Animal Microbiomes as a Source of Novel Antibiotic-Producing Strains
Abstract
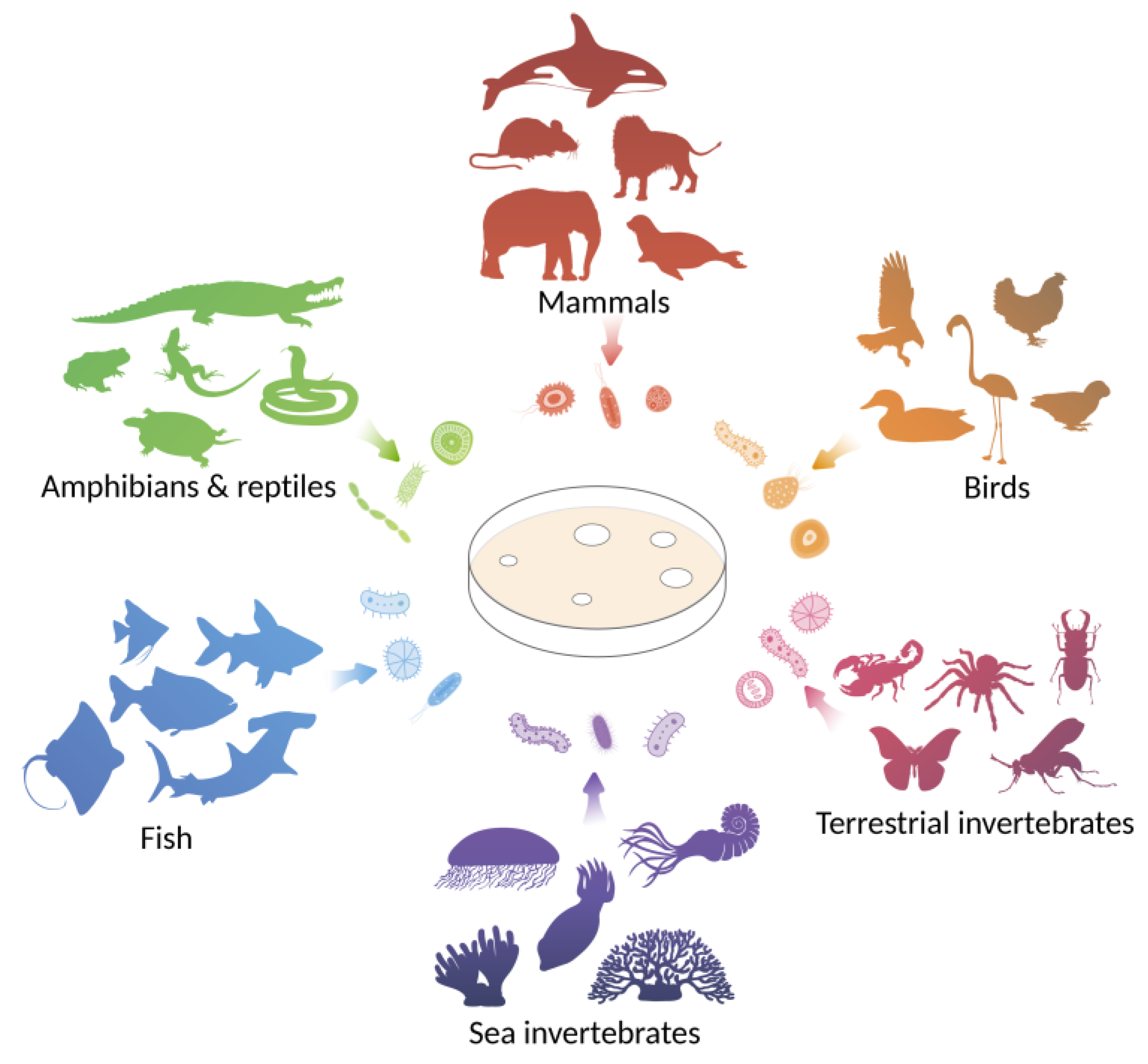
:1. Introduction
2. Marine Invertebrates
3. Terrestrial Invertebrates
3.1. Nematodes
3.2. Insects
4. Fish
5. Amphibians and Reptiles
6. Birds
7. Mammals
8. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Lewis, K. The Science of Antibiotic Discovery. Cell 2020, 2, 29–45. [Google Scholar] [CrossRef]
- Ling, L.L. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Nichols, D. Use of Ichip for High-Throughput In Situ Cultivation of ‘Uncultivable’ Microbial Species. Appl. Environ. Microbiol. 2020, 76, 2445. [Google Scholar] [CrossRef]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Bauman, K.D.; Butler, K.S.; Moore, B.S.; Chekan, J.R. Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 2021, 38, 2100–2129. [Google Scholar] [CrossRef]
- Terekhov, S.S.; Smirnov, I.V.; Stepanova, A.V.; Bobik, T.V.; Mokrushina, Y.A.; Ponomarenko, N.A.; Belogurov, A.A., Jr.; Rubtsova, M.P.; Kartseva, O.V.; Gomzikova, M.O.; et al. Microfluidic droplet platform for ultrahigh-throughput single-cell screening of biodiversity. Proc. Natl. Acad. Sci. USA 2017, 114, 2550–2555. [Google Scholar] [CrossRef]
- Scanlon, T.C.; Dostal, S.M.; Griswold, K.E. A High-Throughput Screen for Antibiotic Drug Discovery. Biotechnol. Bioeng. 2014, 111, 232. [Google Scholar] [CrossRef]
- Mahler, L.; Niehs, S.P.; Martin, K.; Weber, T.; Scherlach, K.; Hertweck, C.; Roth, M.; Rosenbaum, M.A. Highly parallelized droplet cultivation and prioritization of antibiotic producers from natural microbial communities. Elife 2021, 10, e64774. [Google Scholar] [CrossRef]
- Nuti, N.; Rottmann, P.; Stucki, A.; Koch, P.; Panke, S.; Dittrich, P.S. A Multiplexed Cell-Free Assay to Screen for Antimicrobial Peptides in Double Emulsion Droplets. Angew. Chem. Int. Ed. Engl. 2022, 61, e202114632. [Google Scholar] [CrossRef]
- Zada, S.; Sajjad, W.; Rafiq, M.; Ali, S.; Hu, Z.; Wang, H.; Cai, R. Cave Microbes as a Potential Source of Drugs Development in the Modern Era. Microb. Ecol. 2022, 84, 676–687. [Google Scholar] [CrossRef]
- Jose, P.A.; Maharshi, A.; Jha, B. Actinobacteria in natural products research: Progress and prospects. Microbiol. Res. 2021, 246, 126708. [Google Scholar] [CrossRef]
- Xu, D.; Han, L.; Li, C.; Cao, Q.; Zhu, D.; Barrett, N.H.; Harmody, D.; Chen, J.; Zhu, H.; McCarthy, P.J.; et al. Bioprospecting Deep-Sea Actinobacteria for Novel Anti-infective Natural Products. Front. Microbiol. 2018, 30, 787. [Google Scholar] [CrossRef]
- Malard, L.A.; Guisan, A. Into the microbial niche. Trends Ecol. Evol. 2023, 38, 936–945. [Google Scholar] [CrossRef]
- Mullis, M.M.; Rambo, I.M.; Baker, B.J.; Reese, B.K. Diversity, Ecology, and Prevalence of Antimicrobials in Nature. Front. Microbiol. 2019, 10, 2518. [Google Scholar] [CrossRef]
- Baquero, F.; Coque, T.M.; Galán, J.C.; Martinez, J.L. The Origin of Niches and Species in the Bacterial World. Front. Microbiol. 2021, 12, 657986. [Google Scholar] [CrossRef]
- Esser, D.; Lange, J.; Marinos, G.; Sieber, M.; Best, L.; Prasse, D.; Bathia, J.; Rühlemann, M.C.; Boersch, K.; Jaspers, C.; et al. Functions of the Microbiota for the Physiology of Animal Metaorganisms. J. Innate Immun. 2019, 11, 393–404. [Google Scholar] [CrossRef]
- Buchanan, G.O.; Williams, P.G.; Feling, R.H.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Sporolides A and B: Structurally unprecedented halogenated macrolides from the marine actinomycete Salinispora tropica. Org. Lett. 2005, 7, 2731–2734. [Google Scholar] [CrossRef]
- Bech, P.K.; Lysdal, K.L.; Gram, L.; Bentzon-Tilia, M.; Strube, M.L. Marine Sediments Hold an Untapped Potential for Novel Taxonomic and Bioactive Bacterial Diversity. mSystems 2020, 5, e00782-20. [Google Scholar] [CrossRef]
- Macintyre, L.; Zhang, T.; Viegelmann, C.; Martinez, I.J.; Cheng, C.; Dowdells, C.; Abdelmohsen, U.R.; Gernert, C.; Hentschel, U.; Edrada-Ebel, R. Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar. Drugs 2014, 12, 3416–3448. [Google Scholar] [CrossRef]
- Thomas-Poulsen, M.; van Overbeek, L.; Newman, D.J. Predominately Uncultured Microbes as Sources of Bioactive Agents. Front. Microbiol. 2016, 18, 1832. [Google Scholar]
- Rosenberg, E.; Zilber-Rosenberg, I. Microbes Drive Evolution of Animals and Plants: The Hologenome Concept. mBio 2016, 7, e01395. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Theis, K.R. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS Biol. 2015, 13, e1002226. [Google Scholar] [CrossRef]
