Interplay between the Chaperone System and Gut Microbiota Dysbiosis in Systemic Lupus Erythematosus Pathogenesis: Is Molecular Mimicry the Missing Link between Those Two Factors?
Abstract
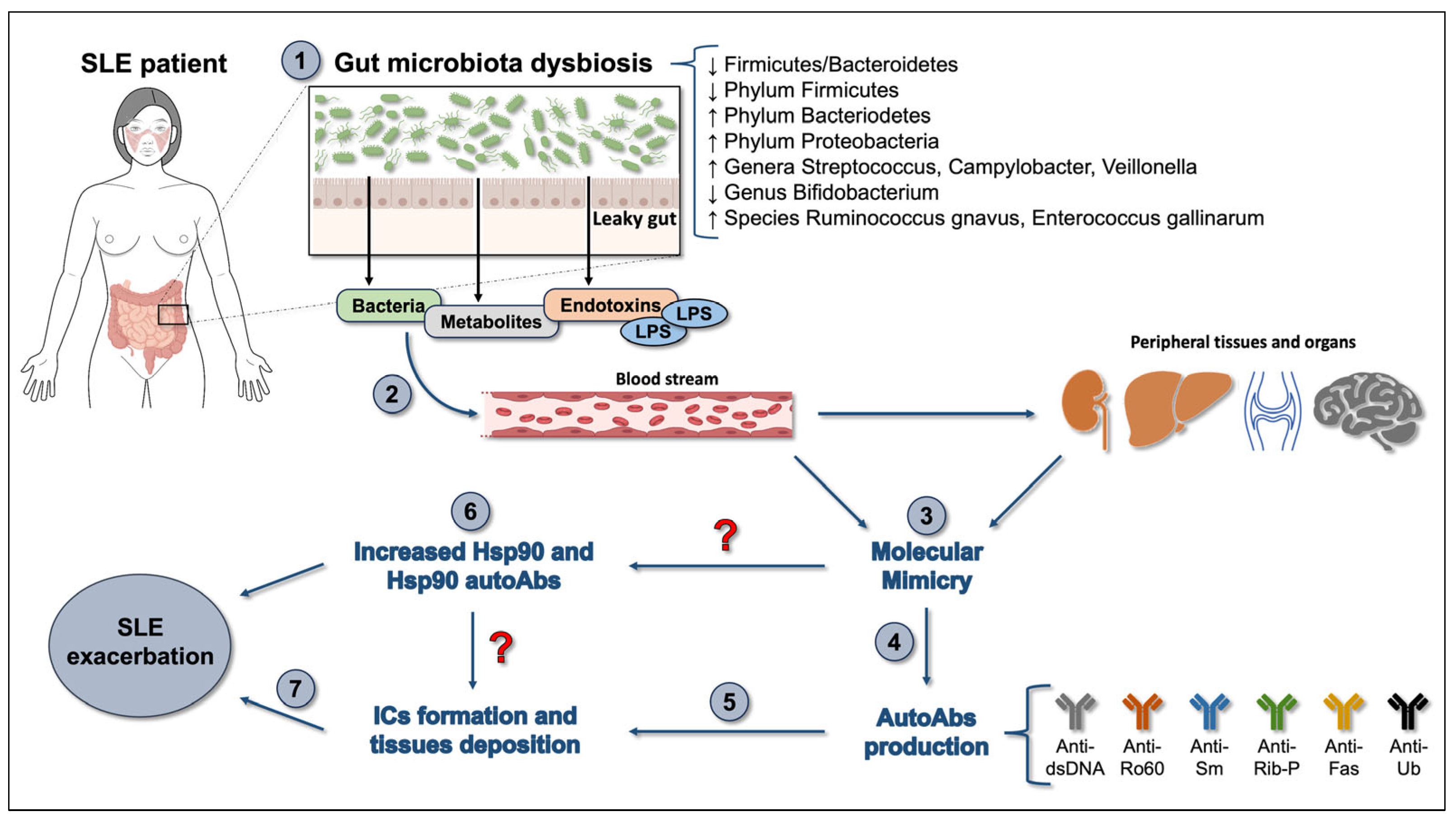
:1. Introduction
2. The Chaperone System and SLE
3. The Gut Microbiota in SLE
4. Molecular Mimicry, Hsps, and Gut Microbiota Dysbiosis in SLE
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Macario, A.J.L.; Conway de Macario, E. Chaperone proteins and chaperonopathies. In Stress: Physiology, Biochemistry, and Pathology. Handbook of Stress Series; Fink, G., Ed.; Elsevier/Academic Press: Cambridge, MA, USA, 2019; Volume 3, Chapter 12; pp. 135–152. [Google Scholar]
- Carlisle, C.; Prill, K.; Pilgrim, D. Chaperones and the proteasome system: Regulating the construction and demolition of striated muscle. Int. J. Mol. Sci. 2017, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk between mammalian autophagy and the ubiquitin-proteasome system. Front. Cell. Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Margulis, B.; Tsimokha, A.; Zubova, S.; Guzhova, I. Molecular chaperones and proteolytic machineries regulate protein homeostasis in aging cells. Cells 2020, 9, 1308. [Google Scholar] [CrossRef] [PubMed]
- Macario, A.J.L.; Conway de Macario, E.; Cappello, F. The Chaperonopathies. Diseases with Defective Molecular Chaperones; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Willison, K.R. The structure and evolution of eukaryotic chaperonin-containing TCP-1 and its mechanism that folds actin into a protein spring. Biochem. J. 2018, 475, 3009–3034. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, V.; Buchner, J. Functional principles and regulation of molecular chaperones. Adv. Protein Chem. Struct. Biol. 2019, 114, 1–60. [Google Scholar] [PubMed]
- Gestaut, D.; Roh, S.H.; Ma, B.; Pintilie, G.; Joachimiak, L.A.; Leitner, A.; Walzthoeni, T.; Aebersold, R.; Chiu, W.; Frydman, J. The chaperonin TRiC/CCT associates with prefoldin through a conserved electrostatic interface essential for cellular proteostasis. Cell 2019, 177, 751–765.e15. [Google Scholar] [CrossRef] [PubMed]
- Havalová, H.; Ondrovičová, G.; Keresztesová, B.; Bauer, J.A.; Pevala, V.; Kutejová, E.; Kunová, N. Mitochondrial HSP70 chaperone system-the influence of post-translational modifications and involvement in human diseases. Int. J. Mol. Sci. 2021, 22, 8077. [Google Scholar] [CrossRef] [PubMed]
- Macario, A.J.L.; Conway de Macario, E. Sick chaperones, cellular stress, and disease. N. Engl. J. Med. 2005, 353, 1489–1501. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Vitale, A.M.; Fucarino, A.; Scalia, F.; Vergilio, G.; Conway de Macario, E.; Macario, A.J.L.; Caruso Bavisotto, C.; Pitruzzella, A. The challenging riddle about the janus-type role of Hsp60 and related extracellular vesicles and miRNAs in carcinogenesis and the promises of its solution. Appl. Sci. 2021, 11, 1175. [Google Scholar] [CrossRef]
- Paladino, L.; Vitale, A.M.; Santonocito, R.; Pitruzzella, A.; Cipolla, C.; Graceffa, G.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; Rappa, F. Molecular Chaperones and Thyroid Cancer. Int. J. Mol. Sci. 2021, 22, 4196. [Google Scholar] [CrossRef]
