Comprehensive Analysis of the Catalase (CAT) Gene Family and Expression Patterns in Rubber Tree (Hevea brasiliensis) under Various Abiotic Stresses and Multiple Hormone Treatments
Abstract
:1. Introduction
2. Results
2.1. Identification of the CAT Gene Family in Rubber Tree
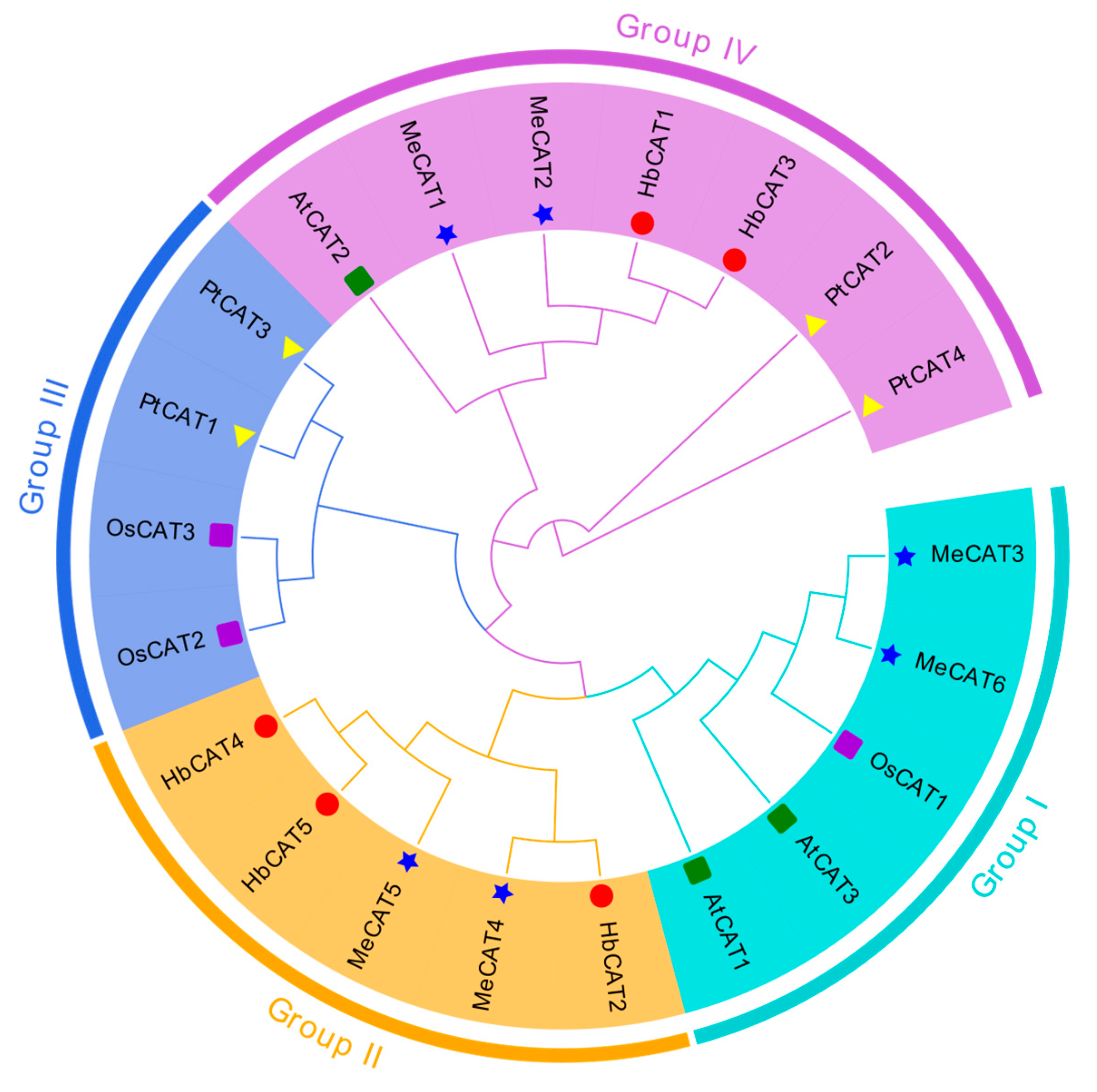
2.2. Phylogenetic Analysis of CAT Genes
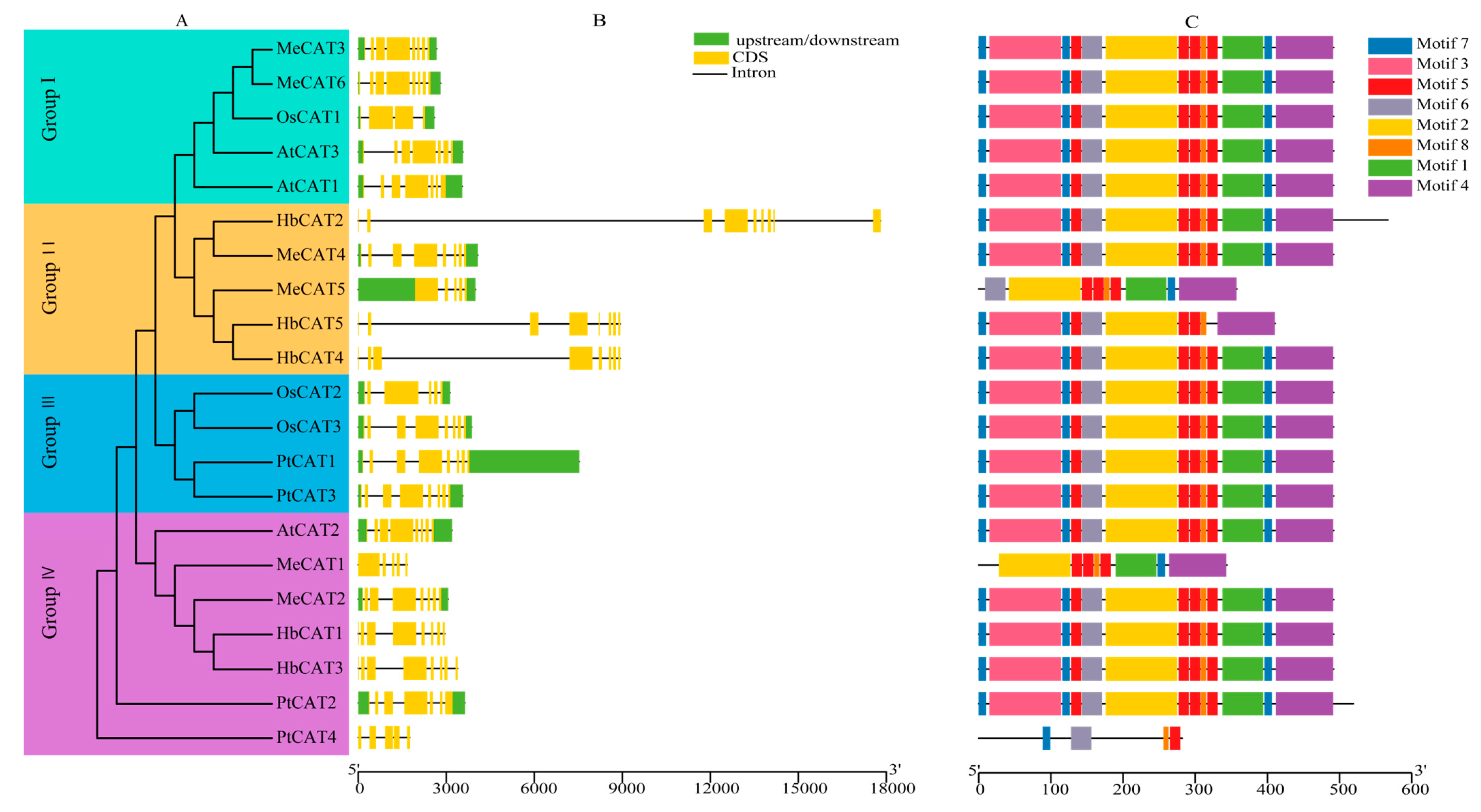
2.3. Gene Structures and Conserved Motifs of CATs
2.4. Chromosomal Localization and Synteny Analysis of CAT Genes
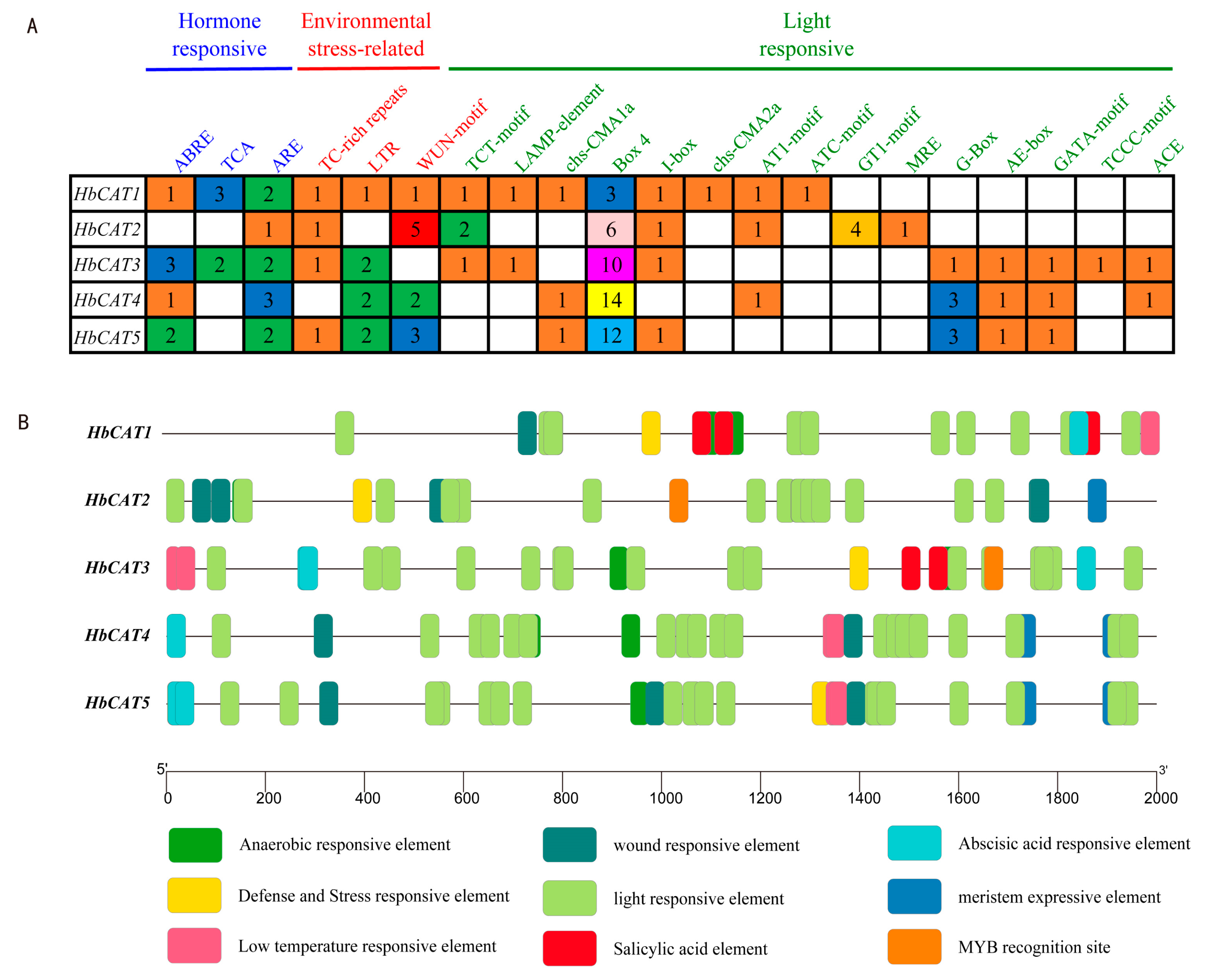
2.5. Analysis of Cis-Elements in the Promoters of HbCAT Genes
2.6. Functional Gene Ontology (GO) Annotation of HbCAT Genes
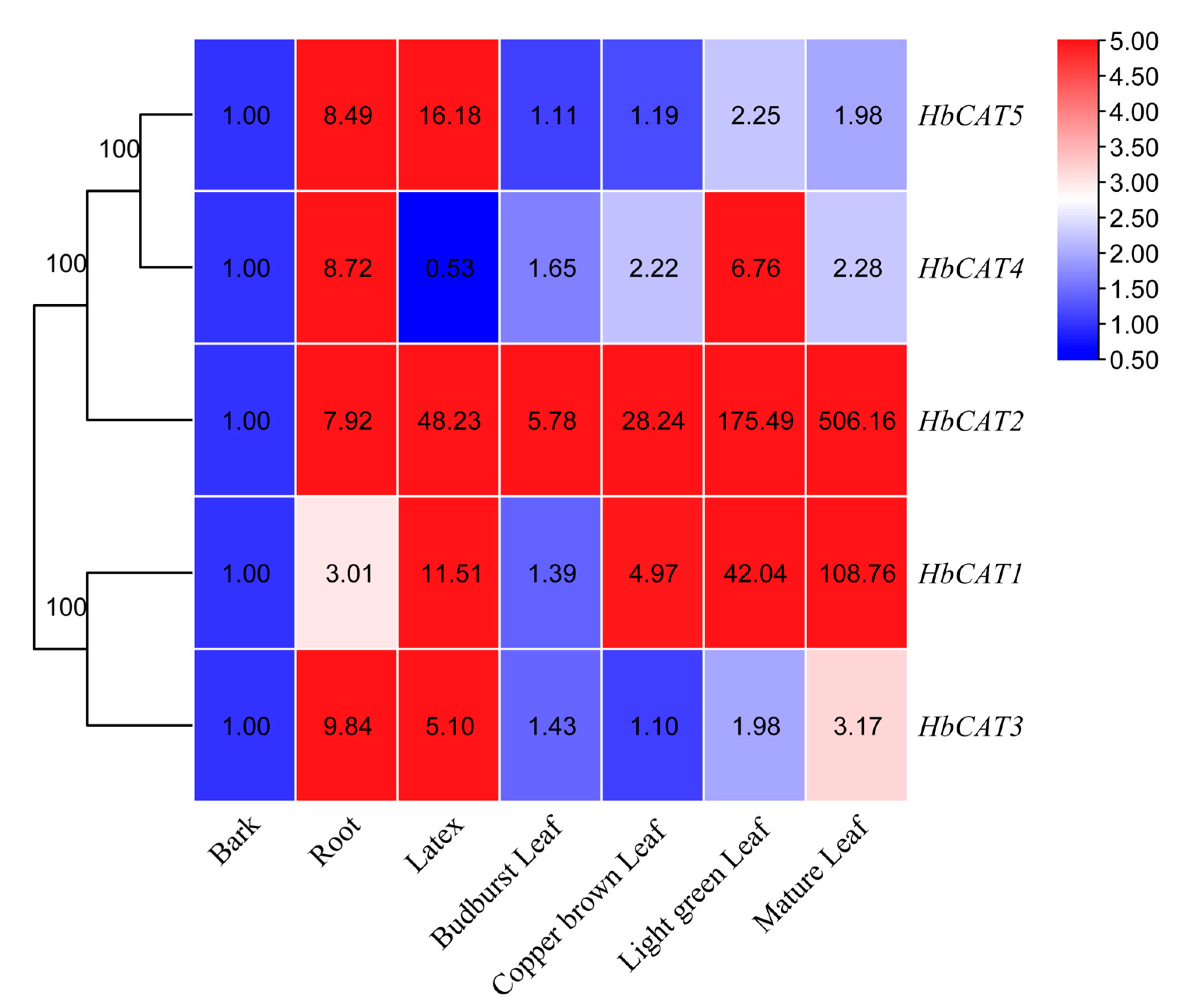
2.7. Expression Patterns of HbCAT Genes in Different Tissues
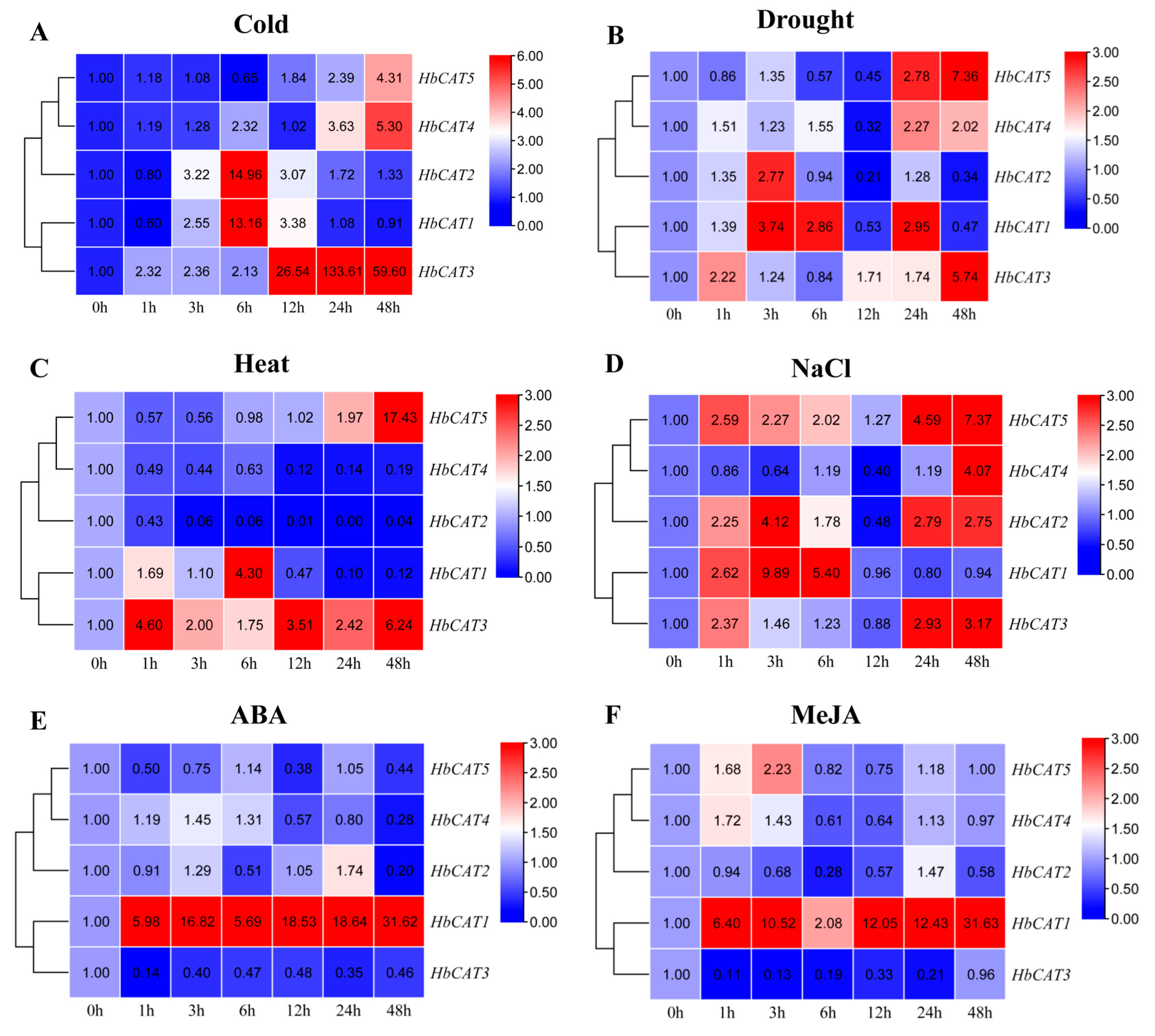
2.8. Expression Patterns of HbCAT Genes under Different Abiotic Stresses and Exogenous Hormone Treatments
3. Discussion
4. Materials and Methods
4.1. Identification of HbCAT Genes in Rubber Tree
4.2. Phylogenetic Tree, Gene Structures, and Conserved Motifs
4.3. Chromosomal Localization, Gene Duplication, and Synteny Analysis
4.4. Cis-Acting Elements Analysis
4.5. GO Enrichment Analysis of HbCAT Genes
4.6. Plant Materials and Stress Treatments
4.7. RNA Extraction and RT-qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2013, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent developments in enzymatic antioxidant defence mechanism in plants with special reference to abiotic stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Raihan, M.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2016, 90, 856–867. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Parvin, K.; Bhuiyan, T.F.; Anee, T.I.; Nahar, K.; Hossen, M.S.; Zulfiqar, F.; Alam, M.M.; Fujita, M. Regulation of ROS metabolism in plants under environmental stress: A review of recent experimental evidence. Int. J. Mol. Sci. 2020, 21, 8695. [Google Scholar] [CrossRef]
- Alam, N.B.; Ghosh, A. Comprehensive analysis and transcript profiling of Arabidopsis thaliana and Oryza sativa catalase gene family suggests their specific roles in development and stress responses. Plant Physiol. Biochem. 2018, 123, 54–64. [Google Scholar] [CrossRef]
