Human and Murine Toll-like Receptor-Driven Disease in Systemic Lupus Erythematosus
Abstract
1. Introduction
2. Animal Mouse Models of SLE and LN and the Involvement of TLRs
2.1. Spontaneous Mouse Models of SLE and LN
2.2. Other Genetic Mice Models Mimicking the Pathogenesis of SLE and LN
2.3. Inducible SLE Mouse Models
2.4. Acceleration of Spontaneous Lupus Models and Humanized Mouse
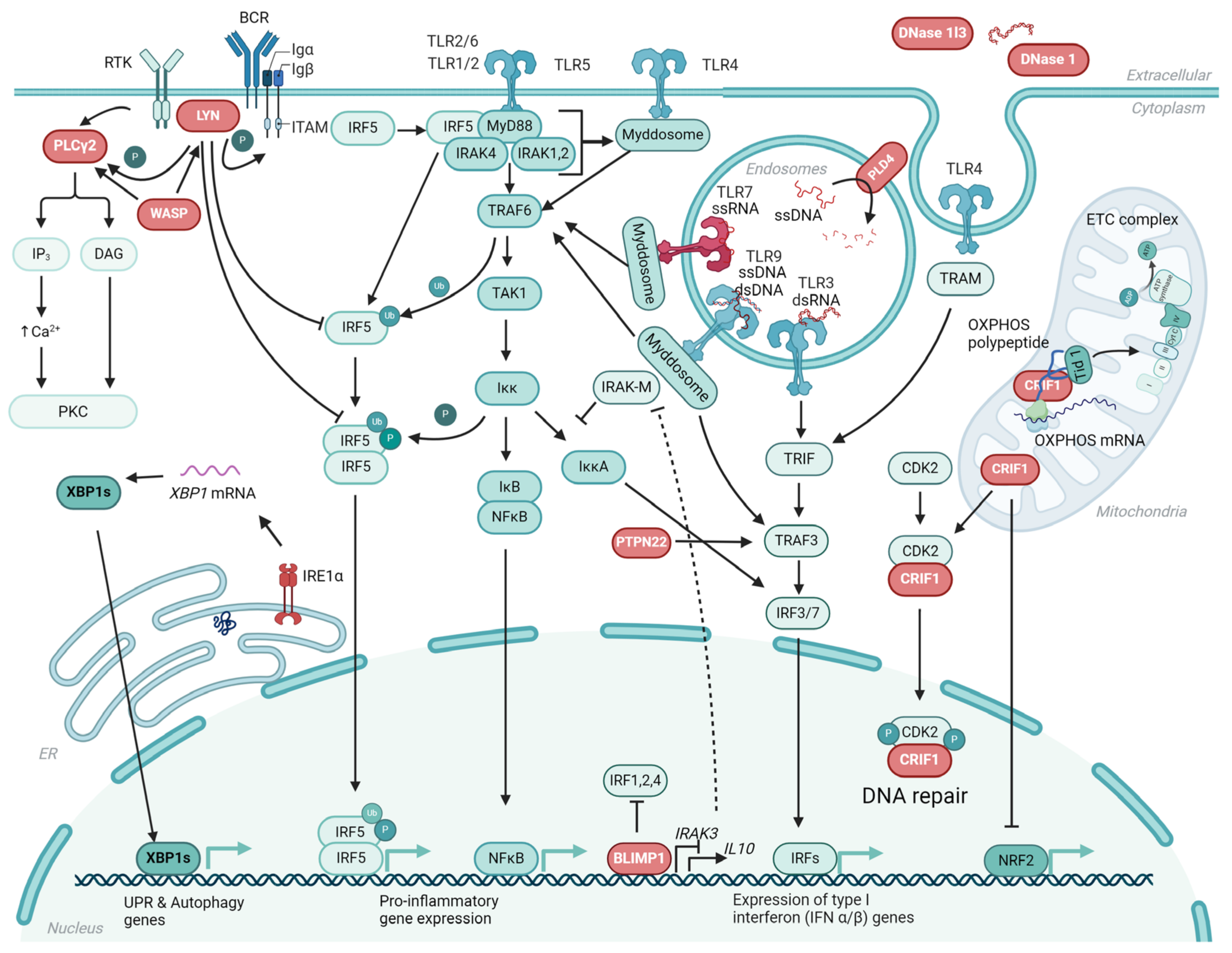
3. TLR Signaling in SLE—An Update on Recent Findings
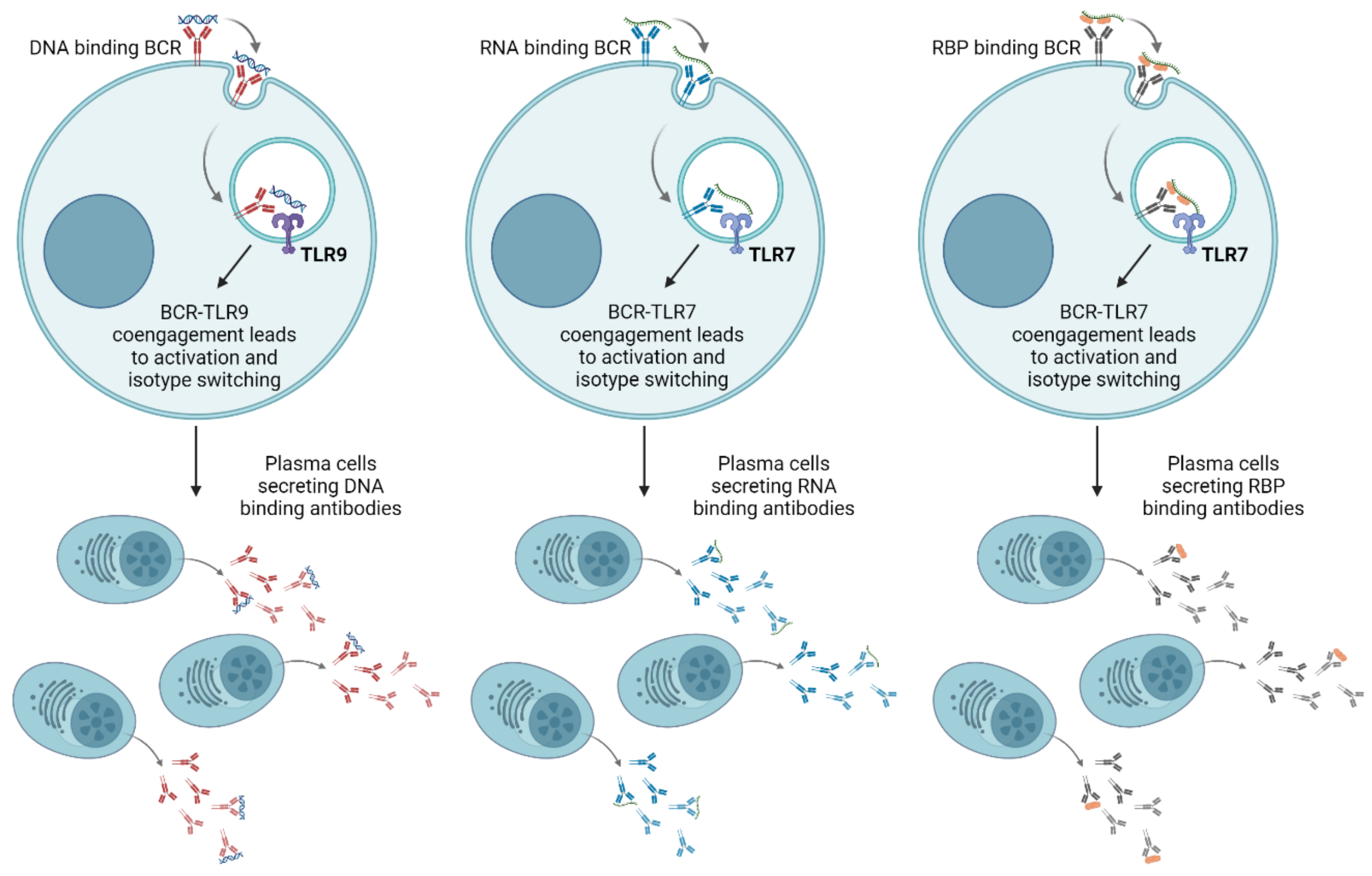
3.1. TLR Driven Autoantibody Production and Tolerance Breakage
3.2. Diverse Effects of Different TLRs on SLE Pathogenesis—TLR7 As the Main Driver of Disease
3.3. Regulatory Functions of TLR8 and TLR9
3.4. Regulation of TLR Signaling—Endosomal Trafficking and Glycosylation
3.5. Diverse Effects of TLR Signaling on Autoantibody Repertoire in Different Mouse Models of SLE
3.6. Age-Associated B Cells as the Main Source of TLR Driven Autoantibody Production
3.7. Targeting TLRs in the Treatment of SLE
4. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Aringer, M.; Costenbader, K.; Daikh, D.; Brinks, R.; Mosca, M.; Ramsey-Goldman, R.; Smolen, J.S.; Wofsy, D.; Boumpas, D.T.; Kamen, D.L.; et al. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann. Rheum. Dis. 2019, 78, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Dema, B.; Charles, N. Autoantibodies in SLE: Specificities, Isotypes and Receptors. Antibodies 2016, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Nowling, T.K.; Gilkeson, G.S. Mechanisms of tissue injury in lupus nephritis. Arthritis Res. Ther. 2011, 13, 250. [Google Scholar] [CrossRef] [PubMed]
- Duan, T.; Du, Y.; Xing, C.; Wang, H.Y.; Wang, R.-F. Toll-Like Receptor Signaling and Its Role in Cell-Mediated Immunity. Front. Immunol. 2022, 13, 812774. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Ikegawa, M.; Ori, D.; Akira, S. Decoding Toll-like receptors: Recent insights and perspectives in innate immunity. Immunity 2024, 57, 649–673. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.J.; Li, F.A.; Sheu, J.J.; Li, S.C.; Weng, S.W.; Shen, F.C.; Chang, Y.H.; Chen, H.Y.; Liou, C.W.; Lin, T.K.; et al. A Study on MDA5 Signaling in Splenic B Cells from an Imiquimod-Induced Lupus Mouse Model with Proteomics. Cells 2022, 11, 3350. [Google Scholar] [CrossRef] [PubMed]
- Moore, E.; Putterman, C. Are lupus animal models useful for understanding and developing new therapies for human SLE? J. Autoimmun. 2020, 112, 102490. [Google Scholar] [CrossRef]
- Du, Y.; Sanam, S.; Kate, K.; Mohan, C. Animal models of lupus and lupus nephritis. Curr. Pharm. Des. 2015, 21, 2320–2349. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Titov, A.A.; Morel, L. An update on lupus animal models. Curr. Opin. Rheumatol. 2017, 29, 434–441. [Google Scholar] [CrossRef]
- Richard, M.L.; Gilkeson, G. Mouse models of lupus: What they tell us and what they don’t. Lupus Sci. Med. 2018, 5, e000199. [Google Scholar] [CrossRef]
- Halkom, A.; Wu, H.; Lu, Q. Contribution of mouse models in our understanding of lupus. Int. Rev. Immunol. 2020, 39, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Katikaneni, D.; Morel, L.; Scindia, Y. Animal models of lupus nephritis: The past, present and a future outlook. Autoimmunity 2024, 57, 2319203. [Google Scholar] [CrossRef] [PubMed]
- Helyer, B.J.; Howie, J.B. Renal disease associated with positive lupus erythematosus tests in a cross-bred strain of mice. Nature 1963, 197, 197. [Google Scholar] [CrossRef] [PubMed]
- Dubois, E.L.; Horowitz, R.E.; Demopoulos, H.B.; Teplitz, R. NZB/NZW mice as a model of systemic lupus erythematosus. JAMA 1966, 195, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Morel, L. Mapping lupus susceptibility genes in the NZM2410 mouse model. Adv. Immunol. 2012, 115, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Vidal, S.; Gelpi, C.; Rodriguez-Sanchez, J.L. (SWR × SJL)F1 mice: A new model of lupus-like disease. J. Exp. Med. 1994, 179, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Andrews, B.S.; Eisenberg, R.A.; Theofilopoulos, A.N.; Izui, S.; Wilson, C.B.; McConahey, P.J.; Murphy, E.D.; Roths, J.B.; Dixon, F.J. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J. Exp. Med. 1978, 148, 1198–1215. [Google Scholar] [CrossRef]
- Bolland, S.; Yim, Y.S.; Tus, K.; Wakeland, E.K.; Ravetch, J.V. Genetic modifiers of systemic lupus erythematosus in FcgammaRIIB(-/-) mice. J. Exp. Med. 2002, 195, 1167–1174. [Google Scholar] [CrossRef]
- Ondee, T.; Surawut, S.; Taratummarat, S.; Hirankarn, N.; Palaga, T.; Pisitkun, P.; Pisitkun, T.; Leelahavanichkul, A. Fc Gamma Receptor IIB Deficient Mice: A Lupus Model with Increased Endotoxin Tolerance-Related Sepsis Susceptibility. Shock 2017, 47, 743–752. [Google Scholar] [CrossRef]
- Han, J.H.; Umiker, B.R.; Kazimirova, A.A.; Fray, M.; Korgaonkar, P.; Selsing, E.; Imanishi-Kari, T. Expression of an anti-RNA autoantibody in a mouse model of SLE increases neutrophil and monocyte numbers as well as IFN-I expression. Eur. J. Immunol. 2014, 44, 215–226. [Google Scholar] [CrossRef]
- Deane, J.A.; Pisitkun, P.; Barrett, R.S.; Feigenbaum, L.; Town, T.; Ward, J.M.; Flavell, R.A.; Bolland, S. Control of Toll-like Receptor 7 Expression Is Essential to Restrict Autoimmunity and Dendritic Cell Proliferation. Immunity 2007, 27, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Tizaoui, K.; Terrazzino, S.; Cargnin, S.; Lee, K.H.; Gauckler, P.; Li, H.; Shin, J.I.; Kronbichler, A. The role of PTPN22 in the pathogenesis of autoimmune diseases: A comprehensive review. Semin. Arthritis Rheum. 2021, 51, 513–522. [Google Scholar] [CrossRef]
- Wang, Y.; Shaked, I.; Stanford, S.M.; Zhou, W.; Curtsinger, J.M.; Mikulski, Z.; Shaheen, Z.R.; Cheng, G.; Sawatzke, K.; Campbell, A.M.; et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity 2013, 39, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Petersen, F.; Yu, X. The role of PTPN22 in autoimmunity: Learning from mice. Autoimmun. Rev. 2014, 13, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Wilber, A.; O’Connor, T.P.; Lu, M.L.; Karimi, A.; Schneider, M.C. Dnase1l3 deficiency in lupus-prone MRL and NZB/W F1 mice. Clin. Exp. Immunol. 2003, 134, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Engavale, M.; Hernandez, C.J.; Infante, A.; LeRoith, T.; Radovan, E.; Evans, L.; Villarreal, J.; Reilly, C.M.; Sutton, R.B.; Keyel, P.A. Deficiency of macrophage-derived Dnase1L3 causes lupus-like phenotypes in mice. J. Leukoc. Biol. 2023, 114, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Sisirak, V.; Sally, B.; D’Agati, V.; Martinez-Ortiz, W.; Ozcakar, Z.B.; David, J.; Rashidfarrokhi, A.; Yeste, A.; Panea, C.; Chida, A.S.; et al. Digestion of Chromatin in Apoptotic Cell Microparticles Prevents Autoimmunity. Cell 2016, 166, 88–101. [Google Scholar] [CrossRef]