- Gaston, J.S. Are heat shock proteins involved in autoimmunity? Int. J. Clin. Lab. Res. 1992, 22, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H. Heat shock proteins and autoimmunity: A critical appraisal. Int. Arch. Allergy Immunol. 1994, 103, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Rajaiah, R.; Moudgil, K.D. Heat-shock proteins can promote as well as regulate autoimmunity. Autoimmun. Rev. 2009, 8, 388–393. [Google Scholar] [CrossRef] [PubMed]
- van Eden, W.; Jansen, M.A.A.; Ludwig, I.; van Kooten, P.; van der Zee, R.; Broere, F. The enigma of heat shock proteins in immune tolerance. Front. Immunol. 2017, 8, 1599. [Google Scholar] [CrossRef] [PubMed]
- Androvitsanea, A.; Stylianou, K.; Drosataki, E.; Petrakis, I. The pathophysiological role of heat shock response in autoimmunity: A literature review. Cells 2021, 10, 2626. [Google Scholar] [CrossRef]
- Cappello, F.; Marino Gammazza, A.; Palumbo Piccionello, A.; Campanella, C.; Pace, A.; Conway de Macario, E.; Macario, A.J.L. Hsp60 chaperonopathies and chaperonotherapy: Targets and agents. Expert Opin. Ther. Targets 2014, 18, 185–208. [Google Scholar] [CrossRef] [PubMed]
- Tukaj, S.; Kaminski, M. Heat shock proteins in the therapy of autoimmune diseases: Too simple to be true? Cell Stress Chaperones 2019, 24, 475–479. [Google Scholar] [CrossRef]
- Zummo, L.; Vitale, A.M.; Caruso Bavisotto, C.; De Curtis, M.; Garbelli, R.; Giallonardo, A.T.; Di Bonaventura, C.; Fanella, M.; Conway de Macario, E.; Cappello, F.; et al. Molecular chaperones and miRNAs in epilepsy: Pathogenic implications and therapeutic prospects. Int. J. Mol. Sci. 2021, 22, 8601. [Google Scholar] [CrossRef] [PubMed]
- Latchman, D.S.; Isenberg, D.A. The role of Hsp90 in SLE. Autoimmunity 1994, 19, 211–218. [Google Scholar] [CrossRef]
- Kuper, B.C.; Failla, S. Systemic lupus erythematosus: A multisystem autoimmune disorder. Nurs. Clin. N. Am. 2000, 35, 253–265. [Google Scholar] [CrossRef]
- Rahman, A.; Isenberg, D.A. Systemic lupus erythematosus. N. Engl. J. Med. 2008, 358, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Cojocaru, M.; Cojocaru, I.M.; Silosi, I.; Vrabie, C.D. Manifestations of systemic lupus erythematosus. Maedica 2011, 6, 330–336. [Google Scholar] [PubMed]
- Doria, A.; Iaccarino, L.; Ghirardello, A.; Zampieri, S.; Arienti, S.; Sarzi-Puttini, P.; Atzeni, F.; Piccoli, A.; Todesco, S. Long-term prognosis and causes of death in systemic lupus erythematosus. Am. J. Med. 2006, 119, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Ramírez Sepúlveda, J.I.; Bolin, K.; Mofors, J.; Leonard, D.; Svenungsson, E.; Jönsen, A.; Bengtsson, C.; DISSECT Consortium; Nordmark, G.; Rantapää Dahlqvist, S.; et al. Sex differences in clinical presentation of systemic lupus erythematosus. Biol. Sex Differ. 2019, 10, 60. [Google Scholar] [CrossRef]
- Wolf, B.; Blaschke, C.R.K.; Mungaray, S.; Weselman, B.T.; Stefanenko, M.; Fedoriuk, M.; Bai, H.; Rodgers, J.; Palygin, O.; Drake, R.R.; et al. Metabolic markers and association of biological sex in lupus nephritis. Int. J. Mol. Sci. 2023, 24, 16490. [Google Scholar] [CrossRef] [PubMed]