- Du, Y.Y.; Wang, P.C.; Chen, J.; Song, C.P. Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Ye, Q.; Wu, Z.; Zhang, Q.; Wang, L.; Liu, J.; Hu, X.; Guo, D.; Wang, X.; Zhang, Z.; et al. Analysis of CAT gene family and functional identification of OsCAT3 in rice. Genes 2023, 14, 138. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.I.; Lee, H.; An, C.S. Differential expression of three catalase genes in the small radish (Rhaphanus sativus L. var. sativus). Mol. Cells 2007, 24, 37–44. [Google Scholar] [PubMed]
- Hu, L.; Yang, Y.; Jiang, L.; Liu, S. The catalase gene family in cucumber: Genome-wide identification and organization. Genet. Mol. Biol. 2016, 39, 408–415. [Google Scholar] [CrossRef]
- Aleem, M.; Aleem, S.; Sharif, I.; Aleem, M.; Shahzad, R.; Khan, M.I.; Batool, A.; Sarwar, G.; Farooq, J.; Iqbal, A.; et al. Whole-genome identification of APX and CAT gene families in cultivated and wild soybeans and their regulatory function in plant development and stress response. Antioxidants 2022, 11, 1626. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cheng, Y.; Chen, D.; Liu, D.; Hu, M.; Dong, J.; Zhang, X.; Song, L.; Shen, F. The catalase gene family in cotton: Genome-wide characterization and bioinformatics analysis. Cells 2019, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, L.; Yun, L.; Ji, L.; Li, G.; Ji, M.; Shi, Y.; Zheng, X. Catalase (CAT) gene family in wheat (Triticum aestivum L.): Evolution, expression pattern and function analysis. Int. J. Mol. Sci. 2022, 23, 542. [Google Scholar] [CrossRef]
- Raza, A.; Su, W.; Gao, A.; Mehmood, S.S.; Hussain, M.A.; Nie, W.; Lv, Y.; Zou, X.; Zhang, X. Catalase (CAT) gene family in rapeseed (Brassica napus L.): Genome-wide analysis, identification, and expression pattern in response to multiple hormones and abiotic stress conditions. Int. J. Mol. Sci. 2021, 22, 4281. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, Y.; Zou, W.; Pan, Y.B.; Lin, P.; Xu, L.; Grisham, M.P.; Ding, Q.; Su, Y.; Que, Y. Genome-wide characterization of sugarcane catalase gene family identifies a ScCAT1 gene associated disease resistance. Int. J. Mol. Sci. 2023, 232, 123398. [Google Scholar] [CrossRef]