- Weisenburger, T.; von Neubeck, B.; Schneider, A.; Ebert, N.; Schreyer, D.; Acs, A.; Winkler, T.H. Epistatic Interactions Between Mutations of Deoxyribonuclease 1-Like 3 and the Inhibitory Fc Gamma Receptor IIB Result in Very Early and Massive Autoantibodies Against Double-Stranded DNA. Front. Immunol. 2018, 9, 1551. [Google Scholar] [CrossRef] [PubMed]
- Korn, M.A.; Steffensen, M.; Brandl, C.; Royzman, D.; Daniel, C.; Winkler, T.H.; Nitschke, L. Epistatic effects of Siglec-G and DNase1 or DNase1l3 deficiencies in the development of systemic lupus erythematosus. Front. Immunol. 2023, 14, 1095830. [Google Scholar] [CrossRef] [PubMed]
- Napirei, M.; Karsunky, H.; Zevnik, B.; Stephan, H.; Mannherz, H.G.; Moroy, T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat. Genet. 2000, 25, 177–181. [Google Scholar] [CrossRef]
- Kenny, E.F.; Raupach, B.; Abu Abed, U.; Brinkmann, V.; Zychlinsky, A. Dnase1-deficient mice spontaneously develop a systemic lupus erythematosus-like disease. Eur. J. Immunol. 2019, 49, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Hibbs, M.L.; Tarlinton, D.M.; Armes, J.; Grail, D.; Hodgson, G.; Maglitto, R.; Stacker, S.A.; Dunn, A.R. Multiple defects in the immune system of Lyn-deficient mice, culminating in autoimmune disease. Cell 1995, 83, 301–311. [Google Scholar] [CrossRef]
- Ban, T.; Sato, G.R.; Nishiyama, A.; Akiyama, A.; Takasuna, M.; Umehara, M.; Suzuki, S.; Ichino, M.; Matsunaga, S.; Kimura, A.; et al. Lyn Kinase Suppresses the Transcriptional Activity of IRF5 in the TLR-MyD88 Pathway to Restrain the Development of Autoimmunity. Immunity 2016, 45, 319–332. [Google Scholar] [CrossRef]
- Park, J.S.; Yang, S.; Hwang, S.H.; Choi, J.; Kwok, S.K.; Kong, Y.Y.; Youn, J.; Cho, M.L.; Park, S.H. B Cell-Specific Deletion of CR6-Interacting Factor 1 Drives Lupus-like Autoimmunity by Activation of Interleukin-17, Interleukin-6, and Pathogenic Follicular Helper T Cells in a Mouse Model. Arthritis Rheumatol. 2022, 74, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Lietke, D.S. CRIF1 and Its Function in Anti-Viral Immunity; Ludwig Maximilian University of Munich: Munich, Germany, 2017. [Google Scholar]
- Lee, K.; Park, J.; Tanno, H.; Georgiou, G.; Diamond, B.; Kim, S.J. Peripheral T cell activation, not thymic selection, expands the T follicular helper repertoire in a lupus-prone murine model. Proc. Natl. Acad. Sci. USA 2023, 120, e2309780120. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Schatzle, S.; Ahmed, S.S.; Haap, W.; Jang, S.H.; Gregersen, P.K.; Georgiou, G.; Diamond, B. Increased cathepsin S in Prdm1(-/-) dendritic cells alters the T(FH) cell repertoire and contributes to lupus. Nat. Immunol. 2017, 18, 1016–1024. [Google Scholar] [CrossRef]
- Kim, V.; Lee, K.; Tian, H.; Jang, S.H.; Diamond, B.; Kim, S.J. IL-17-producing follicular Th cells enhance plasma cell differentiation in lupus-prone mice. JCI Insight 2022, 7, e157332. [Google Scholar] [CrossRef]
- Ko, Y.A.; Chan, Y.H.; Liu, C.H.; Liang, J.J.; Chuang, T.H.; Hsueh, Y.P.; Lin, Y.L.; Lin, K.I. Blimp-1-Mediated Pathway Promotes Type I IFN Production in Plasmacytoid Dendritic Cells by Targeting to Interleukin-1 Receptor-Associated Kinase M. Front. Immunol. 2018, 9, 1828. [Google Scholar] [CrossRef]
- Reuschle, Q.; Van Heddegem, L.; Bosteels, V.; Moncan, M.; Depauw, S.; Wadier, N.; Marechal, S.; De Nolf, C.; Delgado, V.; Messai, Y.; et al. Loss of function of XBP1 splicing activity of IRE1alpha favors B cell tolerance breakdown. J. Autoimmun. 2024, 142, 103152. [Google Scholar] [CrossRef]
- Murga, M.; Fernandez-Capetillo, O.; Field, S.J.; Moreno, B.; Borlado, L.R.; Fujiwara, Y.; Balomenos, D.; Vicario, A.; Carrera, A.C.; Orkin, S.H.; et al. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity 2001, 15, 959–970. [Google Scholar] [CrossRef]
- Marin-Vidalled, M.J.; Bolivar, A.; Zubiaga, A.; Lopez-Hoyos, M. The combined effect of BCL-2 over-expression and E2F2 deficiency induces an autoimmune syndrome in non-susceptible mouse strain C57BL/6. Autoimmunity 2010, 43, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, L.; Wu, C.; Sun, S.; Pan, J.H. E2F2 directly regulates the STAT1 and PI3K/AKT/NF-kappaB pathways to exacerbate the inflammatory phenotype in rheumatoid arthritis synovial fibroblasts and mouse embryonic fibroblasts. Arthritis Res. Ther. 2018, 20, 225. [Google Scholar] [CrossRef] [PubMed]
- Akizuki, S.; Ishigaki, K.; Kochi, Y.; Law, S.M.; Matsuo, K.; Ohmura, K.; Suzuki, A.; Nakayama, M.; Iizuka, Y.; Koseki, H.; et al. PLD4 is a genetic determinant to systemic lupus erythematosus and involved in murine autoimmune phenotypes. Ann. Rheum. Dis. 2019, 78, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.L.; Blane, T.R.; Thinnes, T.C.; Gerlt, E.; Marshak-Rothstein, A.; Huang, D.; Nemazee, D. Disease in the Pld4thss/thss Model of Murine Lupus Requires TLR9. Immunohorizons 2023, 7, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.L.; Huang, D.; Huber, C.; Martensson, A.; Tardif, V.; Skog, P.D.; Blane, T.R.; Thinnes, T.C.; Osborn, K.; Chong, H.S.; et al. PLD3 and PLD4 are single-stranded acid exonucleases that regulate endosomal nucleic-acid sensing. Nat. Immunol. 2018, 19, 942–953. [Google Scholar] [CrossRef]
- Yu, P.; Constien, R.; Dear, N.; Katan, M.; Hanke, P.; Bunney, T.D.; Kunder, S.; Quintanilla-Martinez, L.; Huffstadt, U.; Schroder, A.; et al. Autoimmunity and inflammation due to a gain-of-function mutation in phospholipase C gamma 2 that specifically increases external Ca2+ entry. Immunity 2005, 22, 451–465. [Google Scholar] [CrossRef]
- Becker-Herman, S.; Meyer-Bahlburg, A.; Schwartz, M.A.; Jackson, S.W.; Hudkins, K.L.; Liu, C.; Sather, B.D.; Khim, S.; Liggitt, D.; Song, W.; et al. WASp-deficient B cells play a critical, cell-intrinsic role in triggering autoimmunity. J. Exp. Med. 2011, 208, 2033–2042. [Google Scholar] [CrossRef]
- Jackson, S.W.; Scharping, N.E.; Kolhatkar, N.S.; Khim, S.; Schwartz, M.A.; Li, Q.-Z.; Hudkins, K.L.; Alpers, C.E.; Liggitt, D.; Rawlings, D.J. Opposing Impact of B Cell–Intrinsic TLR7 and TLR9 Signals on Autoantibody Repertoire and Systemic Inflammation. J. Immunol. 2014, 192, 4525–4532. [Google Scholar] [CrossRef]
- Jiang, Y.; Xiang, Y.; Lin, C.; Zhang, W.; Yang, Z.; Xiang, L.; Xiao, Y.; Chen, L.; Ran, Q.; Li, Z. Multifunctions of CRIF1 in cancers and mitochondrial dysfunction. Front. Oncol. 2022, 12, 1009948. [Google Scholar] [CrossRef]
- Nadeau, S.; Martins, G.A. Conserved and Unique Functions of Blimp1 in Immune Cells. Front. Immunol. 2021, 12, 805260. [Google Scholar] [CrossRef]
- Brodie, E.J.; Infantino, S.; Low, M.S.Y.; Tarlinton, D.M. Lyn, Lupus, and (B) Lymphocytes, a Lesson on the Critical Balance of Kinase Signaling in Immunity. Front. Immunol. 2018, 9, 401. [Google Scholar] [CrossRef]
- Jackson, J.T.; Mulazzani, E.; Nutt, S.L.; Masters, S.L. The role of PLCgamma2 in immunological disorders, cancer, and neurodegeneration. J. Biol. Chem. 2021, 297, 100905. [Google Scholar] [CrossRef]