- Remya, T.; Thomas, A.; Kavlekar, D.P.; Lokabharathi, P.A. Marine Drugs from Sponge-Microbe Association-A Review. Mar. Drugs 2010, 8, 1417–1468. [Google Scholar]
- Matroodi, S.; Siitonen, V.; Baral, B.; Yamada, K.; Akhgari, A.; Metsä-Ketelä, M. Genotyping-Guided Discovery of Persiamycin A From Sponge-Associated Halophilic Streptomonospora sp. PA3. Front. Microbiol. 2020, 11, 1237. [Google Scholar] [CrossRef]
- Rao, K.V.; Santarsiero, B.D.; Mesecar, A.D.; Schinazi, R.F.; Tekwani, B.L.; Hamann, M.T. New Manzamine Alkaloids with Activity against Infectious and Tropical Parasitic Diseases from an Indonesian Sponge. J. Nat. Prod. 2003, 66, 823–828. [Google Scholar] [CrossRef]
- Waters, A.L.; Peraud, O.; Kasanah, N.; Sims, J.W.; Kothalawala, N.; Anderson, M.A.; Abbas, S.H.; Rao, K.V.; Jupally, V.R.; Kelly, M.; et al. An analysis of the sponge Acanthostrongylophora igens’ microbiome yields an actinomycete that produces the natural product manzamine A. Front. Mar. Sci. 2014, 1, 54. [Google Scholar] [CrossRef]
- Li, W.; Ding, L.; Li, J.; Wen, H.; Liu, Y.; Tan, S.; Yan, X.; Shi, Y.; Lin, W.; Lin, H.-W.; et al. Novel Antimycin Analogues with Agricultural Antifungal Activities from the Sponge-Associated Actinomycete Streptomyces sp. NBU3104. J. Agric. Food Chem. 2022, 70, 8309–8316. [Google Scholar] [CrossRef]
- MacIntyre, L.W.; Charles, M.J.; Haltli, B.A.; Marchbank, D.H.; Kerr, R.G. An Ichip-Domesticated Sponge Bacterium Produces an N-Acyltyrosine Bearing an α-Methyl Substituent. Org. Lett. 2019, 21, 7768–7771. [Google Scholar] [CrossRef]
- Nagai, K.; Kamigiri, K.; Arao, N.; Suzumura, K.; Kawano, Y.; Yamaoka, M.; Zhang, H.; Watanabe, M.; Suzuki, K. YM-266183 and YM-266184, Novel Thiopeptide Antibiotics Produced by Bacillus cereus Isolated from a Marine Sponge, I. Taxonomy, Fermentation, Isolation, Physico-chemical Properties and Biological Properties. J. Antibiot. 2003, 56, 123–128. [Google Scholar] [CrossRef]
- Pabel, C.T.; Vater, J.; Wilde, C.; Franke, P.; Hofemeister, J.; Adler, B.; Bringmann, G.; Hacker, J.; Hentschel, U. Antimicrobial Activities and Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry of Bacillus Isolates from the Marine Sponge Aplysina aerophoba. Mar. Biotechnol. 2003, 5, 424–434. [Google Scholar] [CrossRef]
- Graça, A.P.; Bondoso, J.; Gaspar, H.; Xavier, J.R.; Monteiro, M.C.; de la Cruz, M.; Oves-Costales, D.; Vicente, F.; Lage, O.M. Antimicrobial Activity of Heterotrophic Bacterial Communities from the Marine Sponge Erylus discophorus (Astrophorida, Geodiidae). PLoS ONE 2013, 8, 78992. [Google Scholar] [CrossRef]
- Sertan-De Guzman, A.A.; Predicala, R.Z.; Bernardo, E.B.; Neilan, B.A.; Elardo, S.P.; Mangalindan, G.C.; Tasdemir, D.; Ireland, C.M.; Barraquio, W.L.; Concepcion, G.P. Pseudovibrio denitrificans strain Z143-1, a heptylprodigiosin-producing bacterium isolated from a Philippine tunicate. FEMS Microbiol. Lett. 2007, 277, 188–196. [Google Scholar] [CrossRef]
- Palomo, S.; González, I.; de la Cruz, M.; Martín, J.; Tormo, J.R.; Anderson, M.; Hill, R.T.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-Derived Kocuria and Micrococcus spp. as Sources of the New Thiazolyl Peptide Antibiotic Kocurin. Mar. Drugs 2013, 11, 1071. [Google Scholar] [CrossRef]
- Martí, J.; da S Sousa, T.; Crespo, G.; Palomo, S.; González, I.; Tormo, J.R.; de la Cruz, M.; Anderson, M.; Hill, R.T.; Vicente, F.; et al. Kocurin, the True Structure of PM181104, an Anti-Methicillin-Resistant Staphylococcus aureus (MRSA) Thiazolyl Peptide from the Marine-Derived Bacterium Kocuria palustris. Mar. Drugs 2013, 11, 387–398. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, H.; Han, X.; Lin, W.; Yan, X. Antimicrobial screening and active compound isolation from marine bacterium NJ6-3-1 associated with the sponge Hymeniacidon perleve. World J. Microbiol. Biotechnol. 2005, 21, 201–206. [Google Scholar] [CrossRef]
- Singh, M.P.; Kong, F.; Janso, J.E.; Arias, D.A.; Suarez, P.A.; Bernan, V.S.; Petersen, P.J.; Weiss, W.J.; Carter, G.; Greenstein, M. Novel α-Pyrones Produced by a Marine Pseudomonas sp. F92S91 Taxonomy and Biological Activities. J. Antibiot. 2003, 56, 1033–1044. [Google Scholar] [CrossRef]