- Balestrasse, K.B.; Yannarelli, G.G.; Noriega, G.O.; Batlle, A.; Tomaro, M.L. Heme oxygenase and catalase gene expression in nodules and roots of soybean plants subjected to cadmium stress. Biometals 2008, 21, 433–441. [Google Scholar] [CrossRef]
- Yong, B.; Wang, X.; Xu, P.; Zheng, H.; Fei, X.; Hong, Z.; Ma, Q.; Miao, Y.; Yuan, X.; Jiang, Y.; et al. Isolation and abiotic stress resistance analyses of a catalase gene from Ipomoea batatas (L.) Lam. BioMed Res. Int. 2017, 2017, 6847532. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-H.; Su, Y.; Guo, J.; Ling, H.; Chen, S.; Wang, S.; Xu, L.; Allan, A.C.; Que, Y. Isolation of a novel peroxisomal catalase gene from sugarcane, which is responsive to biotic and abiotic stresses. PLoS ONE 2014, 9, e84426. [Google Scholar] [CrossRef]
- Bueso, E.; Alejandro, S.; Carbonell, P.; Perez-Amador, M.A.; Fayos, J.; Bellés, J.M.; Rodriguez, P.L.; Serrano, R. The lithium tolerance of the Arabidopsis cat2 mutant reveals a cross-talk between oxidative stress and ethylene. Plant J. 2007, 52, 1052–1065. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.B.; Liu, C.; Tang, D.Y.; Yan, L.; Wang, D.; Yang, Y.Z.; Gui, J.S.; Zhao, X.Y.; Li, L.G.; Tang, X.D.; et al. The receptor-like sytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Takahashi, S. Molecular mechanisms of natural rubber biosynthesis. Annu. Rev. Biochem. 2020, 89, 821–851. [Google Scholar] [CrossRef]
- Cornish, H.M.K. Alternative sources of natural rubber. Appl. Microbiol. Biotechnol. 2000, 53, 355–365. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Poirier, Y. Establishment of new crops for the production of natural rubber. Trends Biotechnol. 2007, 25, 522–529. [Google Scholar] [CrossRef]
- Men, X.; Wang, F.; Chen, G.Q.; Zhang, H.B.; Xian, M. Biosynthesis of natural rubber: Current state and perspectives. Int. J. Mol. Sci. 2018, 20, 50. [Google Scholar] [CrossRef]
- Supriya, R.; Priyadarshan, P.M. Genomic technologies for Hevea breeding. Adv. Genet. 2019, 104, 1–73. [Google Scholar] [CrossRef]