- Rey-Suarez, I.; Wheatley, B.A.; Koo, P.; Bhanja, A.; Shu, Z.; Mochrie, S.; Song, W.; Shroff, H.; Upadhyaya, A. WASP family proteins regulate the mobility of the B cell receptor during signaling activation. Nat. Commun. 2020, 11, 439. [Google Scholar] [CrossRef] [PubMed]
- Junjappa, R.P.; Patil, P.; Bhattarai, K.R.; Kim, H.R.; Chae, H.J. IRE1alpha Implications in Endoplasmic Reticulum Stress-Mediated Development and Pathogenesis of Autoimmune Diseases. Front. Immunol. 2018, 9, 1289. [Google Scholar] [CrossRef]
- Park, S.M.; Kang, T.I.; So, J.S. Roles of XBP1s in Transcriptional Regulation of Target Genes. Biomedicines 2021, 9, 791. [Google Scholar] [CrossRef]
- Freitas, E.C.; de Oliveira, M.S.; Monticielo, O.A. Pristane-induced lupus: Considerations on this experimental model. Clin. Rheumatol. 2017, 36, 2403–2414. [Google Scholar] [CrossRef]
- Satoh, M.; Richards, H.B.; Shaheen, V.M.; Yoshida, H.; Shaw, M.; Naim, J.O.; Wooley, P.H.; Reeves, W.H. Widespread susceptibility among inbred mouse strains to the induction of lupus autoantibodies by pristane. Clin. Exp. Immunol. 2000, 121, 399–405. [Google Scholar] [CrossRef]
- Bender, A.T.; Wu, Y.; Cao, Q.; Ding, Y.; Oestreicher, J.; Genest, M.; Akare, S.; Ishizaka, S.T.; Mackey, M.F. Assessment of the translational value of mouse lupus models using clinically relevant biomarkers. Transl. Res. 2014, 163, 515–532. [Google Scholar] [CrossRef]
- Postal, M.; Vivaldo, J.F.; Fernandez-Ruiz, R.; Paredes, J.L.; Appenzeller, S.; Niewold, T.B. Type I interferon in the pathogenesis of systemic lupus erythematosus. Curr. Opin. Immunol. 2020, 67, 87–94. [Google Scholar] [CrossRef]
- Savarese, E.; Steinberg, C.; Pawar, R.D.; Reindl, W.; Akira, S.; Anders, H.J.; Krug, A. Requirement of Toll-like receptor 7 for pristane-induced production of autoantibodies and development of murine lupus nephritis. Arthritis Rheum. 2008, 58, 1107–1115. [Google Scholar] [CrossRef]
- Urbonaviciute, V.; Starke, C.; Pirschel, W.; Pohle, S.; Frey, S.; Daniel, C.; Amann, K.; Schett, G.; Herrmann, M.; Voll, R.E. Toll-like receptor 2 is required for autoantibody production and development of renal disease in pristane-induced lupus. Arthritis Rheum. 2013, 65, 1612–1623. [Google Scholar] [CrossRef]
- Summers, S.A.; Hoi, A.; Steinmetz, O.M.; O’Sullivan, K.M.; Ooi, J.D.; Odobasic, D.; Akira, S.; Kitching, A.R.; Holdsworth, S.R. TLR9 and TLR4 are required for the development of autoimmunity and lupus nephritis in pristane nephropathy. J. Autoimmun. 2010, 35, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Bossaller, L.; Christ, A.; Pelka, K.; Nündel, K.; Chiang, P.I.; Pang, C.; Mishra, N.; Busto, P.; Bonegio, R.G.; Schmidt, R.E.; et al. TLR9 Deficiency Leads to Accelerated Renal Disease and Myeloid Lineage Abnormalities in Pristane-Induced Murine Lupus. J. Immunol. 2016, 197, 1044–1053. [Google Scholar] [CrossRef] [PubMed]
- Giordano, D.; Kuley, R.; Draves, K.E.; Elkon, K.B.; Giltiay, N.V.; Clark, E.A. B cell-activating factor (BAFF) from dendritic cells, monocytes and neutrophils is required for B cell maturation and autoantibody production in SLE-like autoimmune disease. Front. Immunol. 2023, 14, 1050528. [Google Scholar] [CrossRef] [PubMed]
- Yoo, E.J.; Oh, K.H.; Piao, H.; Kang, H.J.; Jeong, G.W.; Park, H.; Lee, C.J.; Ryu, H.; Yang, S.H.; Kim, M.G.; et al. Macrophage transcription factor TonEBP promotes systemic lupus erythematosus and kidney injury via damage-induced signaling pathways. Kidney Int. 2023, 104, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, F.; Tagaya, Y.; Ozato, K.; Asada, H. Essential Requirement for IFN Regulatory Factor 7 in Autoantibody Production but Not Development of Nephritis in Murine Lupus. J. Immunol. 2016, 197, 2167–2176. [Google Scholar] [CrossRef] [PubMed]
- Motwani, M.; McGowan, J.; Antonovitch, J.; Gao, K.M.; Jiang, Z.; Sharma, S.; Baltus, G.A.; Nickerson, K.M.; Marshak-Rothstein, A.; Fitzgerald, K.A. cGAS-STING Pathway Does Not Promote Autoimmunity in Murine Models of SLE. Front. Immunol. 2021, 12, 605930. [Google Scholar] [CrossRef]
- Corzo, C.A.; Varfolomeev, E.; Setiadi, A.F.; Francis, R.; Klabunde, S.; Senger, K.; Sujatha-Bhaskar, S.; Drobnick, J.; Do, S.; Suto, E.; et al. The kinase IRAK4 promotes endosomal TLR and immune complex signaling in B cells and plasmacytoid dendritic cells. Sci. Signal 2020, 13, eaaz1053. [Google Scholar] [CrossRef]
- Lin, W.; Seshasayee, D.; Lee, W.P.; Caplazi, P.; McVay, S.; Suto, E.; Nguyen, A.; Lin, Z.; Sun, Y.; DeForge, L.; et al. Dual B cell immunotherapy is superior to individual anti-CD20 depletion or BAFF blockade in murine models of spontaneous or accelerated lupus. Arthritis Rheumatol. 2015, 67, 215–224. [Google Scholar] [CrossRef]
- Gardet, A.; Chou, W.C.; Reynolds, T.L.; Velez, D.B.; Fu, K.; Czerkowicz, J.M.; Bajko, J.; Ranger, A.M.; Allaire, N.; Kerns, H.M.; et al. Pristane-Accelerated Autoimmune Disease in (SWR X NZB) F1 Mice Leads to Prominent Tubulointerstitial Inflammation and Human Lupus Nephritis-Like Fibrosis. PLoS ONE 2016, 11, e0164423. [Google Scholar] [CrossRef]
- Satoh, M.; Weintraub, J.P.; Yoshida, H.; Shaheen, V.M.; Richards, H.B.; Shaw, M.; Reeves, W.H. Fas and Fas ligand mutations inhibit autoantibody production in pristane-induced lupus. J. Immunol. 2000, 165, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Yokogawa, M.; Takaishi, M.; Nakajima, K.; Kamijima, R.; Fujimoto, C.; Kataoka, S.; Terada, Y.; Sano, S. Epicutaneous application of toll-like receptor 7 agonists leads to systemic autoimmunity in wild-type mice: A new model of systemic Lupus erythematosus. Arthritis Rheumatol. 2014, 66, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Gleichmann, E.; Gleichmann, H. Pathogenesis of graft-versus-host reactions (GVHR) and GVH-like diseases. J. Invest. Dermatol. 1985, 85, 115s–120s. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Flynn, R.; Price, A.; Ranger, A.; Browning, J.L.; Taylor, P.A.; Ritz, J.; Antin, J.H.; Murphy, W.J.; Luznik, L.; et al. Donor B-cell alloantibody deposition and germinal center formation are required for the development of murine chronic GVHD and bronchiolitis obliterans. Blood 2012, 119, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Bracken, S.J.; Suthers, A.N.; DiCioccio, R.A.; Su, H.; Anand, S.; Poe, J.C.; Jia, W.; Visentin, J.; Basher, F.; Jordan, C.Z.; et al. Heightened TLR7 signaling primes BCR-activated B cells in chronic graft-versus-host disease for effector functions. Blood Adv. 2024, 8, 667–680. [Google Scholar] [CrossRef] [PubMed]
