, X.; Zhou, C. Early Diagnostic Potential of APC hypermethylation in Esophageal Cancer. Cancer Manag. Res. 2018, 10, 181–198. [Google Scholar] [CrossRef]
- Restucci, B.; Martano, M.; DE Vico, G.; Lo Muzio, L.; Maiolino, P. Expression of E-Cadherin, Beta-Catenin and APC Protein in Canine Colorectal Tumours. Anticancer Res. 2009, 29, 2919–2925. [Google Scholar]
- Petrova, Y.I.; Schecterson, L.; Gumbiner, B.M. Roles for E-Cadherin Cell Surface Regulation in Cancer. Mol. Biol. Cell 2016, 27, 3233–3244. [Google Scholar] [CrossRef]
- Pinheiro, H.; Bordeira-Carrico, R.; Seixas, S.; Carvalho, J.; Senz, J.; Oliveira, P.; Inacio, P.; Gusmao, L.; Rocha, J.; Huntsman, D.; et al. Allele-Specific CDH1 Downregulation and Hereditary Diffuse Gastric Cancer. Hum. Mol. Genet. 2010, 19, 943–952. [Google Scholar] [CrossRef]
- Shenoy, S. CDH1 (E-Cadherin) Mutation and Gastric Cancer: Genetics, Molecular Mechanisms and Guidelines for Management. Cancer Manag. Res. 2019, 11, 10477–10486. [Google Scholar] [CrossRef]
- Pecchillo Cimmino, T.; Ammendola, R.; Cattaneo, F.; Esposito, G. NOX Dependent ROS Generation and Cell Metabolism. Int. J. Mol. Sci. 2023, 24, 2086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Hormi-Carver, K.; Zhang, X.; Spechler, S.J.; Souza, R.F. In Benign Barrett’s Epithelial Cells, Acid Exposure Generates Reactive Oxygen Species That Cause DNA Double-Strand Breaks. Cancer Res. 2009, 69, 9083–9089. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The Fundamental Role of Epigenetic Events in Cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Jammula, S.; Katz-Summercorn, A.C.; Li, X.; Linossi, C.; Smyth, E.; Killcoyne, S.; Biasci, D.; Subash, V.V.; Abbas, S.; Blasko, A.; et al. Identification of Subtypes of Barrett’s Esophagus and Esophageal Adenocarcinoma Based on DNA Methylation Profiles and Integration of Transcriptome and Genome Data. Gastroenterology 2020, 158, 1682–1697.e1. [Google Scholar] [CrossRef]
- Alvarez, H.; Opalinska, J.; Zhou, L.; Sohal, D.; Fazzari, M.J.; Yu, Y.; Montagna, C.; Montgomery, E.A.; Canto, M.; Dunbar, K.B.; et al. Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis. PLoS Genet. 2011, 7, e1001356. [Google Scholar] [CrossRef]
- Boldrin, E.; Curtarello, M.; Dallan, M.; Alfieri, R.; Realdon, S.; Fassan, M.; Saggioro, D. Detection of LINE-1 Hypomethylation in CfDNA of Esophageal Adenocarcinoma Patients. Int. J. Mol. Sci. 2020, 21, 1547. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Bhagat, T.D.; Yang, X.; Song, J.H.; Cheng, Y.; Agarwal, R.; Abraham, J.M.; Ibrahim, S.; Bartenstein, M.; Hussain, Z.; et al. Hypomethylation of Noncoding DNA Regions and Overexpression of the Long Noncoding RNA, AFAP1-AS1, in Barrett’s Esophagus and Esophageal Adenocarcinoma. Gastroenterology 2013, 144, 956–966.e4. [Google Scholar] [CrossRef]
- Gilbert, N. Biophysical Regulation of Local Chromatin Structure. Curr. Opin. Genet. Dev. 2019, 55, 66–75. [Google Scholar] [CrossRef]
- Tessarz, P.; Kouzarides, T. Histone Core Modifications Regulating Nucleosome Structure and Dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef]
- Grady, W.M.; Yu, M.; Markowitz, S.D.; Hutchinson, F. Emerging Use for Biomarkers of Cancer. Gastroenterology 2022, 160, 690–709. [Google Scholar] [CrossRef]