- Venkatachalam, P.; Jayashree, R.; Rekha, K.; Sushmakumari, S.; Sobha, S.; Jayasree, P.K.; Kala, R.G.; Thulaseedharan, A. Rubber tree (Hevea brasiliensis Muell. Arg). Methods Mol. Biol. 2006, 344, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Carr, M.K.V. The water relations of rubber (Hevea brasiliensis): A Review. Exp. Agric. 2011, 48, 176–193. [Google Scholar] [CrossRef]
- Junaidi; Nuringtyas, T.R.; Clément Vidal, A.; Flori, A.; Syafaah, A.; Oktavia, F.; Ismawanto, S.; Aji, M.; Subandiyah, S.; Montoro, P. Analysis of reduced and oxidized antioxidants in Hevea brasiliensis latex reveals new insights into the regulation of antioxidants in response to harvesting stress and tapping panel dryness. Heliyon 2022, 8, e09840. [Google Scholar] [CrossRef] [PubMed]
- Gébelin, V.; Leclercq, J.; Hu, S.; Tang, C.; Montoro, P. Regulation of MIR genes in response to abiotic stress in Hevea brasiliensis. Int. J. Mol. Sci. 2013, 14, 19587–19604. [Google Scholar] [CrossRef] [PubMed]
- Zhai, D.L.; Yu, H.; Chen, S.C.; Ranjitkar, S.; Xu, J. Responses of rubber leaf phenology to climatic variations in southwest China. Int. J. Biometeorol. 2019, 63, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Du, L. The DUB family in populus: Identification, characterization, evolution and expression patterns. BMC Genom. 2021, 22, 541. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lin, Y.C.; Li, Q.; Shi, R.; Lin, C.Y.; Chen, H.; Chuang, L.; Qu, G.Z.; Sederoff, R.R.; Chiang, V.L. A robust chromatin immunoprecipitation protocol for studying transcription factor–DNA interactions and histone modifications in wood-forming tissue. Nat. Protoc. 2014, 9, 2180–2193. [Google Scholar] [CrossRef]
- Lin, Y.C.; Li, W.; Chen, H.; Li, Q.; Sun, Y.H.; Shi, R.; Lin, C.Y.; Wang, J.P.; Chen, H.C.; Chuang, L.; et al. A simple improved-throughput xylem protoplast system for studying wood formation. Nat. Protoc. 2014, 9, 2194–2205. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Mohamed Sathik, M.B.; Luke, L.P.; Rajamani, A.; Kuruvilla, L.; Sumesh, K.V.; Thomas, M. De novo transcriptome analysis of abiotic stress-responsive transcripts of Hevea brasiliensis. Mol. Breed. 2018, 38, 32. [Google Scholar] [CrossRef]