- Woods, M.; Zou, Y.R.; Davidson, A. Defects in germinal center selection in SLE. Front. Immunol. 2015, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Voll, R.E.; Kalden, J.R. Etiopathogenesis of systemic lupus erythematosus. Immunol. Today 2000, 21, 424–426. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.S.; Brown, E.E.; Kimberly, R.P.; Langefeld, C.D. Genetic factors predisposing to systemic lupus erythematosus and lupus nephritis. Semin. Nephrol. 2010, 30, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Kamen, D.L. Environmental influences on systemic lupus erythematosus expression. Rheum. Dis. Clin. N. Am. 2014, 40, 401–412. [Google Scholar] [CrossRef]
- Pan, L.; Lu, M.P.; Wang, J.H.; Xu, M.; Yang, S.R. Immunological pathogenesis and treatment of systemic lupus erythematosus. World J. Pediatr. 2020, 16, 19–30. [Google Scholar] [CrossRef]
- Woo, J.M.P.; Parks, C.G.; Jacobsen, S.; Costenbader, K.H.; Bernatsky, S. The role of environmental exposures and gene-environment interactions in the etiology of systemic lupus erythematous. J. Intern. Med. 2022, 291, 755–778. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012, 3, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Garabatos, N.; Santamaria, P. Gut microbial antigenic mimicry in autoimmunity. Front. Immunol. 2022, 13, 873607. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, V.; Latchman, D.; Isenberg, D. Heat shock proteins and systemic lupus erythematosus. Lupus 1991, 1, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Stephanou, A.; Latchman, D.S.; Isenberg, D.A. The regulation of heat shock proteins and their role in systemic lupus erythematosus. Semin. Arthritis Rheum. 1998, 28, 155–162. [Google Scholar] [CrossRef]
- Deguchi, Y.; Negoro, S.; Kishimoto, S. Heat-shock protein synthesis by human peripheral mononuclear cells from SLE patients. Biochem. Biophys. Res. Commun. 1987, 148, 1063–1068. [Google Scholar] [CrossRef]
- Norton, P.M.; Isenberg, D.A.; Latchman, D.S. Elevated levels of the 90 kd heat shock protein in a proportion of SLE patients with active disease. J. Autoimmun. 1989, 2, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, V.B.; McCallum, S.; Norton, P.; Twomey, B.M.; Erkeller-Yuksel, F.; Lydyard, P.; Isenberg, D.A.; Latchman, D.S. Differential heat shock protein overexpression and its clinical relevance in systemic lupus erythematosus. Ann. Rheum. Dis. 1993, 52, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Twomey, B.M.; Dhillon, V.B.; McCallum, S.; Isenberg, D.A.; Latchman, D.S. Elevated levels of the 90 kD heat shock protein in patients with systemic lupus erythematosus are dependent upon enhanced transcription of the hsp90 beta gene. J. Autoimmun. 1993, 6, 495–506. [Google Scholar] [CrossRef]
- Faulds, G.B.; Isenberg, D.A.; Latchman, D.S. The tissue specific elevation in synthesis of the 90 kDa heat shock protein precedes the onset of disease in lupus prone MRL/lpr mice. J. Rheumatol. 1994, 21, 234–238. [Google Scholar]
- Tsagalis, G.C.; Nikolopoulou, N.; Sotsiou, F.; Hadjiconstantinou, V. The expression of heat shock proteins 27 and 70 in lupus nephritis. Hosp. Chron. 2006, 1, 125–129. [Google Scholar]
- Minota, S.; Winfield, J.B. IgG anti-lymphocyte antibodies in systemic lupus erythematosus react with surface molecules shared by peripheral T cells and a primitive T cell line. J. Immunol. 1987, 138, 1750–1756. [Google Scholar] [CrossRef] [PubMed]