- Damaskos, C.; Garmpis, N.; Valsami, S.; Kontos, M.; Spartalis, E.; Kalampokas, T.; Kalampokas, E.; Athanasiou, A.; Moris, D.; Daskalopoulou, A.; et al. Histone Deacetylase Inhibitors: An Attractive Therapeutic Strategy Against Breast Cancer. Anticancer Res. 2017, 37, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Schizas, D.; Mastoraki, A.; Naar, L.; Spartalis, E.; Tsilimigras, D.I.; Karachaliou, G.S.; Bagias, G.; Moris, D. Concept of Histone Deacetylases in Cancer: Reflections on Esophageal Carcinogenesis and Treatment. World J. Gastroenterol. 2018, 24, 4635–4642. [Google Scholar] [CrossRef]
- Langer, R.; Mutze, K.; Becker, K.; Feith, M.; Ott, K.; Höfler, H.; Keller, G. Expression of Class I Histone Deacetylases (HDAC1 and HDAC2) in Oesophageal Adenocarcinomas: An Immunohistochemical Study. J. Clin. Pathol. 2010, 63, 994–998. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Song, B.; Wang, Y.; Kudo, K.; Gavin, E.J.; Xi, Y.; Ju, J. MiR-192 Regulates Dihydrofolate Reductase and Cellular Proliferation through the P53-MicroRNA Circuit. Clin. Cancer Res. 2008, 14, 8080–8086. [Google Scholar] [CrossRef]
- Sun, J.; Fan, Z.; Lu, S.; Yang, J.; Hao, T.; Huo, Q. MiR-192 Suppresses the Tumorigenicity of Prostate Cancer Cells by Targeting and Inhibiting Nin One Binding Protein. Int. J. Mol. Med. 2016, 37, 485–492. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. MicroRNAs as Oncogenes and Tumor Suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fassan, M.; Volinia, S.; Palatini, J.; Pizzi, M.; Baffa, R.; De Bernard, M.; Battaglia, G.; Parente, P.; Croce, C.M.; Zaninotto, G.; et al. MicroRNA Expression Profiling in Human Barrett’s Carcinogenesis. Int. J. Cancer 2011, 129, 1661–1670. [Google Scholar] [CrossRef]
- Li, X.; Kleeman, S.; Coburn, S.B.; Fumagalli, C.; Perner, J.; Jammula, S.; Pfeiffer, R.M.; Orzolek, L.; Hao, H.; Taylor, P.R.; et al. Selection and Application of Tissue MicroRNAs for Nonendoscopic Diagnosis of Barrett’s Esophagus. Gastroenterology 2018, 155, 771–783.e3. [Google Scholar] [CrossRef]
- Revilla-Nuin, B.; Parrilla, P.; Lozano, J.J.; De Haro, L.F.M.; Ortiz, A.; Martínez, C.; Munitiz, V.; De Angulo, D.R.; Bermejo, J.; Molina, J.; et al. Predictive Value of MicroRNAs in the Progression of Barrett Esophagus to Adenocarcinoma in a Long-Term Follow-up Study. Ann. Surg. 2013, 257, 886–893. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, X. Serum Exosomal MiRNAs Expression as Novel Biomarkers for Detection of Esophageal Adenocarcinoma: 441. Off. J. Am. Coll. Gastroenterol. ACG 2016, 111, S198–S199. [Google Scholar] [CrossRef]
- Wijnhoven, B.P.L.; Hussey, D.J.; Watson, D.J.; Tsykin, A.; Smith, C.M.; Michael, M.Z. MicroRNA Profiling of Barrett’s Oesophagus and Oesophageal Adenocarcinoma. Br. J. Surg. 2010, 97, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ji, F.; Liu, G.; Wang, W.; Li, Z.; Yue, Y.; Wang, Z. Upregulation of Circulating Mir130a Is Correlated with Development of Barrett’s Esophagus and Esophageal Adenocarcinoma. OncoTargets Ther. 2019, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Fassan, M.; Realdon, S.; Cascione, L.; Hahne, J.C.; Munari, G.; Guzzardo, V.; Arcidiacono, D.; Lampis, A.; Brignola, S.; Dal Santo, L.; et al. Circulating MicroRNA Expression Profiling Revealed MiR-92a-3p as a Novel Biomarker of Barrett’s Carcinogenesis. Pathol. Res. Pract. 2020, 216, 152907. [Google Scholar] [CrossRef]
- Yao, C.; Li, Y.; Luo, L.; Xiong, Q.; Zhong, X.; Xie, F.; Feng, P. Identification of MiRNAs and Genes for Predicting Barrett’s Esophagus Progressing to Esophageal Adenocarcinoma Using MiRNAmRNA Integrated Analysis. PLoS ONE 2021, 16, e0260353. [Google Scholar] [CrossRef]