- Campos Mantello, C.; Boatwright, L.; da Silva, C.C.; Scaloppi, E.J.; de Souza Goncalves, P.; Barbazuk, W.B.; Pereira de Souza, A. Deep expression analysis reveals distinct cold-response strategies in rubber tree (Hevea brasiliensis). BMC Genom. 2019, 20, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.; Meenakumari, T.; Lakshmanan, R. Long term performance of certain ortets and hybrid clones of Hevea brasiliensis in a high altitude region in Kerala, India. J. Plant. Crops 2020, 48, 82–90. [Google Scholar] [CrossRef]
- Anjum, N.A.; Sharma, P.; Gill, S.S.; Hasanuzzaman, M.; Khan, E.A.; Kachhap, K.; Mohamed, A.A.; Thangavel, P.; Devi, G.D.; Vasudhevan, P.; et al. Catalase and ascorbate peroxidase—Representative H2O2-detoxifying heme enzymes in plants. Environ. Sci. Pollut. Res. 2016, 23, 19002–19029. [Google Scholar] [CrossRef] [PubMed]
- Sepasi Tehrani, H.; Moosavi-Movahedi, A.A. Catalase and its mysteries. Prog. Biophys. Mol. Biol. 2018, 140, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Azpilicueta, C.E.; Pena, L.B.; Tomaro, M.L.; Gallego, S.M. Modifications in catalase activity and expression in developing sunflower seedlings under cadmium stress. Redox Rep. 2013, 13, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.; Lin, C.C.; Lett, C.; Karpinska, B.; Wright, M.H.; Foyer, C.H. Catalase: A critical node in the regulation of cell fate. Free Radic. Biol. Med. 2023, 199, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Noctor, G.; Baker, A. Plant catalases: Peroxisomal redox guardians. Arch. Biochem. Biophys. 2012, 525, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Cross, L.L.; Ebeed, H.T.; Baker, A. Peroxisome biogenesis, protein targeting mechanisms and PEX gene functions in plants. Biochim. Biophys. Acta 2016, 1863, 850–862. [Google Scholar] [CrossRef]
- Kamigaki, A.; Mano, S.; Terauchi, K.; Nishi, Y.; Tachibe-Kinoshita, Y.; Nito, K.; Kondo, M.; Hayashi, M.; Nishimura, M.; Esaka, M. Identification of peroxisomal targeting signal of pumpkin catalase and the binding analysis with PTS1 receptor. Plant J. 2003, 33, 161–175. [Google Scholar] [CrossRef]
- Oshima, Y.; Kamigaki, A.; Nakamori, C.; Mano, S.; Hayashi, M.; Nishimura, M.; Esaka, M. Plant catalase is imported into peroxisomes by PEX5p but is distinct from typical PTS1 import. Plant Cell Physiol. 2008, 49, 671–677. [Google Scholar] [CrossRef]