- Sugden, S.; Holert, J.; Cardenas, E.; Mohn, W.W.; Stein, L.Y. Microbiome of the freshwater sponge Ephydatia muelleri shares compositional and functional similarities with those of marine sponges. ISME J. 2022, 16, 2503–2512. [Google Scholar] [CrossRef]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Osset, M.; Pérez-Victoria, I.; Martín, J.; de Pedro, N.; de la Cruz, M.; Díaz, C.; Vicente, F.; Reyes, F.; et al. Lobophorin K, a New Natural Product with Cytotoxic Activity Produced by Streptomyces sp. M-207 Associated with the Deep-Sea Coral Lophelia pertusa. Mar. Drugs 2017, 15, 144. [Google Scholar]
- Lin, Z.; Koch, M.; Pond, C.D.; Mabeza, G.; Seronay, R.A.; Concepcion, G.P.; Barrows, L.R.; Olivera, B.M.; Schmidt, E.W. Lobophorins with antimycobacterial activity from a turrid mollusk-associated Streptomyces sp. J. Antibiot. 2014, 67, 121. [Google Scholar] [CrossRef]
- Wibowo, J.T.; Kellermann, M.Y.; Köck, M.; Putra, M.Y.; Murniasih, T.; Mohr, K.I.; Wink, J.; Praditya, D.F.; Steinmann, E.; Schupp, P.J. Anti-Infective and Antiviral Activity of Valinomycin and Its Analogues from a Sea Cucumber-Associated Bacterium, Streptomyces sp. SV 21. Mar. Drugs 2021, 19, 81. [Google Scholar] [CrossRef]
- Schmidt, E.W. The secret to a successful relationship: Lasting chemistry between ascidians and their symbiotic bacteria. Invertebr. Biol. 2015, 134, 88–102. [Google Scholar] [CrossRef]
- Kwan, J.C.; Tianero, M.D.; Donia, M.S.; Wyche, T.P.; Bugni, T.S.; Schmidt, E.W. Host control of symbiont natural product chemistry in cryptic populations of the tunicate Lissoclinum patella. PLoS ONE 2014, 9, e95850. [Google Scholar] [CrossRef]
- Erwin, P.; Pineda, M.; Webster, N.; Turon, X.; López-Legentil, S. Down under the tunic: Bacterial biodiversity hotspots and widespread ammonia-oxidizing archaea in coral reef ascidians. ISME J. 2014, 8, 575–588. [Google Scholar] [CrossRef]
- Dou, X.; Dong, B. Origins and Bioactivities of Natural Compounds Derived from Marine Ascidians and Their Symbionts. Mar. Drugs 2019, 17, 670. [Google Scholar] [CrossRef]
- Gromek, S.M.; Suria, A.M.; Fullmer, M.S.; Garcia, J.L.; Gogarten, J.P.; Nyholm, S.V.; Balunas, M.J. Leisingera sp. JC1, a Bacterial Isolate from Hawaiian Bobtail Squid Eggs, Produces Indigoidine and Differentially Inhibits Vibrios. Front. Microbiol. 2016, 7, 1342. [Google Scholar] [CrossRef]
- Dreyer, J.; Malan, A.P.; Dicks, L.M.T. Bacteria of the Genus Xenorhabdus, a Novel Source of Bioactive Compounds. Front. Microbiol. 2018, 9, 3177. [Google Scholar] [CrossRef]
- Eleftherianos, I.G. Novel antibiotic compounds produced by the insect pathogenic bacterium Photorhabdus. Recent. Pat. Antiinfect. Drug Discov. 2009, 4, 81–89. [Google Scholar] [CrossRef]
- Zhou, Q.; Grundmann, F.; Kaiser, M.; Schiell, M.; Gaudriault, S.; Batzer, A.; Kurz, M.; Bode, H.B. Structure and biosynthesis of xenoamicins from entomopathogenic Xenorhabdus. Chemistry 2013, 19, 16772–16779. [Google Scholar] [CrossRef]
- McInerney, B.V.; Taylor, W.C.; Lacey, M.J.; Akhurst, R.J.; Gregson, R.P. Biologically Active Metabolites from Xenorhabdus spp., Part 2. Benzopyran-1-one Derivatives with Gastroprotective Activity. J. Nat. Prod. 1991, 54, 785–795. [Google Scholar] [CrossRef]
- Gualtieri, M.; Aumelas, A.; Thaler, J.O. Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J. Antibiot. 2009, 62, 295–302. [Google Scholar] [CrossRef]
- Thaler, J.O.; Baghdiguian, S.; Boemare, N. Purification and characterization of xenorhabdicin, a phage tail-like bacteriocin, from the lysogenic strain F1 of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 1995, 61, 2049. [Google Scholar] [CrossRef]
- Park, H.B.; Perez, C.E.; Perry, E.K.; Crawford, J.M. Activating and Attenuating the Amicoumacin Antibiotics. Molecules 2016, 21, 824. [Google Scholar] [CrossRef]
- Ffrench-Constant, R.; Waterfield, N.; Daborn, P.; Joyce, S.; Bennett, H.; Au, C.; Dowling, A.; Boundy, S.; Reynolds, S.; Clarke, D. Photorhabdus: Towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol. Rev. 2003, 26, 433–456. [Google Scholar] [CrossRef]
- Li, J.; Chen, G.; Wu, H.; Webster, J.M. Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl. Environ. Microbiol. 1995, 61, 4329. [Google Scholar] [CrossRef]
- Imai, Y.; Meyer, K.J.; Iinishi, A.; Favre-Godal, Q.; Green, R.; Manuse, S.; Caboni, M.; Mori, M.; Niles, S.; Ghiglieri, M.; et al. A new antibiotic selectively kills Gram-negative pathogens. Nature 2019, 576, 459. [Google Scholar] [CrossRef]