- Wu, X.; Ajani, J.A.; Gu, J.; Chang, D.W.; Tan, W.; Hildebrandt, M.A.T.; Huang, M.; Wang, K.K.; Hawk, E. MicroRNA Expression Signatures during Malignant Progression from Barrett’s Esophagus to Esophageal Adenocarcinoma. Cancer Prev. Res. 2013, 6, 196–205. [Google Scholar] [CrossRef]
- Slaby, O.; Srovnal, J.; Radova, L.; Gregar, J.; Juracek, J.; Luzna, P.; Svoboda, M.; Hajduch, M.; Ehrmann, J. Dynamic Changes in MicroRNA Expression Profiles Reflect Progression of Barrett’s Esophagus to Esophageal Adenocarcinoma. Carcinogenesis 2015, 36, 521–527. [Google Scholar] [CrossRef]
- Leidner, R.S.; Ravi, L.; Leahy, P.; Chen, Y.; Bednarchik, B.; Streppel, M.; Canto, M.; Wang, J.S.; Maitra, A.; Willis, J.; et al. The MicroRNAs, MiR-31 and MiR-375, as Candidate Markers in Barrett’s Esophageal Carcinogenesis. Genes Chromosomes Cancer 2012, 51, 473–479. [Google Scholar] [CrossRef]
- Hezova, R.; Kovarikova, A.; Srovnal, J.; Zemanova, M.; Harustiak, T.; Ehrmann, J.; Hajduch, M.; Svoboda, M.; Sachlova, M.; Slaby, O. Diagnostic and Prognostic Potential of MiR-21, MiR-29c, MiR-148 and MiR-203 in Adenocarcinoma and Squamous Cell Carcinoma of Esophagus. Diagn. Pathol. 2015, 10, 42. [Google Scholar] [CrossRef]
- Drahos, J.; Schwameis, K.; Orzolek, L.D.; Hao, H.; Birner, P.; Taylor, P.R.; Pfeiffer, R.M.; Schoppmann, S.F.; Cook, M.B. MicroRNA Profiles of Barrett’s Esophagus and Esophageal Adenocarcinoma: Differences in Glandular Non-Native Epithelium. Cancer Epidemiol. Biomark. Prev. 2016, 25, 429–437. [Google Scholar] [CrossRef]
- Zhang, M.; Zhuang, Q.; Cui, L. MiR-194 Inhibits Cell Proliferation and Invasion via Repression of RAP2B in Bladder Cancer. Biomed. Pharmacother. 2016, 80, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Yen, Y.-T.; Yang, J.-C.; Chang, J.-B.; Tsai, S.-C. Down-Regulation of MiR-194-5p for Predicting Metastasis in Breast Cancer Cells. Int. J. Mol. Sci. 2021, 23, 325. [Google Scholar] [CrossRef]
- Bus, P.; Kestens, C.; Ten Kate, F.J.W.; Peters, W.; Drenth, J.P.H.; Roodhart, J.M.L.; Siersema, P.D.; van Baal, J.W.P.M. Profiling of Circulating MicroRNAs in Patients with Barrett’s Esophagus and Esophageal Adenocarcinoma. J. Gastroenterol. 2016, 51, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Hino, K.; Tsuchiya, K.; Fukao, T.; Kiga, K.; Okamoto, R.; Kanai, T.; Watanabe, M. Inducible Expression of MicroRNA-194 Is Regulated by HNF-1α during Intestinal Epithelial Cell Differentiation. Rna 2008, 14, 1433–1442. [Google Scholar] [CrossRef] [PubMed]
- Cabibi, D.; Caruso, S.; Bazan, V.; Castiglia, M.; Bronte, G.; Ingrao, S.; Fanale, D.; Cangemi, A.; Calò, V.; Listì, A.; et al. Analysis of Tissue and Circulating MicroRNA Expression during Metaplastic Transformation of the Esophagus. Oncotarget 2016, 7, 47821–47830. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhen, J.; Xu, X.; Zhen, K.; Zhu, B.; Pan, R.; Zhao, C. MiR-215 Functions as a Tumor Suppressor and Directly Targets ZEB2 in Human Non-Small Cell Lung Cancer. Oncol. Lett. 2021, 22, 600. [Google Scholar] [CrossRef]
- Li, N.; Zhang, Q.-Y.; Zou, J.-L.; Li, Z.-W.; Tian, T.-T.; Dong, B.; Liu, X.-J.; Ge, S.; Zhu, Y.; Gao, J.; et al. MiR-215 Promotes Malignant Progression of Gastric Cancer by Targeting RUNX1. Oncotarget 2016, 7, 4817–4828. [Google Scholar] [CrossRef]