- Yan, J.; Ma, Z.; Xu, X.; Guo, A. Evolution, functional divergence and conserved exon-intron structure of bHLH-PAS gene family. Mol. Genet. Genom. 2014, 289, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Mihelčić, M.; Šmuc, T.; Supek, F. Patterns of diverse gene functions in genomic neighborhoods predict gene function and phenotype. Sci. Rep. 2019, 9, 19537. [Google Scholar] [CrossRef] [PubMed]
- Frugoli, J.A.; McPeek, M.A.; Thomas, T.L.; McClung, C.R. Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics 1998, 149, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Cao, Q.A.; Li, Y.P.; He, M.Q.; Liu, X.G. Advances in cis-element and natural variation-mediated transcriptional regulation and applications in gene editing of major crops. J. Exp. Bot. 2023, 74, 5441–5457. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Wang, L.; Wu, X.; Gao, W.; Zhao, M.; Zhang, J.; Huang, C. Differential expression patterns of pleurotus ostreatus catalase genes during developmental stages and under heat stress. Genes 2017, 8, 335. [Google Scholar] [CrossRef]
- Chiang, C.; Chen, S.; Chen, L.; Chiang, M.; Chien, H.; Lin, K. Expression of the broccoli catalase gene (BoCAT) enhances heat tolerance in transgenic Arabidopsis. J. Plant Biochem. Biotechnol. 2014, 23, 266–277. [Google Scholar] [CrossRef]
- Ghorbel, M.; Zribi, I.; Besbes, M.; Bouali, N.; Brini, F. Catalase gene family in durum wheat: Genome-wide analysis and expression profiling in response to multiple abiotic stress conditions. Plants 2023, 12, 2720. [Google Scholar] [CrossRef]
- Liu, J.; Shi, C.; Shi, C.C.; Li, W.; Zhang, Q.J.; Zhang, Y.; Li, K.; Lu, H.F.; Shi, C.; Zhu, S.T.; et al. The chromosome-based rubber tree genome provides new insights into spurge genome evolution and rubber biosynthesis. Mol. Plant 2020, 13, 336–350. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Kong, G.; Chao, J.; Yin, T.; Tian, H.; Ya, H.; He, L.; Zhang, H. Genome-wide identification of the rubber tree superoxide dismutase (SOD) gene family and analysis of its expression under abiotic stress. PeerJ 2022, 10, e14251. [Google Scholar] [CrossRef] [PubMed]