- Van Moll, L.; De Smet, J.; Cos, P.; Van Campenhout, L. Microbial symbionts of insects as a source of new antimicrobials: A review. Crit. Rev. Microbiol. 2021, 47, 562–579. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Carlson, C.M.; Ortega, H.E.; Thomas, C.; Ananiev, G.E.; Barns, K.J.; Book, A.J.; Cagnazzo, J.; Carlos, C.; Flanigan, W.; et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019, 10, 516. [Google Scholar] [CrossRef]
- Qin, Z.; Munnoch, J.T.; Devine, R.; Holmes, N.A.; Seipke, R.F.; Wilkinson, K.A.; Wilkinson, B.; Hutchings, M.I. Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chem. Sci. 2017, 8, 3218–3227. [Google Scholar] [CrossRef]
- Van Arnam, E.B.; Ruzzini, A.C.; Sit, C.S.; Horn, H.; Pinto-Tomás, A.A.; Currie, C.R.; Clardy, J. Selvamicin, an atypical antifungal polyene from two alternative genomic contexts. Proc. Natl. Acad. Sci. USA 2016, 113, 12940–12945. [Google Scholar] [CrossRef]
- Kim, K.H.; Ramadhar, T.R.; Beemelmanns, C.; Cao, S.; Poulsen, M.; Currie, C.R.; Clardy, J. Natalamycin A, an ansamycin from a termite-associated Streptomyces sp. Chem. Sci. 2014, 5, 4333–4338. [Google Scholar] [CrossRef]
- Klassen, J.L.; Lee, S.R.; Poulsen, M.; Beemelmanns, C.; Kim, K.H. Efomycins K and L From a Termite-Associated Streptomyces sp. M56 and Their Putative Biosynthetic Origin. Front. Microbiol. 2019, 10, 1739. [Google Scholar] [CrossRef]
- Nirma, C.; Eparvier, V.; Stien, D. Antibacterial ilicicolinic acids C and D and ilicicolinal from Neonectria discophora SNB-CN63 isolated from a termite nest. J. Nat. Prod. 2015, 78, 159–162. [Google Scholar] [CrossRef]
- Scott, J.J.; Oh, D.C.; Yuceer, M.C.; Klepzig, K.D.; Clardy, J.; Currie, C.R. Bacterial protection of beetle-fungus mutualism. Science 2008, 322, 63. [Google Scholar] [CrossRef]
- Blodgett, J.A.V.; Oh, D.C.; Cao, S.; Currie, C.R.; Kolter, R.; Clardy, J. Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 11692–11697. [Google Scholar] [CrossRef]
- Ganley, J.G.; Carr, G.; Ioerger, T.R.; Sacchettini, J.C.; Clardy, J.; Derbyshire, E.R. Discovery of Antimicrobial Lipodepsipeptides Produced by a Serratia sp. within Mosquito Microbiomes. ChemBioChem 2018, 19, 1590–1594. [Google Scholar] [CrossRef]
- Flórez, L.V.; Scherlach, K.; Miller, I.J.; Rodrigues, A.; Kwan, J.C.; Hertweck, C.; Kaltenpoth, M. An antifungal polyketide associated with horizontally acquired genes supports symbiont-mediated defense in Lagria villosa beetles. Nat. Commun. 2018, 9, 2478. [Google Scholar] [CrossRef]
- An, J.S.; Hong, S.H.; Somers, E.; Lee, J.; Kim, B.Y.; Woo, D.; Kim, S.W.; Hong, H.J.; Jo, S.I.; Shin, J.; et al. Lenzimycins A and B, Metabolites with Antibacterial Properties from Brevibacillus sp. Associated with the Dung Beetle Onthophagus lenzii. Front. Microbiol. 2020, 11, 2705. [Google Scholar] [CrossRef]
- Schmidt, V.T.; Smith, K.F.; Melvin, D.W.; Amaral-Zettler, L.A. Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol. Ecol. 2015, 24, 2537–2550. [Google Scholar] [CrossRef]
- Schmidt, V.; Amaral-Zettler, L.; Davidson, J.; Summerfelt, S.; Good, C. Influence of Fishmeal-Free Diets on Microbial Communities in Atlantic Salmon (Salmo salar) Recirculation Aquaculture Systems. Appl. Environ. Microbiol. 2016, 82, 4470–4481. [Google Scholar] [CrossRef]
- Sanchez, L.M.; Wong, W.R.; Riener, R.M.; Schulze, C.J.; Linington, R.G. Examining the fish microbiome: Vertebrate-derived bacteria as an environmental niche for the discovery of unique marine natural products. PLoS ONE 2012, 7, e35398. [Google Scholar] [CrossRef]
- Alshammari, E.; Patel, M.; Sachidanandan, M.; Kumar, P.; Adnan, M. Potential Evaluation and Health Fostering Intrinsic Traits of Novel Probiotic Strain Enterococcus durans F3 Isolated from the Gut of Fresh Water Fish Catla catla. Food Sci. Anim. Resour. 2019, 39, 844. [Google Scholar] [CrossRef]
- Tan, H.Y.; Chen, S.W.; Hu, S.Y. Improvements in the growth performance, immunity, disease resistance, and gut microbiota by the probiotic Rummeliibacillus stabekisii in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2019, 92, 265–275. [Google Scholar] [CrossRef]
- Elsadek, M.M.; Wang, S.; Wu, Z.; Wang, J.; Wang, X.; Zhang, Y.; Yu, M.; Guo, Z.; Wang, Q.; Wang, G.; et al. Characterization of Bacillus spp. isolated from the intestines of Rhynchocypris lagowskii as a potential probiotic and their effects on fish pathogens. Microb. Pathog. 2023, 180, 106163. [Google Scholar] [CrossRef]
- Khalid, F.; Khalid, A.; Fu, Y.; Hu, Q.; Zheng, Y.; Khan, S.; Wang, Z. Potential of Bacillus velezensis as a probiotic in animal feed: A review. J. Microbiol. 2021, 59, 627–633. [Google Scholar] [CrossRef]