- Erkeller-Yüksel, F.M.; Isenberg, D.A.; Dhillon, V.B.; Latchman, D.S.; Lydyard, P.M. Surface expression of heat shock protein 90 by blood mononuclear cells from patients with systemic lupus erythematosus. J. Autoimmun. 1992, 5, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Minota, S.; Koyasu, S.; Yahara, I.; Winfield, J. Autoantibodies to the heat-shock protein Hsp90 in systemic lupus erythematosus. J. Clin. Investig. 1988, 81, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Conroy, S.E.; Faulds, G.B.; Williams, W.; Latchman, D.S.; Isenberg, D.A. Detection of autoantibodies to the 90 kDa heat shock protein in systemic lupus erythematosus and other autoimmune diseases. Br. J. Rheumatol. 1994, 33, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Conroy, S.E.; Tucker, L.; Latchman, D.S.; Isenberg, D.A. Incidence of anti Hsp 90 and 70 antibodies in children with SLE, juvenile dermatomyositis and juvenile chronic arthritis. Clin. Exp. Rheumatol. 1996, 14, 99–104. [Google Scholar] [PubMed]
- Kenderov, A.; Minkova, V.; Mihailova, D.; Giltiay, N.; Kyurkchiev, S.; Kehayov, I.; Kazatchkine, M.; Kaveri, S.; Pashov, A. Lupus-specific kidney deposits of HSP90 are associated with altered IgG idiotypic interactions of anti-HSP90 autoantibodies. Clin. Exp. Immunol. 2002, 129, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Ripley, B.J.; Isenberg, D.A.; Latchman, D.S. Elevated levels of the 90 kDa heat shock protein (Hsp90) in SLE correlate with levels of IL-6 and autoantibodies to hsp90. J. Autoimmun. 2001, 17, 341–346. [Google Scholar] [CrossRef]
- Linker-Israeli, M.; Deans, R.J.; Wallace, D.J.; Prehn, J.; Ozeri-Chen, T.; Klinenberg, J.R. Elevated levels of endogenous IL-6 in systemic lupus erythematosus. A putative role in pathogenesis. J. Immunol. 1991, 147, 117–123. [Google Scholar] [CrossRef]
- Park, Y.B.; Lee, S.K.; Kim, D.S.; Lee, J.; Lee, C.H.; Song, C.H. Elevated interleukin-10 levels correlated with disease activity in systemic lupus erythematosus. Clin. Exp. Rheumatol. 1998, 16, 283–288. [Google Scholar]
- Mercader-Salvans, J.; García-González, M.; Gómez-Bernal, F.; Quevedo-Abeledo, J.C.; de Vera-González, A.; González-Delgado, A.; López-Mejías, R.; Martín-González, C.; González-Gay, M.Á.; Ferraz-Amaro, I. Relationship between Disease Characteristics and Circulating Interleukin 6 in a Well-characterized cohort of patients with systemic lupus erythematosus. Int J. Mol. Sci. 2023, 24, 14006. [Google Scholar] [CrossRef] [PubMed]
- Stephanou, A.; Amin, V.; Isenberg, D.A.; Akira, S.; Kishimoto, T.; Latchman, D.S. Interleukin 6 activates heat-shock protein 90β gene expression. Biochem. J. 1997, 321, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Ripley, B.J.; Stephanou, A.; Isenberg, D.A.; Latchman, D.S. Interleukin-10 activates heat-shock protein 90β gene expression. Immunology 1999, 97, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Stephanou, A.; Conroy, S.; Isenberg, D.A.; Maione, D.; Poli, V.; Ciliberto, G.; Latchman, D.S. Elevation of IL-6 in transgenic mice results in increased levels of the 90 kDa heat shock protein (Hsp90) and the production of anti-Hsp90 antibodies. J. Autoimmun. 1998, 11, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Shukla, H.D.; Pitha, P.M. Role of Hsp90 in systemic lupus erythematosus and its clinical relevance. Autoimmune Dis. 2012, 2012, 728605. [Google Scholar] [CrossRef] [PubMed]