- Garimella, M.G.; He, C.; Chen, G.; Li, Q.Z.; Huang, X.; Karlsson, M.C.I. The B cell response to both protein and nucleic acid antigens displayed on apoptotic cells are dependent on endosomal pattern recognition receptors. J. Autoimmun. 2021, 117, 102582. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Fujishiro, M.; Yoshida, Y.; Kataoka, Y.; Sakuma, S.; Nishi, T.; Ikeda, K.; Morimoto, S.; Takamori, K.; Sekigawa, I. Exposure of female NZBWF1 mice to imiquimod-induced lupus nephritis at an early age via a unique mechanism that differed from spontaneous onset. Clin. Exp. Immunol. 2022, 208, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Terrell, M.; Brown, J.; Castellanos Garcia, A.; Elshikha, A.; Morel, L. TLR7/TLR8 activation and susceptibility genes synergize to breach gut barrier in a mouse model of lupus. Front. Immunol. 2023, 14, 1187145. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Bethunaickan, R.; Huang, W.; Ramanujam, M.; Madaio, M.P.; Davidson, A. IFN-alpha confers resistance of systemic lupus erythematosus nephritis to therapy in NZB/W F1 mice. J. Immunol. 2011, 187, 1506–1513. [Google Scholar] [CrossRef]
- Pollard, K.M.; Escalante, G.M.; Huang, H.; Haraldsson, K.M.; Hultman, P.; Christy, J.M.; Pawar, R.D.; Mayeux, J.M.; Gonzalez-Quintial, R.; Baccala, R.; et al. Induction of Systemic Autoimmunity by a Xenobiotic Requires Endosomal TLR Trafficking and Signaling from the Late Endosome and Endolysosome but Not Type I IFN. J. Immunol. 2017, 199, 3739–3747. [Google Scholar] [CrossRef]
- Gill, R.F.; Mathieu, P.A.; Lash, L.H.; Rosenspire, A.J. Naturally occurring autoimmune disease in (NZB X NZW) F1 mice is correlated with suppression of MZ B cell development due to aberrant B Cell Receptor (BCR) signaling, which is exacerbated by exposure to inorganic mercury. Toxicol. Sci. 2023, 197, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Favor, O.K.; Chauhan, P.S.; Pourmand, E.; Edwards, A.M.; Wagner, J.G.; Lewandowski, R.P.; Heine, L.K.; Harkema, J.R.; Lee, K.S.S.; Pestka, J.J. Lipidome modulation by dietary omega-3 polyunsaturated fatty acid supplementation or selective soluble epoxide hydrolase inhibition suppresses rough LPS-accelerated glomerulonephritis in lupus-prone mice. Front. Immunol. 2023, 14, 1124910. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Hayashi, T. Synthetic CpG oligodeoxynucleotides accelerate the development of lupus nephritis during preactive phase in NZB x NZWF1 mice. Lupus 2003, 12, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liao, S.; Zhou, H.; Yang, L.; Guo, F.; Chen, S.; Li, A.; Pan, Q.; Yang, C.; Liu, H.F.; et al. Humanized Mouse Models of Systemic Lupus Erythematosus: Opportunities and Challenges. Front. Immunol. 2021, 12, 816956. [Google Scholar] [CrossRef] [PubMed]
- Maria, N.I.; Papoin, J.; Raparia, C.; Sun, Z.; Josselsohn, R.; Lu, A.; Katerji, H.; Syeda, M.M.; Polsky, D.; Paulson, R.; et al. Human TLR8 induces inflammatory bone marrow erythromyeloblastic islands and anemia in SLE-prone mice. Life Sci. Alliance 2023, 6, e202302241. [Google Scholar] [CrossRef] [PubMed]
- Cakan, E.; Ah Kioon, M.D.; Garcia-Carmona, Y.; Glauzy, S.; Oliver, D.; Yamakawa, N.; Vega Loza, A.; Du, Y.; Schickel, J.N.; Boeckers, J.M.; et al. TLR9 ligand sequestration by chemokine CXCL4 negatively affects central B cell tolerance. J. Exp. Med. 2023, 220, e20230944. [Google Scholar] [CrossRef] [PubMed]
- Schaper, F.; de Leeuw, K.; Horst, G.; Maas, F.; Bootsma, H.; Heeringa, P.; Limburg, P.C.; Westra, J. Autoantibodies to box A of high mobility group box 1 in systemic lupus erythematosus. Clin. Exp. Immunol. 2017, 188, 412–419. [Google Scholar] [CrossRef]
- Abdulahad, D.A.; Westra, J.; Bijzet, J.; Limburg, P.C.; Kallenberg, C.G.; Bijl, M. High mobility group box 1 (HMGB1) and anti-HMGB1 antibodies and their relation to disease characteristics in systemic lupus erythematosus. Arthritis Res. Ther. 2011, 13, R71. [Google Scholar] [CrossRef]
- Wirestam, L.; Schierbeck, H.; Skogh, T.; Gunnarsson, I.; Ottosson, L.; Erlandsson-Harris, H.; Wettero, J.; Sjowall, C. Antibodies against High Mobility Group Box protein-1 (HMGB1) versus other anti-nuclear antibody fine-specificities and disease activity in systemic lupus erythematosus. Arthritis Res. Ther. 2015, 17, 338. [Google Scholar] [CrossRef]
- Tanaka, A.; Ito, T.; Kibata, K.; Inagaki-Katashiba, N.; Amuro, H.; Nishizawa, T.; Son, Y.; Ozaki, Y.; Nomura, S. Serum high-mobility group box 1 is correlated with interferon-alpha and may predict disease activity in patients with systemic lupus erythematosus. Lupus 2019, 28, 1120–1127. [Google Scholar] [CrossRef]
- Urbonaviciute, V.; Furnrohr, B.G.; Meister, S.; Munoz, L.; Heyder, P.; De Marchis, F.; Bianchi, M.E.; Kirschning, C.; Wagner, H.; Manfredi, A.A.; et al. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: Implications for the pathogenesis of SLE. J. Exp. Med. 2008, 205, 3007–3018. [Google Scholar] [CrossRef]
- Das, N.; Dewan, V.; Grace, P.M.; Gunn, R.J.; Tamura, R.; Tzarum, N.; Watkins, L.R.; Wilson, I.A.; Yin, H. HMGB1 Activates Proinflammatory Signaling via TLR5 Leading to Allodynia. Cell Rep. 2016, 17, 1128–1140. [Google Scholar] [CrossRef]
- Ma, K.; Li, J.; Wang, X.; Lin, X.; Du, W.; Yang, X.; Mou, F.; Fang, Y.; Zhao, Y.; Hong, X.; et al. TLR4(+)CXCR4(+) plasma cells drive nephritis development in systemic lupus erythematosus. Ann. Rheum. Dis. 2018, 77, 1498–1506. [Google Scholar] [CrossRef]
- Li, S.J.; Ruan, D.D.; Wu, W.Z.; Wu, M.; Wu, Q.Y.; Wang, H.L.; Ji, Y.Y.; Zhang, Y.P.; Lin, X.F.; Fang, Z.T.; et al. Potential regulatory role of the Nrf2/HMGB1/TLR4/NF-kappaB signaling pathway in lupus nephritis. Pediatr. Rheumatol. Online J. 2023, 21, 130. [Google Scholar] [CrossRef]
- Huang, Q.; Shen, S.; Qu, H.; Huang, Y.; Wu, D.; Jiang, H.; Yuan, C. Expression of HMGB1 and TLR4 in neuropsychiatric systemic lupus erythematosus patients with seizure disorders. Ann. Transl. Med. 2020, 8, 9. [Google Scholar] [CrossRef]
- Elloumi, N.; Tahri, S.; Fakhfakh, R.; Abida, O.; Mahfoudh, N.; Hachicha, H.; Marzouk, S.; Bahloul, Z.; Masmoudi, H. Role of innate immune receptors TLR4 and TLR2 polymorphisms in systemic lupus erythematosus susceptibility. Ann. Hum. Genet. 2022, 86, 137–144. [Google Scholar] [CrossRef]
- Lee, Y.H.; Lee, H.S.; Choi, S.J.; Ji, J.D.; Song, G.G. Associations between TLR polymorphisms and systemic lupus erythematosus: A systematic review and meta-analysis. Clin. Exp. Rheumatol. 2012, 30, 262–265. [Google Scholar]
- Sanchez, E.; Orozco, G.; Lopez-Nevot, M.A.; Jimenez-Alonso, J.; Martin, J. Polymorphisms of toll-like receptor 2 and 4 genes in rheumatoid arthritis and systemic lupus erythematosus. Tissue Antigens 2004, 63, 54–57. [Google Scholar] [CrossRef]
- Alajoleen, R.M.; Oakland, D.N.; Estaleen, R.; Shakeri, A.; Lu, R.; Appiah, M.; Sun, S.; Neumann, J.; Kawauchi, S.; Cecere, T.E.; et al. Tlr5 deficiency exacerbates lupus-like disease in the MRL/lpr mouse model. Front. Immunol. 2024, 15, 1359534. [Google Scholar] [CrossRef]
- Patole, P.S.; Pawar, R.D.; Lech, M.; Zecher, D.; Schmidt, H.; Segerer, S.; Ellwart, A.; Henger, A.; Kretzler, M.; Anders, H.J. Expression and regulation of Toll-like receptors in lupus-like immune complex glomerulonephritis of MRL-Fas(lpr) mice. Nephrol. Dial. Transpl. 2006, 21, 3062–3073. [Google Scholar] [CrossRef]