- Lowrey, L.; Woodhams, D.C.; Tacchi, L.; Salinas, I. Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl. Environ. Microbiol. 2015, 81, 6915–6925. [Google Scholar] [CrossRef]
- De Assis, A.B.; Barreto, C.C.; Navas, C.A. Skin microbiota in frogs from the Brazilian Atlantic Forest: Species, forest type, and potential against pathogens. PLoS ONE 2017, 12, e0179628. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–5453. [Google Scholar] [CrossRef]
- Eibach, D.; Nagel, M.; Lorenzen, S.; Hogan, B.; Belmar Campos, C.; Aepfelbacher, M.; Sarpong, N.; May, J. Extended-spectrum β-lactamase-producing Enterobacteriaceae among geckos (Hemidactylus brookii) in a Ghanaian hospital. Clin. Microbiol. Infect. 2019, 25, 1048–1050. [Google Scholar] [CrossRef]
- Li, C.-F.; Tang, H.-L.; Chiou, C.-S.; Tung, K.-C.; Lu, M.-C.; Lai, Y.-C. Draft genome sequence of CTX-M-type β-lactamase-producing Klebsiella quasipneumoniae subsp. similipneumoniae isolated from a Box turtle. J. Glob. Antimicrob. Resist. 2018, 12, 235–236. [Google Scholar] [CrossRef]
- Quintana, G.; Niederle, M.V.; Minahk, C.J.; Picariello, G.; Nader-Macías, M.E.F.; Pasteris, S.E. Nisin Z produced by Lactococcus lactis from bullfrog hatchery is active against Citrobacter freundii, a red-leg syndrome related pathogen. World J. Microbiol. Biotechnol. 2017, 33, 186. [Google Scholar] [CrossRef]
- Pasteris, S.E.; Pingitore, E.V.e.r.a.; Ale, C.E.; Nader-Macías, M.E. Characterization of a bacteriocin produced by Lactococcus lactis subsp. lactis CRL 1584 isolated from a Lithobates catesbeianus hatchery. World J. Microbiol. Biotechnol. 2014, 30, 1053–1062. [Google Scholar] [CrossRef]
- Woodhams, D.C.; LaBumbard, B.C.; Barnhart, K.L.; Becker, M.H.; Bletz, M.C.; Escobar, L.A.; Flechas, S.V.; Forman, M.E.; Iannetta, A.A.; Joyce, M.D.; et al. Prodigiosin, Violacein, and Volatile Organic Compounds Produced by Widespread Cutaneous Bacteria of Amphibians Can Inhibit Two Batrachochytrium Fungal Pathogens. Microb. Ecol. 2018, 75, 1049–1062. [Google Scholar] [CrossRef]
- Lim, S.; Bhak, J.; Jeon, S.; Mun, W.; Bhak, J.; Choi, S.Y.; Mitchell, R.J. The Kiss of Death: Serratia marcescens Antibacterial Activities against Staphylococcus aureus Requires Both de novo Prodigiosin Synthesis and Direct Contact. Microbiol. Spectr. 2022, 10, e00607-22. [Google Scholar] [CrossRef]
- Asadi, S.; Soleimani, N.; Babadi, Z.K.; Ebrahimipour, G.H. Isolation and identification of the bacterium producing antitumor and antimicrobial compounds derived from Iranian swamp frog (Rana ridibunda) skin. Iran. J. Microbiol. 2021, 13, 372–380. [Google Scholar] [CrossRef]
- Akbar, N.; Khan, N.A.; Sagathevan, K.; Iqbal, M.; Tawab, A.; Siddiqui, R. Gut bacteria of Cuora amboinensis (turtle) produce broad-spectrum antibacterial molecules. Sci. Rep. 2019, 9, 17012. [Google Scholar] [CrossRef]
- Khan, N.A.; Soopramanien, M.; Maciver, S.K.; Anuar, T.S.; Sagathevan, K.; Siddiqui, R. Crocodylus porosus Gut Bacteria: A Possible Source of Novel Metabolites. Molecules 2021, 26, 4999. [Google Scholar] [CrossRef]
- Marcelino, V.R.; Wille, M.; Hurt, A.C.; González-Acuña, D.; Klaassen, M.; Schlub, T.E.; Eden, J.S.; Shi, M.; Iredell, J.R.; Sorrell, T.C.; et al. Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. 2019, 17, 31. [Google Scholar] [CrossRef]
- Zou, A.; Nadeau, K.; Xiong, X.; Wang, P.W.; Copeland, J.K.; Lee, J.Y.; Pierre, J.S.; Ty, M.; Taj, B.; Brumell, J.H.; et al. Systematic profiling of the chicken gut microbiome reveals dietary supplementation with antibiotics alters expression of multiple microbial pathways with minimal impact on community structure. Microbiome 2022, 10, 127. [Google Scholar] [CrossRef]
- Neveling, D.P.; Dicks, L.M.T. Probiotics: An Antibiotic Replacement Strategy for Healthy Broilers and Productive Rearing. Probiotics Antimicrob. Proteins 2021, 13, 1–11. [Google Scholar] [CrossRef]
- Bodawatta, K.H.; Hird, S.M.; Grond, K.; Poulsen, M.; Jønsson, K.A. Avian gut microbiomes taking flight. Trends Microbiol. 2022, 30, 268–280. [Google Scholar] [CrossRef]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken Gut Microbiota: Importance and Detection Technology. Front. Vet. Sci. 2018, 5, 254. [Google Scholar] [CrossRef]
- Pan, D.; Yu, Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef]
- Diaz Carrasco, J.M.; Casanova, N.A.; Fernández Miyakawa, M.E. Microbiota, Gut Health and Chicken Productivity: What Is the Connection? Microorganisms 2019, 7, 374. [Google Scholar] [CrossRef]