- Yin, T.; Han, P.; Xi, D.; Yu, W.; Zhu, L.; Du, C.; Yang, N.; Liu, X.; Zhang, H. Genome-wide identification, characterization, and expression profile of NBS-LRR gene family in sweet orange (Citrus sinensis). Gene 2023, 854, 147117. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y. The Arabidopsis information resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003, 31, 224–228. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P.W.; Kayastha, A.M.; Singh, V.K. MFPPI—Multi fasta protparam interface. Bioinformation 2016, 12, 74–77. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Contreras-Moreira, B.; Naamati, G.; Rosello, M.; Allen, J.E.; Hunt, S.E.; Muffato, M.; Gall, A.; Flicek, P. Scripting analyses of genomes in ensembl plants. Methods Mol. Biol. 2022, 2443, 27–55. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.d.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J.; Tamura, K. eggNOG-mapper v2: Functional annotation, orthology assignments, and somain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Li, Y.; Yu, W.; Chen, Y.; Yang, S.; Wu, S.; Chao, J.; Wang, X.; Tian, W. Genome-wide identification and characterization of Heat-Shock transcription factors in rubber tree. Forests 2019, 10, 1157. [Google Scholar] [CrossRef]




| Gene Name | Gene ID | Chr | Genomic Position (5′–3′) | CDS Length (bp) | Exon | Protein Length (aa) | Molecular Weight (kDa) | Isoelectric Point (pI) | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| HbCAT1 | GH714_028220 | 2 | 60,823,793–60,826,740 (+) | 1479 | 8 | 492 | 56.75 | 7.60 | Peroxisome |
| HbCAT2 | GH714_016778 | 5 | 41,479,542–41,497,345 (−) | 1704 | 9 | 567 | 65.13 | 6.84 | Cytoplasm |
| HbCAT3 | GH714_034861 | 14 | 3,781,996–3,785,382 (−) | 1479 | 8 | 492 | 56.89 | 7.24 | Cytoplasm |
| HbCAT4 | GH714_007303 | 14 | 16,753,136–16,762,068 (−) | 1479 | 8 | 492 | 47.15 | 6.45 | Cytoplasm |
| HbCAT5 | GH714_007170 | 14 | 17,423,940–17,432,871 (−) | 1236 | 8 | 411 | 56.81 | 6.82 | Cytoplasm |
| Group A | Group B | Ks | Ka | Ka/Ks | Duplicated Type | Selection Pressure | ||
|---|---|---|---|---|---|---|---|---|
| Gene Name | Gene ID | Gene Name | Gene ID | |||||
| HbCAT1 | GH714_028220 | HbCAT2 | GH714_016778 | 0.083 | 1.519 | 0.054 | Segmental duplication | Purifying selection |