- Elloumi, N.; Fakhfakh, R.; Abida, O.; Ayadi, L.; Marzouk, S.; Hachicha, H.; Fourati, M.; Bahloul, Z.; Mhiri, M.N.; Kammoun, K.; et al. Relevant genetic polymorphisms and kidney expression of Toll-like receptor (TLR)-5 and TLR-9 in lupus nephritis. Clin. Exp. Immunol. 2017, 190, 328–339. [Google Scholar] [CrossRef]
- Rupasree, Y.; Naushad, S.M.; Varshaa, R.; Mahalakshmi, G.S.; Kumaraswami, K.; Rajasekhar, L.; Kutala, V.K. Application of Various Statistical Models to Explore Gene-Gene Interactions in Folate, Xenobiotic, Toll-Like Receptor and STAT4 Pathways that Modulate Susceptibility to Systemic Lupus Erythematosus. Mol. Diagn. Ther. 2016, 20, 83–95. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, L.; Luo, C.; Tang, W.; Dai, R.; An, Y.; Tang, X. Clinical characteristics of early-onset paediatric systemic lupus erythematosus in a single centre in China. Rheumatology 2023, 62, 3373–3381. [Google Scholar] [CrossRef]
- Oldenburg, M.; Kruger, A.; Ferstl, R.; Kaufmann, A.; Nees, G.; Sigmund, A.; Bathke, B.; Lauterbach, H.; Suter, M.; Dreher, S.; et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 2012, 337, 1111–1115. [Google Scholar] [CrossRef]
- Li, X.D.; Chen, Z.J. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. Elife 2012, 1, e00102. [Google Scholar] [CrossRef]
- Lee, S.M.; Yip, T.F.; Yan, S.; Jin, D.Y.; Wei, H.L.; Guo, R.T.; Peiris, J.S.M. Recognition of Double-Stranded RNA and Regulation of Interferon Pathway by Toll-Like Receptor 10. Front. Immunol. 2018, 9, 516. [Google Scholar] [CrossRef]
- Rodrigues, C.R.; Balachandran, Y.; Aulakh, G.K.; Singh, B. TLR10: An intriguing Toll-like receptor with many unanswered questions. J. Innate Immun. 2024, 16, 96–104. [Google Scholar] [CrossRef]
- Caielli, S.; Wan, Z.; Pascual, V. Systemic Lupus Erythematosus Pathogenesis: Interferon and Beyond. Annu. Rev. Immunol. 2023, 41, 533–560. [Google Scholar] [CrossRef]
- Scherlinger, M.; Guillotin, V.; Truchetet, M.E.; Contin-Bordes, C.; Sisirak, V.; Duffau, P.; Lazaro, E.; Richez, C.; Blanco, P. Systemic lupus erythematosus and systemic sclerosis: All roads lead to platelets. Autoimmun. Rev. 2018, 17, 625–635. [Google Scholar] [CrossRef]
- Robert, M.; Scherlinger, M. Platelets are a major player and represent a therapeutic opportunity in systemic lupus erythematosus. Jt. Bone Spine 2024, 91, 105622. [Google Scholar] [CrossRef]
- Linge, P.; Fortin, P.R.; Lood, C.; Bengtsson, A.A.; Boilard, E. The non-haemostatic role of platelets in systemic lupus erythematosus. Nat. Rev. Rheumatol. 2018, 14, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Scherlinger, M.; Richez, C.; Tsokos, G.C.; Boilard, E.; Blanco, P. The role of platelets in immune-mediated inflammatory diseases. Nat. Rev. Immunol. 2023, 23, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Baroni Pietto, M.C.; Glembotsky, A.C.; Lev, P.R.; Marin Oyarzun, C.R.; De Luca, G.; Gomez, G.; Collado, M.V.; Charo, N.; Cellucci, A.S.; Heller, P.G.; et al. Toll-like receptor expression and functional behavior in platelets from patients with systemic lupus erythematosus. Immunobiology 2024, 229, 152782. [Google Scholar] [CrossRef]
- Hornung, V.; Rothenfusser, S.; Britsch, S.; Krug, A.; Jahrsdöfer, B.; Giese, T.; Endres, S.; Hartmann, G. Quantitative Expression of Toll-Like Receptor 1–10 mRNA in Cellular Subsets of Human Peripheral Blood Mononuclear Cells and Sensitivity to CpG Oligodeoxynucleotides1. J. Immunol. 2002, 168, 4531–4537. [Google Scholar] [CrossRef]
- Demaria, O.; Pagni, P.P.; Traub, S.; de Gassart, A.; Branzk, N.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Flavell, R.A.; Alexopoulou, L. TLR8 deficiency leads to autoimmunity in mice. J. Clin. Investig. 2010, 120, 3651–3662. [Google Scholar] [CrossRef] [PubMed]
- Tilstra, J.S.; John, S.; Gordon, R.A.; Leibler, C.; Kashgarian, M.; Bastacky, S.; Nickerson, K.M.; Shlomchik, M.J. B cell-intrinsic TLR9 expression is protective in murine lupus. J. Clin. Investig. 2020, 130, 3172–3187. [Google Scholar] [CrossRef] [PubMed]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-Specific Recognition of Single-Stranded RNA via Toll-like Receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.L. Signaling in B cells via Toll-like receptors. Curr. Opin. Immunol. 2005, 17, 230–236. [Google Scholar] [CrossRef]
- Wen, L.; Zhang, B.; Wu, X.; Liu, R.; Fan, H.; Han, L.; Zhang, Z.; Ma, X.; Chu, C.-Q.; Shi, X. Toll-like receptors 7 and 9 regulate the proliferation and differentiation of B cells in systemic lupus erythematosus. Front. Immunol. 2023, 14, 1093208. [Google Scholar] [CrossRef]
- Rubin, S.J.S.; Bloom, M.S.; Robinson, W.H. B cell checkpoints in autoimmune rheumatic diseases. Nat. Rev. Rheumatol. 2019, 15, 303–315. [Google Scholar] [CrossRef]
- Leadbetter, E.A.; Rifkin, I.R.; Hohlbaum, A.M.; Beaudette, B.C.; Shlomchik, M.J.; Marshak-Rothstein, A. Chromatin–IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 2002, 416, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Viglianti, G.A.; Lau, C.M.; Hanley, T.M.; Miko, B.A.; Shlomchik, M.J.; Marshak-Rothstein, A. Activation of Autoreactive B Cells by CpG dsDNA. Immunity 2003, 19, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.R.; Shupe, J.; Nickerson, K.; Kashgarian, M.; Flavell, R.A.; Shlomchik, M.J. Toll-like Receptor 7 and TLR9 Dictate Autoantibody Specificity and Have Opposing Inflammatory and Regulatory Roles in a Murine Model of Lupus. Immunity 2006, 25, 417–428. [Google Scholar] [CrossRef]
- Lau, C.M.; Broughton, C.; Tabor, A.S.; Akira, S.; Flavell, R.A.; Mamula, M.J.; Christensen, S.R.; Shlomchik, M.J.; Viglianti, G.A.; Rifkin, I.R.; et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J. Exp. Med. 2005, 202, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Gerth, A.J.; Peng, S.L. CpG DNA redirects class-switching towards ‘Th1-like’ Ig isotype production via TLR9 and MyD88. Eur. J. Immunol. 2004, 34, 1483–1487. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Qiao, X.; Cerutti, A. CpG DNA Induces IgG Class Switch DNA Recombination by Activating Human B Cells through an Innate Pathway That Requires TLR9 and Cooperates with IL-101. J. Immunol. 2004, 173, 4479–4491. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, M.; Fukuyama, H.; McGaha, T.L.; Aderem, A.; Ravetch, J.V. TLR9/MyD88 signaling is required for class switching to pathogenic IgG2a and 2b autoantibodies in SLE. J. Exp. Med. 2006, 203, 553–561. [Google Scholar] [CrossRef]
- Eckl-Dorna, J.; Batista, F.D. BCR-mediated uptake of antigen linked to TLR9 ligand stimulates B-cell proliferation and antigen-specific plasma cell formation. Blood 2009, 113, 3969–3977. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C.E.; Zhou, Y.; Chupp, D.P.; Yan, H.; Fisher, A.D.; Simon, R.; Zan, H.; Xu, Z.; Casali, P. Intrinsic B cell TLR-BCR linked coengagement induces class-switched, hypermutated, neutralizing antibody responses in absence of T cells. Sci. Adv. 2023, 9, eade8928. [Google Scholar] [CrossRef]