- Pilasombut, K.; Sakpuaram, T.; Wajjwalku, W.; Nitisinprasert, S.; Swetwiwathana, A.; Zendo, T.; Fujita, K.; Nakayama, J.; Sonomoto, K. Purification and amino acid sequence of a bacteriocins produced by Lactobacillus salivarius K7 isolated from chicken intestine. Songklanakarin J. Sci. Technol. 2006, 28, 121–131. [Google Scholar]
- Stern, N.J.; Svetoch, E.A.; Eruslanov, B.V.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Pokhilenko, V.D.; Levchuk, V.P.; Svetoch, O.E.; Seal, B.S. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 2006, 50, 3111–3116. [Google Scholar] [CrossRef]
- Svetoch, E.A.; Eruslanov, B.V.; Levchuk, V.P.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Stepanshin, J.; Dyatlov, I.; Seal, B.S.; Stern, N.J. Isolation of Lactobacillus salivarius 1077 (NRRL B-50053) and characterization of its bacteriocin, including the antimicrobial activity spectrum. Appl. Environ. Microbiol. 2011, 77, 2749–2754. [Google Scholar] [CrossRef]
- Messaoudi, S.; Kergourlay, G.; Rossero, A.; Ferchichi, M.; Prévost, H.; Drider, D.; Manai, M.; Dousset, X. Identification of lactobacilli residing in chicken ceca with antagonism against Campylobacter. Int. Microbiol. 2011, 14, 103–110. [Google Scholar]
- Messaoudi, S.; Manai, M.; Kergourlay, G.; Prévost, H.; Connil, N.; Chobert, J.M.; Dousset, X. Lactobacillus salivarius: Bacteriocin and probiotic activity. Food Microbiol. 2013, 36, 296–304. [Google Scholar] [CrossRef]
- Arbulu, S.; Jiménez, J.J.; Gútiez, L.; Campanero, C.; Del Campo, R.; Cintas, L.M.; Herranz, C.; Hernández, P.E. Evaluation of bacteriocinogenic activity, safety traits and biotechnological potential of fecal lactic acid bacteria (LAB), isolated from Griffon Vultures (Gyps fulvus subsp. fulvus). BMC Microbiol. 2016, 16, 228. [Google Scholar] [CrossRef]
- Javůrková, V.G.; Kreisinger, J.; Procházka, P.; Požgayová, M.; Ševčíková, K.; Brlík, V.; Adamík, P.; Heneberg, P.; Porkert, J. Unveiled feather microcosm: Feather microbiota of passerine birds is closely associated with host species identity and bacteriocin-producing bacteria. ISME J. 2019, 13, 2363–2376. [Google Scholar] [CrossRef]
- Martín-Platero, A.M.; Valdivia, E.; Ruíz-Rodríguez, M.; Soler, J.J.; Martín-Vivaldi, M.; Maqueda, M.; Martínez-Bueno, M. Characterization of Antimicrobial Substances Produced by Enterococcus faecalis MRR 10-3, Isolated from the Uropygial Gland of the Hoopoe (Upupa epops). Appl. Environ. Microbiol. 2006, 72, 4245–4249. [Google Scholar] [CrossRef]
- Ruiz-Rodríguez, M.; Martínez-Bueno, M.; Martín-Vivaldi, M.; Valdivia, E.; Soler, J.J. Bacteriocins with a broader antimicrobial spectrum prevail in enterococcal symbionts isolated from the hoopoe’s uropygial gland. FEMS Microbiol. Ecol. 2013, 85, 495–502. [Google Scholar] [CrossRef]
- Yang, F.; Hou, C.; Zeng, X.; Qiao, S. The use of lactic Acid bacteria as a probiotic in Swine diets. Pathogens 2015, 4, 34–45. [Google Scholar] [CrossRef]
- Gupta, M.; Pattanaik, A.K.; Singh, A.; Sharma, S.; Jadhav, S.E.; Kumar, A.; Verma, A.K. Functional and probiotic characterization of Ligilactobacillus salivarius CPN60 isolated from calf faeces and its appraisal in rats. J. Biosci. Bioeng. 2021, 132, 575–584. [Google Scholar] [CrossRef]
- Garsa, A.K.; Choudhury, P.K.; Puniya, A.K.; Dhewa, T.; Malik, R.K.; Tomar, S.K. Bovicins: The Bacteriocins of Streptococci and Their Potential in Methane Mitigation. Probiotics Antimicrob. Proteins 2019, 11, 1403–1413. [Google Scholar] [CrossRef]
- Ochoa, J.L.; Sanchez, L.M.; Koo, B.M.; Doherty, J.S.; Rajendram, M.; Huang, K.C.; Gross, C.A.; Linington, R.G. Marine Mammal Microbiota Yields Novel Antibiotic with Potent Activity Against Clostridium difficile. ACS Infect. Dis. 2018, 4, 59–67. [Google Scholar] [CrossRef]
- Mousa, W.K.; Athar, B.; Merwin, N.J.; Magarvey, N.A. Antibiotics and specialized metabolites from the human microbiota. Nat. Prod. Rep. 2017, 34, 1302–1331. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, E.; Mayer, M.J.; Cotter, P.D.; Narbad, A. Gut microbiota as a source of novel antimicrobials. Gut Microbes 2019, 10, 1–21. [Google Scholar] [CrossRef]
- Tagg, J.R. Streptococcal Bacteriocin-Like Inhibitory Substances: Some Personal Insights into the Bacteriocin-Like Activities Produced by Streptococci Good and Bad. Probiotics Antimicrob. Proteins 2009, 1, 60–66. [Google Scholar] [CrossRef]