- Bansal, A.; Lee, I.H.; Hong, X.; Mathur, S.C.; Tawfik, O.; Rastogi, A.; Buttar, N.; Visvanathan, M.; Sharma, P.; Christenson, L.K. Discovery and Validation of Barrett’s Esophagus MicroRNA Transcriptome by Next Generation Sequencing. PLoS ONE 2013, 8, e54240. [Google Scholar] [CrossRef]
- Fassan, M.; Volinia, S.; Palatini, J.; Pizzi, M.; Fernandez-Cymering, C.; Balistreri, M.; Realdon, S.; Battaglia, G.; Souza, R.; Odze, R.D.; et al. MicroRNA Expression Profiling in the Histological Subtypes of Barrett’s Metaplasia. Clin. Transl. Gastroenterol. 2013, 4, e34-7. [Google Scholar] [CrossRef]
- Inokuchi, K.; Ochiya, T.; Matsuzaki, J. Extracellular Mirnas for the Management of Barrett’s Esophagus and Esophageal Adenocarcinoma: A Systematic Review. J. Clin. Med. 2021, 10, 117. [Google Scholar] [CrossRef]
- Sonkoly, E.; Lovén, J.; Xu, N.; Meisgen, F.; Wei, T.; Brodin, P.; Jaks, V.; Kasper, M.; Shimokawa, T.; Harada, M.; et al. MicroRNA-203 Functions as a Tumor Suppressor in Basal Cell Carcinoma. Oncogenesis 2012, 1, e3. [Google Scholar] [CrossRef]
- Zhang, A.; Lakshmanan, J.; Motameni, A.; Harbrecht, B.G. MicroRNA-203 Suppresses Proliferation in Liver Cancer Associated with PIK3CA, P38 MAPK, c-Jun, and GSK3 Signaling. Mol. Cell Biochem. 2018, 441, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Li, X.; Zhang, Y.; Guo, Y.; Zhou, J.; Gao, K.; Dai, J.; Hu, G.; Lv, L.; Du, J. The MicroRNA Feedback Regulation of P63 in Cancer Progression. Oncotarget 2015, 6, 8434–8453. [Google Scholar] [CrossRef]
- Yuan, Y.; Zeng, Z.-Y.; Liu, X.-H.; Gong, D.-J.; Tao, J.; Cheng, H.-Z.; Huang, S.-D. MicroRNA-203 Inhibits Cell Proliferation by Repressing ΔNp63 Expression in Human Esophageal Squamous Cell Carcinoma. BMC Cancer 2011, 11, 57. [Google Scholar] [CrossRef]
- Qi, Q.; Ling, Y.; Zhu, M.; Zhang, Y.; Zhou, L.; Wan, M.; Bao, Y.; Liu, Y. Hypermethylation and Low Expression of MiR-203 in Patients with Esophageal Cancer in Chinese Population. Int. J. Clin. Exp. Pathol. 2016, 9, 6245–6251. [Google Scholar]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The MiR-200 Family and MiR-205 Regulate Epithelial to Mesenchymal Transition by Targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Hezova, R.; Kovarikova, A.; Srovnal, J.; Zemanova, M.; Harustiak, T.; Ehrmann, J.; Hajduch, M.; Sachlova, M.; Svoboda, M.; Slaby, O. MiR-205 Functions as a Tumor Suppressor in Adenocarcinoma and an Oncogene in Squamous Cell Carcinoma of Esophagus. Tumor Biol. 2016, 37, 8007–8018. [Google Scholar] [CrossRef]
- Pan, F.; Mao, H.; Bu, F.; Tong, X.; Li, J.; Zhang, S.; Liu, X.; Wang, L.; Wu, L.; Chen, R.; et al. Sp1-Mediated Transcriptional Activation of MiR-205 Promotes Radioresistance in Esophageal Squamous Cell Carcinoma. Oncotarget 2017, 8, 5735–5752. [Google Scholar] [CrossRef] [PubMed]
- Saller, J.; Jiang, K.; Xiong, Y.; Yoder, S.J.; Neill, K.; Pimiento, J.M.; Pena, L.; Corbett, F.S.; Magliocco, A.; Coppola, D. A MicroRNA Signature Identifies Patients at Risk of Barrett Esophagus Progression to Dysplasia and Cancer. Dig. Dis. Sci. 2022, 67, 516–523. [Google Scholar] [CrossRef]
- Zarrilli, G.; Galuppini, F.; Angerilli, V.; Munari, G.; Sabbadin, M.; Lazzarin, V.; Nicolè, L.; Biancotti, R.; Fassan, M. Mirnas Involved in Esophageal Carcinogenesis and MiRNA- Related Therapeutic Perspectives in Esophageal Carcinoma. Int. J. Mol. Sci. 2021, 22, 3640. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).