- Christensen, S.R.; Kashgarian, M.; Alexopoulou, L.; Flavell, R.A.; Akira, S.; Shlomchik, M.J. Toll-like receptor 9 controls anti-DNA autoantibody production in murine lupus. J. Exp. Med. 2005, 202, 321–331. [Google Scholar] [CrossRef]
- Rahman, A.H.; Eisenberg, R.A. The role of toll-like receptors in systemic lupus erythematosus. Springer Semin. Immunopathol. 2006, 28, 131–143. [Google Scholar] [CrossRef]
- Yi, A.-K.; Chang, M.; Peckham, D.W.; Krieg, A.M.; Ashman, R.F. CpG Oligodeoxyribonucleotides Rescue Mature Spleen B Cells from Spontaneous Apoptosis and Promote Cell Cycle Entry1. J. Immunol. 1998, 160, 5898–5906. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-S.; Chung, S.-T.; Robertson, D.A.; Chelvarajan, R.L.; Bondada, S. CpG oligodeoxynucleotides rescue BKS-2 immature B cell lymphoma from anti-IgM-mediated growth inhibition by up-regulation of egr-1. Int. Immunol. 1999, 11, 871–879. [Google Scholar] [CrossRef][Green Version]
- Nickerson, K.M.; Christensen, S.R.; Shupe, J.; Kashgarian, M.; Kim, D.; Elkon, K.; Shlomchik, M.J. TLR9 Regulates TLR7- and MyD88-Dependent Autoantibody Production and Disease in a Murine Model of Lupus. J. Immunol. 2010, 184, 1840–1848. [Google Scholar] [CrossRef] [PubMed]
- Teichmann, L.L.; Schenten, D.; Medzhitov, R.; Kashgarian, M.; Shlomchik, M.J. Signals via the adaptor MyD88 in B cells and DCs make distinct and synergistic contributions to immune activation and tissue damage in lupus. Immunity 2013, 38, 528–540. [Google Scholar] [CrossRef]
- Tilstra, J.S.; Kim, M.; Gordon, R.A.; Leibler, C.; Cosgrove, H.A.; Bastacky, S.; Nickerson, K.M.; Shlomchik, M.J. B cell–intrinsic Myd88 regulates disease progression in murine lupus. J. Exp. Med. 2023, 220, e20230263. [Google Scholar] [CrossRef]
- Hwang, S.-H.; Lee, H.; Yamamoto, M.; Jones, L.A.; Dayalan, J.; Hopkins, R.; Zhou, X.J.; Yarovinsky, F.; Connolly, J.E.; Curotto de Lafaille, M.A.; et al. B Cell TLR7 Expression Drives Anti-RNA Autoantibody Production and Exacerbates Disease in Systemic Lupus Erythematosus–Prone Mice. J. Immunol. 2012, 189, 5786–5796. [Google Scholar] [CrossRef]
- Satterthwaite, A.B. TLR7 Signaling in Lupus B Cells: New Insights into Synergizing Factors and Downstream Signals. Curr. Rheumatol. Rep. 2021, 23, 80. [Google Scholar] [CrossRef] [PubMed]
- Desnues, B.; Macedo, A.B.; Roussel-Queval, A.; Bonnardel, J.; Henri, S.; Demaria, O.; Alexopoulou, L. TLR8 on dendritic cells and TLR9 on B cells restrain TLR7-mediated spontaneous autoimmunity in C57BL/6 mice. Proc. Natl. Acad. Sci. USA 2014, 111, 1497–1502. [Google Scholar] [CrossRef]
- Scofield, R.H.; Bruner, G.R.; Namjou, B.; Kimberly, R.P.; Ramsey-Goldman, R.; Petri, M.; Reveille, J.D.; Alarcón, G.S.; Vilá, L.M.; Reid, J.; et al. Klinefelter’s syndrome (47,XXY) in male systemic lupus erythematosus patients: Support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008, 58, 2511–2517. [Google Scholar] [CrossRef]
- Souyris, M.; Cenac, C.; Azar, P.; Daviaud, D.; Canivet, A.; Grunenwald, S.; Pienkowski, C.; Chaumeil, J.; Mejía, J.E.; Guéry, J.-C. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 2018, 3, eaap8855. [Google Scholar] [CrossRef] [PubMed]
- Youness, A.; Cenac, C.; Faz-López, B.; Grunenwald, S.; Barrat, F.J.; Chaumeil, J.; Mejía, J.E.; Guéry, J.-C. TLR8 escapes X chromosome inactivation in human monocytes and CD4+ T cells. Biol. Sex. Differ. 2023, 14, 60. [Google Scholar] [CrossRef]
- Wang, T.; Marken, J.; Chen, J.; Tran, V.B.; Li, Q.-Z.; Li, M.; Cerosaletti, K.; Elkon, K.B.; Zeng, X.; Giltiay, N.V. High TLR7 Expression Drives the Expansion of CD19+CD24hiCD38hi Transitional B Cells and Autoantibody Production in SLE Patients. Front. Immunol. 2019, 10, 01243. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhao, J.; Sakurai, D.; Kaufman, K.M.; Edberg, J.C.; Kimberly, R.P.; Kamen, D.L.; Gilkeson, G.S.; Jacob, C.O.; Scofield, R.H.; et al. MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus. PLOS Genet. 2013, 9, e1003336. [Google Scholar] [CrossRef]
- Shen, N.; Fu, Q.; Deng, Y.; Qian, X.; Zhao, J.; Kaufman, K.M.; Wu, Y.L.; Yu, C.Y.; Tang, Y.; Chen, J.-Y.; et al. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 2010, 107, 15838–15843. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-M.; Chang, S.-W.; Wu, Y.-J.J.; Lin, J.-C.; Ho, H.-H.; Chou, T.-C.; Yang, B.; Wu, J.; Chen, J.-Y. Genetic variations in Toll-like receptors (TLRs 3/7/8) are associated with systemic lupus erythematosus in a Taiwanese population. Sci. Rep. 2014, 4, 3792. [Google Scholar] [CrossRef]
- Brown, G.J.; Cañete, P.F.; Wang, H.; Medhavy, A.; Bones, J.; Roco, J.A.; He, Y.; Qin, Y.; Cappello, J.; Ellyard, J.I.; et al. TLR7 gain-of-function genetic variation causes human lupus. Nature 2022, 605, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, K.M.; Wang, Y.; Bastacky, S.; Shlomchik, M.J. Toll-like receptor 9 suppresses lupus disease in Fas-sufficient MRL Mice. PLoS ONE 2017, 12, e0173471. [Google Scholar] [CrossRef] [PubMed]
- Lartigue, A.; Courville, P.; Auquit, I.; François, A.; Arnoult, C.; Tron, F.; Gilbert, D.; Musette, P. Role of TLR9 in Anti-Nucleosome and Anti-DNA Antibody Production in lpr Mutation-Induced Murine Lupus1. J. Immunol. 2006, 177, 1349–1354. [Google Scholar] [CrossRef]
- Santiago-Raber, M.L.; Dunand-Sauthier, I.; Wu, T.; Li, Q.Z.; Uematsu, S.; Akira, S.; Reith, W.; Mohan, C.; Kotzin, B.L.; Izui, S. Critical role of TLR7 in the acceleration of systemic lupus erythematosus in TLR9-deficient mice. J. Autoimmun. 2010, 34, 339–348. [Google Scholar] [CrossRef]
- Stoehr, A.D.; Schoen, C.T.; Mertes, M.M.M.; Eiglmeier, S.; Holecska, V.; Lorenz, A.K.; Schommartz, T.; Schoen, A.-L.; Hess, C.; Winkler, A.; et al. TLR9 in Peritoneal B-1b Cells Is Essential for Production of Protective Self-Reactive IgM To Control Th17 Cells and Severe Autoimmunity. J. Immunol. 2011, 187, 2953–2965. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Wellmann, U.; Kunder, S.; Quintanilla-Martinez, L.; Jennen, L.; Dear, N.; Amann, K.; Bauer, S.; Winkler, T.H.; Wagner, H. Toll-like receptor 9-independent aggravation of glomerulonephritis in a novel model of SLE. Int. Immunol. 2006, 18, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, H.A.; Gingras, S.; Kim, M.; Bastacky, S.; Tilstra, J.S.; Shlomchik, M.J. B cell-intrinsic TLR7 expression drives severe lupus in TLR9-deficient mice. JCI Insight 2023, 8, e172219. [Google Scholar] [CrossRef] [PubMed]
- Celhar, T.; Yasuga, H.; Lee, H.Y.; Zharkova, O.; Tripathi, S.; Thornhill, S.I.; Lu, H.K.; Au, B.; Lim, L.H.K.; Thamboo, T.P.; et al. Toll-Like Receptor 9 Deficiency Breaks Tolerance to RNA-Associated Antigens and Up-Regulates Toll-Like Receptor 7 Protein in Sle1 Mice. Arthritis Rheumatol. 2018, 70, 1597–1609. [Google Scholar] [CrossRef] [PubMed]