- Santagati, M.; Scillato, M.; Patanè, F.; Aiello, C.; Stefani, S. Bacteriocin-producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunol. Med. Microbiol. 2012, 65, 23–31. [Google Scholar] [CrossRef]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2018, 95, fiy241. [Google Scholar] [CrossRef]
- Kim, S.G.; Becattini, S.; Moody, T.U.; Shliaha, P.V.; Littmann, E.R.; Seok, R.; Gjonbalaj, M.; Eaton, V.; Fontana, E.; Amoretti, L.; et al. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 2019, 572, 665–669. [Google Scholar] [CrossRef]
- Coyne, M.J.; Béchon, N.; Matano, L.M.; McEneany, V.L.; Chatzidaki-Livanis, M.; Comstock, L.E. A family of anti-Bacteroidales peptide toxins wide-spread in the human gut microbiota. Nat. Commun. 2019, 10, 3460. [Google Scholar] [CrossRef]
- Sato, Y.; Atarashi, K.; Plichta, D.R.; Arai, Y.; Sasajima, S.; Kearney, S.M.; Suda, W.; Takeshita, K.; Sasaki, T.; Okamoto, S.; et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature 2021, 599, 458–464. [Google Scholar] [CrossRef]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef]
- Terekhov, S.S.; Smirnov, I.V.; Malakhova, M.V.; Samoilov, A.E.; Manolov, A.I.; Nazarov, A.S.; Danilov, D.V.; Dubiley, S.A.; Osterman, I.A.; Rubtsova, M.P.; et al. Ultrahigh-throughput functional profiling of microbiota communities. Proc. Natl. Acad. Sci. USA 2018, 115, 9551–9556. [Google Scholar] [CrossRef]
- Baranova, M.N.; Kudzhaev, A.M.; Mokrushina, Y.A.; Babenko, V.V.; Kornienko, M.A.; Malakhova, M.V.; Yudin, V.G.; Rubtsova, M.P.; Zalevsky, A.; Belozerova, O.A.; et al. Deep Functional Profiling of Wild Animal Microbiomes Reveals Probiotic Bacillus pumilus Strains with a Common Biosynthetic Fingerprint. Int. J. Mol. Sci. 2022, 23, 1168. [Google Scholar] [CrossRef]
- Akbar, N.; Siddiqui, R.; Sagathevan, K.A.; Khan, N.A. Gut bacteria of animals/pests living in polluted environments are a potential source of antibacterials. Appl. Microbiol. Biotechnol. 2019, 103, 3955–3964. [Google Scholar] [CrossRef]
- Pereira, W.A.; Mendonça, C.M.N.; Urquiza, A.V.; Marteinsson, V.Þ.; LeBlanc, J.G.; Cotter, P.D.; Villalobos, E.F.; Romero, J.; Oliveira, R.P.S. Use of Probiotic Bacteria and Bacteriocins as an Alternative to Antibiotics in Aquaculture. Microorganisms 2022, 10, 1705. [Google Scholar] [CrossRef]
- Liao, S.F.; Nyachoti, M. Using probiotics to improve swine gut health and nutrient utilization. Anim. Nutr. 2017, 3, 331–343. [Google Scholar] [CrossRef]
- Liang, L.; Yang, C.; Liu, L.; Mai, G.; Li, H.; Wu, L.; Jin, M.; Chen, Y. Commensal bacteria-derived extracellular vesicles suppress ulcerative colitis through regulating the macrophages polarization and remodeling the gut microbiota. Microb. Cell Fact. 2022, 21, 88. [Google Scholar] [CrossRef]
- El-Saadony, M.T.; Alagawany, M.; Patra, A.K.; Kar, I.; Tiwari, R.; Dawood, M.A.O.; Dhama, K.; Abdel-Latif, H.M.R. The functionality of probiotics in aquaculture: An overview. Fish Shellfish Immunol. 2021, 117, 36–52. [Google Scholar] [CrossRef]
- Moree, W.J.; Phelan, V.V.; Wu, C.H.; Bandeira, N.; Cornett, D.S.; Duggan, B.M.; Dorrestein, P.C. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA 2012, 109, 13811–13816. [Google Scholar] [CrossRef]
- Hall, S.; McDermott, C.; Anoopkumar-Dukie, S.; McFarland, A.J.; Forbes, A.; Perkins, A.V.; Davey, A.K.; Chess-Williams, R.; Kiefel, M.J.; Arora, D.; et al. Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins 2016, 8, 236. [Google Scholar] [CrossRef]
- Schiessl, K.T.; Hu, F.; Jo, J.; Nazia, S.Z.; Wang, B.; Price-Whelan, A.; Min, W.; Dietrich, L.E.P. Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat. Commun. 2019, 10, 762. [Google Scholar] [CrossRef]
- Orazi, G.; Ruoff, K.L.; O’Toole, G.A. Pseudomonas aeruginosa Increases the Sensitivity of Biofilm-Grown Staphylococcus aureus to Membrane-Targeting Antiseptics and Antibiotics. mBio 2019, 10, e01501-19. [Google Scholar] [CrossRef]
- Tran, C.; Cock, I.E.; Chen, X.; Feng, Y. Antimicrobial Bacillus: Metabolites and Their Mode of Action. Antibiotics 2022, 11, 88. [Google Scholar] [CrossRef]
- Bilal, M.; Si, W.; Barbe, F.; Chevaux, E.; Sienkiewicz, O.; Zhao, X. Effects of novel probiotic strains of Bacillus pumilus and Bacillus subtilis on production, gut health, and immunity of broiler chickens raised under suboptimal conditions. Poult. Sci. 2021, 100, 100871. [Google Scholar] [CrossRef]
- Opriessnig, T.; Karuppannan, A.K.; Beckler, D.; Ait-Ali, T.; Cubas-Atienzar, A.; Halbur, P.G. Bacillus pumilus probiotic feed supplementation mitigates Lawsonia intracellularis shedding and lesions. Vet. Res. 2019, 50, 85. [Google Scholar] [CrossRef]