| HbCAT1 | GH714_028220 | HbCAT3 | GH714_034861 | 0.025 | 0.225 | 0.111 | Segmental duplication | Purifying selection |
| HbCAT1 | GH714_028220 | HbCAT4 | GH714_007303 | 0.096 | 1.660 | 0.058 | Segmental duplication | Purifying selection |
| HbCAT2 | GH714_016778 | HbCAT4 | GH714_007303 | 0.038 | 0.351 | 0.107 | Segmental duplication | Purifying selection |
| HbCAT2 | GH714_016778 | HbCAT3 | GH714_034861 | 0.086 | 1.472 | 0.058 | Segmental duplication | Purifying selection |
| HbCAT2 | GH714_016778 | HbCAT5 | GH714_007170 | 0.036 | 0.273 | 0.131 | Segmental duplication | Purifying selection |
| HbCAT3 | GH714_034861 | HbCAT4 | GH714_007303 | 0.101 | 1.417 | 0.071 | Segmental duplication | Purifying selection |
| HbCAT4 | GH714_007303 | HbCAT5 | GH714_007170 | 0.020 | 0.108 | 0.187 | Segmental duplication | Purifying selection |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, W.; Kong, G.; Ya, H.; He, L.; Wu, Y.; Zhang, H. Comprehensive Analysis of the Catalase (CAT) Gene Family and Expression Patterns in Rubber Tree (Hevea brasiliensis) under Various Abiotic Stresses and Multiple Hormone Treatments. Int. J. Mol. Sci. 2024, 25, 70. https://doi.org/10.3390/ijms25010070
Yu W, Kong G, Ya H, He L, Wu Y, Zhang H. Comprehensive Analysis of the Catalase (CAT) Gene Family and Expression Patterns in Rubber Tree (Hevea brasiliensis) under Various Abiotic Stresses and Multiple Hormone Treatments. International Journal of Molecular Sciences. 2024; 25(1):70. https://doi.org/10.3390/ijms25010070
Chicago/Turabian StyleYu, Wencai, Guanghong Kong, Huajin Ya, Ligang He, Yu Wu, and Hanyao Zhang. 2024. "Comprehensive Analysis of the Catalase (CAT) Gene Family and Expression Patterns in Rubber Tree (Hevea brasiliensis) under Various Abiotic Stresses and Multiple Hormone Treatments" International Journal of Molecular Sciences 25, no. 1: 70. https://doi.org/10.3390/ijms25010070
APA StyleYu, W., Kong, G., Ya, H., He, L., Wu, Y., & Zhang, H. (2024). Comprehensive Analysis of the Catalase (CAT) Gene Family and Expression Patterns in Rubber Tree (Hevea brasiliensis) under Various Abiotic Stresses and Multiple Hormone Treatments. International Journal of Molecular Sciences, 25(1), 70. https://doi.org/10.3390/ijms25010070