- Fukui, R.; Saitoh, S.-I.; Kanno, A.; Onji, M.; Shibata, T.; Ito, A.; Onji, M.; Matsumoto, M.; Akira, S.; Yoshida, N.; et al. Unc93B1 Restricts Systemic Lethal Inflammation by Orchestrating Toll-like Receptor 7 and 9 Trafficking. Immunity 2011, 35, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.W.; Shibata, T.; Hwang, M.; Kwon, E.H.; Jang, M.S.; Fukui, R.; Kanno, A.; Jung, D.J.; Jang, M.H.; Miyake, K.; et al. UNC93B1 is essential for the plasma membrane localization and signaling of Toll-like receptor 5. Proc. Natl. Acad. Sci. USA 2014, 111, 7072–7077. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Brinkmann, M.M.; Paquet, M.-E.; Ploegh, H.L. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 2008, 452, 234–238. [Google Scholar] [CrossRef]
- Fukui, R.; Saitoh, S.; Matsumoto, F.; Kozuka-Hata, H.; Oyama, M.; Tabeta, K.; Beutler, B.; Miyake, K. Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing. J. Exp. Med. 2009, 206, 1339–1350. [Google Scholar] [CrossRef] [PubMed]
- Leibler, C.; John, S.; Elsner, R.A.; Thomas, K.B.; Smita, S.; Joachim, S.; Levack, R.C.; Callahan, D.J.; Gordon, R.A.; Bastacky, S.; et al. Genetic dissection of TLR9 reveals complex regulatory and cryptic proinflammatory roles in mouse lupus. Nat. Immunol. 2022, 23, 1457–1469. [Google Scholar] [CrossRef]
- Hanna Kazazian, N.; Wang, Y.; Roussel-Queval, A.; Marcadet, L.; Chasson, L.; Laprie, C.; Desnues, B.; Charaix, J.; Irla, M.; Alexopoulou, L. Lupus Autoimmunity and Metabolic Parameters Are Exacerbated Upon High Fat Diet-Induced Obesity Due to TLR7 Signaling. Front. Immunol. 2019, 10, 02015. [Google Scholar] [CrossRef]
- Fejtkova, M.; Sukova, M.; Hlozkova, K.; Skvarova Kramarzova, K.; Rackova, M.; Jakubec, D.; Bakardjieva, M.; Bloomfield, M.; Klocperk, A.; Parackova, Z.; et al. TLR8/TLR7 dysregulation due to a novel TLR8 mutation causes severe autoimmune hemolytic anemia and autoinflammation in identical twins. Am. J. Hematol. 2022, 97, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Lind, N.A.; Rael, V.E.; Pestal, K.; Liu, B.; Barton, G.M. Regulation of the nucleic acid-sensing Toll-like receptors. Nat. Rev. Immunol. 2022, 22, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-S.; Park, S.; Huh, J.-W.; Lee, Y.-R.; Jung, D.-J.; Yang, C.; Kim, S.H.; Kim, H.M.; Kim, Y.-M. N-glycosylation of UNC93B1 at a Specific Asparagine Residue Is Required for TLR9 Signaling. Front. Immunol. 2022, 13, 875083. [Google Scholar] [CrossRef] [PubMed]
- Majer, O.; Liu, B.; Woo, B.J.; Kreuk, L.S.M.; Van Dis, E.; Barton, G.M. Release from UNC93B1 reinforces the compartmentalized activation of select TLRs. Nature 2019, 575, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Wang, Y.; Yao, K.; Wang, L.; Huang, J.; Xiao, Y.; Chen, H.; Liu, B.; Yang, C.Y.; Zhao, J. Cyclical palmitoylation regulates TLR9 signalling and systemic autoimmunity in mice. Nat. Commun. 2024, 15, 1. [Google Scholar] [CrossRef]
- Majer, O.; Liu, B.; Kreuk, L.S.M.; Krogan, N.; Barton, G.M. UNC93B1 recruits syntenin-1 to dampen TLR7 signalling and prevent autoimmunity. Nature 2019, 575, 366–370. [Google Scholar] [CrossRef]
- Mishra, H.; Schlack-Leigers, C.; Lim, E.L.; Thieck, O.; Magg, T.; Raedler, J.; Wolf, C.; Klein, C.; Ewers, H.; Lee-Kirsch, M.A.; et al. Disrupted degradative sorting of TLR7 is associated with human lupus. Sci. Immunol. 2024, 9, eadi9575. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.; Lim, E.L.; Mokhtari, M.; Kind, B.; Odainic, A.; Lara-Villacanas, E.; Koss, S.; Mages, S.; Menzel, K.; Engel, K.; et al. UNC93B1 variants underlie TLR7-dependent autoimmunity. Sci. Immunol. 2024, 9, eadi9769. [Google Scholar] [CrossRef]
- Sato, R.; Shibata, T.; Tanaka, Y.; Kato, C.; Yamaguchi, K.; Furukawa, Y.; Shimizu, E.; Yamaguchi, R.; Imoto, S.; Miyano, S.; et al. Requirement of glycosylation machinery in TLR responses revealed by CRISPR/Cas9 screening. Int. Immunol. 2017, 29, 347–355. [Google Scholar] [CrossRef]
- Sun, J.; Duffy, K.E.; Ranjith-Kumar, C.T.; Xiong, J.; Lamb, R.J.; Santos, J.; Masarapu, H.; Cunningham, M.; Holzenburg, A.; Sarisky, R.T.; et al. Structural and Functional Analyses of the Human Toll-like Receptor 3: ROLE OF GLYCOSYLATION*. J. Biol. Chem. 2006, 281, 11144–11151. [Google Scholar] [CrossRef]
- Amith, S.R.; Jayanth, P.; Franchuk, S.; Siddiqui, S.; Seyrantepe, V.; Gee, K.; Basta, S.; Beyaert, R.; Pshezhetsky, A.V.; Szewczuk, M.R. Dependence of pathogen molecule-induced Toll-like receptor activation and cell function on Neu1 sialidase. Glycoconj. J. 2009, 26, 1197–1212. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Martínez, I.; Ramos-Martínez, E.; Cerbón, M.; Pérez-Torres, A.; Pérez-Campos Mayoral, L.; Hernández-Huerta, M.T.; Martínez-Cruz, M.; Pérez-Santiago, A.D.; Sánchez-Medina, M.A.; García-Montalvo, I.A.; et al. The Role of B Cell and T Cell Glycosylation in Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2023, 24, 863. [Google Scholar] [CrossRef] [PubMed]
- Cancro, M.P. Age-Associated B Cells. Annu. Rev. Immunol. 2020, 38, 315–340. [Google Scholar] [CrossRef] [PubMed]
- Mouat, I.C.; Goldberg, E.; Horwitz, M.S. Age-associated B cells in autoimmune diseases. Cell. Mol. Life Sci. 2022, 79, 402. [Google Scholar] [CrossRef]
- Jenks, S.A.; Cashman, K.S.; Zumaquero, E.; Marigorta, U.M.; Patel, A.V.; Wang, X.; Tomar, D.; Woodruff, M.C.; Simon, Z.; Bugrovsky, R.; et al. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 2018, 49, 725–739.e726. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, J.; Kumar, V.; Karnell, J.L.; Naiman, B.; Gross, P.S.; Rahman, S.; Zerrouki, K.; Hanna, R.; Morehouse, C.; et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat. Commun. 2018, 9, 1758. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, A.V.; Rubtsova, K.; Fischer, A.; Meehan, R.T.; Gillis, J.Z.; Kappler, J.W.; Marrack, P. Toll-like receptor 7 (TLR7)–driven accumulation of a novel CD11c+ B-cell population is important for the development of autoimmunity. Blood 2011, 118, 1305–1315. [Google Scholar] [CrossRef]
- Aranburu, A.; Höök, N.; Gerasimcik, N.; Corleis, B.; Ren, W.; Camponeschi, A.; Carlsten, H.; Grimsholm, O.; Mårtensson, I.-L. Age-associated B cells expanded in autoimmune mice are memory cells sharing H-CDR3-selected repertoires. Eur. J. Immunol. 2018, 48, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Gu, S.; Han, X.; Ding, H.; Jiang, Y.; Zhang, X.; Yao, C.; Hong, S.; Zhang, J.; Shen, Y.; et al. The transcription factor ZEB2 drives the formation of age-associated B cells. Science 2024, 383, 413–421. [Google Scholar] [CrossRef]
- Naradikian, M.S.; Myles, A.; Beiting, D.P.; Roberts, K.J.; Dawson, L.; Herati, R.S.; Bengsch, B.; Linderman, S.L.; Stelekati, E.; Spolski, R.; et al. Cutting Edge: IL-4, IL-21, and IFN-γ Interact To Govern T-bet and CD11c Expression in TLR-Activated B Cells. J. Immunol. 2016, 197, 1023–1028. [Google Scholar] [CrossRef]