- Hlordzi, V.; Kuebutornye, F.K.A.; Afriyie, G.; Abarike, E.D.; Lu, Y.; Chi, S.; Anokyewaa, M.A. The use of Bacillus species in maintenance of water quality in aquaculture: A review. Aquac. Rep. 2020, 18, 100503. [Google Scholar] [CrossRef]
- Duc le, H.; Hong, H.A.; Barbosa, T.M.; Henriques, A.O.; Cutting, S.M. Characterization of Bacillus Probiotics Available for Human Use. Appl. Environ. Microbiol. 2004, 70, 2161–2171. [Google Scholar] [CrossRef]
- Adnani, N.; Rajski, S.R.; Bugni, T.S. Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod. Rep. 2017, 34, 784–814. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Terekhov, S.S.; Osterman, I.A.; Smirnov, I.V. High-Throughput Screening of Biodiversity for Antibiotic Discovery. Acta Naturae 2018, 10, 23–29. [Google Scholar] [CrossRef]
- Agresti, J.J.; Antipov, E.; Abate, A.R.; Ahn, K.; Rowat, A.C.; Baret, J.C.; Marquez, M.; Klibanov, A.M.; Griffiths, A.D.; Weitz, D.A. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 4004–4009. [Google Scholar] [CrossRef]
| Bacteria | Hosts | ||
|---|---|---|---|
| Sea Invertebrates | |||
| Kocuria palustris | Sponge | Xestospongia sp. | [34,35] |
| Kocuria marina | |||
| Micrococcus yunnanensis | |||
| Micromonospora sp. | Sponge | Acanthostrongylophora ingens | [26,27] |
| Streptomyces sp. NBU3104 | Sponge | [28] | |
| Pseudomonas sp. | Sponge | [37] | |
| Bacillus cereus | Sponge | Halichondria japonica | [30] |
| Streptomyces sp. M-207 | Cnidarian (coral polyp) | Lophelia pertusa | [39] |
| Streptomyces sp. 1053U.I.1a.3b | Mollusk | Lienardia totopotens | [40] |
| Streptomyces cavourensis SV 21 | Echinoderm | Stichopus vastus | [23] |
| Prochloron didemni | Chordate (ascidian) | Lissoclimum patella | [42,43] |
| Terrestrial Invertebrates | |||
| Xenorhabdus doucetiae DSM17909 | Nematode | Steinernema diaprepesi | [49] |
| Xenorhabdus nematophila | Nematode | Steinernema sp. | [51] |
| Xenorbabdus sp. strain Q1, Xenorbabdus nematopbilus All | Nematode | Steinernema sp., Steinernema filtiae | [50] |
| Photorhabdus khaini | Nematode | Heterorhabditis sp. | [56] |
| Streptomyces sp. ISID311 | Insect (ant) | Cyphomyrmex sp. | [58] |
| Streptomyces formicae | Insect (ant) | Tetraponera penzigi | [59] |
| Pseudonocardia sp. | Insect (ant) | Apterostigma | [60] |
| Streptomyces sp. M56 | Insect (termite) | Macrotermes natalensis | [61] |
| [62] | |||
| Neonectria discophora | Insect (termite) | Nasutitermes corniger | [63] |
| Streptomyces sp. | Insect (beetle) | Dendroctonus frontalis | [64] |
| Streptomyces sp. | Insect (beetle) | Dendroctonus frontalis | [65] |
| Serracia marcescens | Insect (fly) | Anopheles stephensi | [66] |
| Burkholderia gladioli Lv-StB | Insect (beetle) | Largia villosa | [67] |
| Brevibacillus sp. | Insect (beetle) | Onthophagus lenzii | [68] |
| Fish | |||
| Paraoerskovia sp. | Fish (cod) | Lotella rhacina | [71] |
| Amphibians and Reptiles | |||
| Pseudomonas aeruginosa CM3 | Reptile (turtle) | Cuora amboinensis | [87] |
| Birds | |||
| Enterococcus faecium M3K31 | Bird (vulture) | Gyps fulvus subsp. fulvus | [101] |
| Enterococcus faecalis | Bird (hornbill) | Upupa epops | [103] |
| Mammals | |||
| Micromonospora auratingra | Mammal (cetacean) | Phocoena Phocoena | [108] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baranova, M.N.; Pilipenko, E.A.; Gabibov, A.G.; Terekhov, S.S.; Smirnov, I.V. Animal Microbiomes as a Source of Novel Antibiotic-Producing Strains. Int. J. Mol. Sci. 2024, 25, 537. https://doi.org/10.3390/ijms25010537
Baranova MN, Pilipenko EA, Gabibov AG, Terekhov SS, Smirnov IV. Animal Microbiomes as a Source of Novel Antibiotic-Producing Strains. International Journal of Molecular Sciences. 2024; 25(1):537. https://doi.org/10.3390/ijms25010537
Chicago/Turabian StyleBaranova, Margarita N., Ekaterina A. Pilipenko, Alexander G. Gabibov, Stanislav S. Terekhov, and Ivan V. Smirnov. 2024. "Animal Microbiomes as a Source of Novel Antibiotic-Producing Strains" International Journal of Molecular Sciences 25, no. 1: 537. https://doi.org/10.3390/ijms25010537
APA StyleBaranova, M. N., Pilipenko, E. A., Gabibov, A. G., Terekhov, S. S., & Smirnov, I. V. (2024). Animal Microbiomes as a Source of Novel Antibiotic-Producing Strains. International Journal of Molecular Sciences, 25(1), 537. https://doi.org/10.3390/ijms25010537