- Nickerson, K.M.; Smita, S.; Hoehn, K.B.; Marinov, A.D.; Thomas, K.B.; Kos, J.T.; Yang, Y.; Bastacky, S.I.; Watson, C.T.; Kleinstein, S.H.; et al. Age-associated B cells are heterogeneous and dynamic drivers of autoimmunity in mice. J. Exp. Med. 2023, 220, e20221346. [Google Scholar] [CrossRef]
- Peng, S.L.; Szabo, S.J.; Glimcher, L.H. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl. Acad. Sci. USA 2002, 99, 5545–5550. [Google Scholar] [CrossRef] [PubMed]
- Woldemeskel, B.A.; Dykema, A.G.; Garliss, C.C.; Cherfils, S.; Smith, K.N.; Blankson, J.N. CD4+ T cells from COVID-19 mRNA vaccine recipients recognize a conserved epitope present in diverse coronaviruses. J. Clin. Investig. 2022, 132, e156083. [Google Scholar] [CrossRef] [PubMed]
- Yasaka, K.; Yamazaki, T.; Sato, H.; Shirai, T.; Cho, M.; Ishida, K.; Ito, K.; Tanaka, T.; Ogasawara, K.; Harigae, H.; et al. Phospholipase D4 as a signature of toll-like receptor 7 or 9 signaling is expressed on blastic T-bet + B cells in systemic lupus erythematosus. Arthritis Res. Ther. 2023, 25, 200. [Google Scholar] [CrossRef] [PubMed]
- Kalliolias, G.D.; Basdra, E.K.; Papavassiliou, A.G. Targeting TLR Signaling Cascades in Systemic Lupus Erythematosus and Rheumatoid Arthritis: An Update. Biomedicines 2024, 12, 138. [Google Scholar] [CrossRef] [PubMed]
- Patinote, C.; Karroum, N.B.; Moarbess, G.; Cirnat, N.; Kassab, I.; Bonnet, P.A.; Deleuze-Masquéfa, C. Agonist and antagonist ligands of toll-like receptors 7 and 8: Ingenious tools for therapeutic purposes. Eur. J. Med. Chem. 2020, 193, 112238. [Google Scholar] [CrossRef]
- Cenac, C.; Ducatez, M.F.; Guéry, J.-C. Hydroxychloroquine inhibits proteolytic processing of endogenous TLR7 protein in human primary plasmacytoid dendritic cells. Eur. J. Immunol. 2022, 52, 54–61. [Google Scholar] [CrossRef]
- Ewald, S.E.; Lee, B.L.; Lau, L.; Wickliffe, K.E.; Shi, G.-P.; Chapman, H.A.; Barton, G.M. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 2008, 456, 658–662. [Google Scholar] [CrossRef]
- Schrezenmeier, E.; Dörner, T. Mechanisms of action of hydroxychloroquine and chloroquine: Implications for rheumatology. Nat. Rev. Rheumatol. 2020, 16, 155–166. [Google Scholar] [CrossRef]
- Guiducci, C.; Gong, M.; Xu, Z.; Gill, M.; Chaussabel, D.; Meeker, T.; Chan, J.H.; Wright, T.; Punaro, M.; Bolland, S.; et al. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature 2010, 465, 937–941. [Google Scholar] [CrossRef]
- Deshmukh, A.; Pereira, A.; Geraci, N.; Tzvetkov, E.; Przetak, M.; Catalina, M.D.; Morand, E.F.; Bender, A.T.; Vaidyanathan, B. Preclinical Evidence for the Glucocorticoid-Sparing Potential of a Dual Toll-Like Receptor 7/8 Inhibitor in Autoimmune Diseases. J. Pharmacol. Exp. Ther. 2024, 388, 751–764. [Google Scholar] [CrossRef]
- Shisha, T.; Posch, M.G.; Lehmann, J.; Feifel, R.; Junt, T.; Hawtin, S.; Schuemann, J.; Avrameas, A.; Danekula, R.; Misiolek, P.; et al. First-in-Human Study of the Safety, Pharmacokinetics, and Pharmacodynamics of MHV370, a Dual Inhibitor of Toll-Like Receptors 7 and 8, in Healthy Adults. Eur. J. Drug Metab. Pharmacokinet. 2023, 48, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Hawtin, S.; André, C.; Collignon-Zipfel, G.; Appenzeller, S.; Bannert, B.; Baumgartner, L.; Beck, D.; Betschart, C.; Boulay, T.; Brunner, H.I.; et al. Preclinical characterization of the Toll-like receptor 7/8 antagonist MHV370 for lupus therapy. Cell Rep. Med. 2023, 4, 101036. [Google Scholar] [CrossRef] [PubMed]
- Tojo, S.; Zhang, Z.; Matsui, H.; Tahara, M.; Ikeguchi, M.; Kochi, M.; Kamada, M.; Shigematsu, H.; Tsutsumi, A.; Adachi, N.; et al. Structural analysis reveals TLR7 dynamics underlying antagonism. Nat. Commun. 2020, 11, 5204. [Google Scholar] [CrossRef]
- Ishizaka, S.T.; Hawkins, L.; Chen, Q.; Tago, F.; Yagi, T.; Sakaniwa, K.; Zhang, Z.; Shimizu, T.; Shirato, M. A novel Toll-like receptor 7/8–specific antagonist E6742 ameliorates clinically relevant disease parameters in murine models of lupus. Eur. J. Pharmacol. 2023, 957, 175962. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Fukui, R.; Tanaka, R.; Motoi, Y.; Kanno, A.; Sato, R.; Yamaguchi, K.; Amano, H.; Furukawa, Y.; Suzuki, H.; et al. Anti-TLR7 Antibody Protects Against Lupus Nephritis in NZBWF1 Mice by Targeting B Cells and Patrolling Monocytes. Front. Immunol. 2021, 12, 777197. [Google Scholar] [CrossRef]
- Achek, A.; Kwon, H.K.; Patra, M.C.; Shah, M.; Hong, R.; Lee, W.H.; Baek, W.Y.; Choi, Y.S.; Kim, G.Y.; Pham, T.L.H.; et al. A peptide derived from the core beta-sheet region of TIRAP decoys TLR4 and reduces inflammatory and autoimmune symptoms in murine models. EBioMedicine 2020, 52, 102645. [Google Scholar] [CrossRef] [PubMed]
- Moroni, G.; Frontini, G.; Ponticelli, C. When and How Is It Possible to Stop Therapy in Patients with Lupus Nephritis: A Narrative Review. Clin. J. Am. Soc. Nephrol. 2021, 16, 1909–1917. [Google Scholar] [CrossRef] [PubMed]
- Le Naour, J.; Kroemer, G. Trial watch: Toll-like receptor ligands in cancer therapy. OncoImmunology 2023, 12, 2180237. [Google Scholar] [CrossRef]
- Srinivasa, A.; Tosounidou, S.; Gordon, C. Increased Incidence of Gastrointestinal Side Effects in Patients Taking Hydroxychloroquine: A Brand-related Issue? J. Rheumatol. 2017, 44, 398. [Google Scholar] [CrossRef]
- Shang, L.; Wang, L.; Shi, X.; Wang, N.; Zhao, L.; Wang, J.; Liu, C. HMGB1 was negatively regulated by HSF1 and mediated the TLR4/MyD88/NF-kappaB signal pathway in asthma. Life Sci. 2020, 241, 117120. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Suarez, J.S.; Minaai, M.; Li, S.; Gaudino, G.; Pass, H.I.; Carbone, M.; Yang, H. HMGB1 as a therapeutic target in disease. J. Cell Physiol. 2021, 236, 3406–3419. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
von Hofsten, S.; Fenton, K.A.; Pedersen, H.L. Human and Murine Toll-like Receptor-Driven Disease in Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2024, 25, 5351. https://doi.org/10.3390/ijms25105351
von Hofsten S, Fenton KA, Pedersen HL. Human and Murine Toll-like Receptor-Driven Disease in Systemic Lupus Erythematosus. International Journal of Molecular Sciences. 2024; 25(10):5351. https://doi.org/10.3390/ijms25105351
Chicago/Turabian Stylevon Hofsten, Susannah, Kristin Andreassen Fenton, and Hege Lynum Pedersen. 2024. "Human and Murine Toll-like Receptor-Driven Disease in Systemic Lupus Erythematosus" International Journal of Molecular Sciences 25, no. 10: 5351. https://doi.org/10.3390/ijms25105351
APA Stylevon Hofsten, S., Fenton, K. A., & Pedersen, H. L. (2024). Human and Murine Toll-like Receptor-Driven Disease in Systemic Lupus Erythematosus. International Journal of Molecular Sciences, 25(10), 5351. https://doi.org/10.3390/ijms25105351