- Han, J.M.; Kwon, N.H.; Lee, J.Y.; Jeong, S.J.; Jung, H.J.; Kim, H.R.; Li, Z.; Kim, S. Identification of gp96 as a novel target for treatment of autoimmune disease in mice. PLoS ONE 2010, 5, e9792. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Shi, F.D.; Cohen, I.R.; Castaldo, G.; Matarese, G.; Quintana, F.J.; La Cava, A. DNA vaccine encoding heat shock protein 90 protects from murine lupus. Arthritis Res. Ther. 2020, 22, 152. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Ferretti, C.; Shi, F.D.; Cohen, I.R.; Quintana, F.J.; La Cava, A. DNA vaccination with Hsp70 protects against systemic lupus erythematosus in (NZB × NZW)F1 mice. Arthritis Rheumatol. 2020, 72, 997–1002. [Google Scholar] [CrossRef]
- Berg, S.I.T.; Knapp, J.; Braunstein, M.; Shirriff, C. The small heat shock protein HSPB5 attenuates the severity of lupus nephritis in lupus-prone mice. Autoimmunity 2022, 55, 192–202. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Hill, D.A.; Artis, D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 2010, 28, 623–667. [Google Scholar] [CrossRef] [PubMed]
- Haverson, K.; Rehakova, Z.; Sinkora, J.; Sver, L.; Bailey, M. Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: A study in germ-free pigs. Vet. Immunol. Immunopathol. 2007, 119, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut microbiota and immune system interactions. Microorganisms 2020, 8, 1587, https://doi.org/10.3390/microorganisms8101587;Erratum in Microorganisms 2020, 8, 2046. [Google Scholar] [CrossRef] [PubMed]
- Santana, P.T.; Rosas, S.L.B.; Ribeiro, B.E.; Marinho, Y.; de Souza, H.S.P. Dysbiosis in inflammatory bowel disease: Pathogenic role and potential therapeutic targets. Int. J. Mol. Sci. 2022, 23, 3464. [Google Scholar] [CrossRef] [PubMed]
- Intili, G.; Paladino, L.; Rappa, F.; Alberti, G.; Plicato, A.; Calabrò, F.; Fucarino, A.; Cappello, F.; Bucchieri, F.; Tomasello, G.; et al. From dysbiosis to neurodegenerative diseases through different communication pathways: An overview. Biology 2023, 12, 195. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, M.; Cao, J.; Li, X.; Fan, D.; Xia, Y.; Lu, X.; Li, J.; Ju, D.; Zhao, H. The dynamic interplay between the gut microbiota and autoimmune diseases. J. Immunol. Res. 2019, 2019, 7546047. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Kwok, S.K.; Choe, J.Y.; Park, S.H. Recent advances in our understanding of the link between the intestinal microbiota and systemic lupus erythematosus. Int. J. Mol. Sci. 2019, 20, 4871. [Google Scholar] [CrossRef]
- Miyauchi, E.; Shimokawa, C.; Steimle, A.; Desai, M.S.; Ohno, H. The impact of the gut microbiome on extra-intestinal autoimmune diseases. Nat. Rev. Immunol. 2023, 23, 9–23. [Google Scholar] [CrossRef]
- Zhang, H.; Liao, X.; Sparks, J.B.; Luo, X.M. Dynamics of gut microbiota in autoimmune lupus. Appl. Environ. Microbiol. 2014, 80, 7551–7760. [Google Scholar] [CrossRef]
- Gomez, A.; Luckey, D.; Taneja, V. The gut microbiome in autoimmunity: Sex matters. Clin. Immunol. 2015, 159, 154–162. [Google Scholar] [CrossRef]
- Mu, Q.; Zhang, H.; Liao, X.; Lin, K.; Liu, H.; Edwards, M.R.; Ahmed, S.A.; Yuan, R.; Li, L.; Cecere, T.E.; et al. Control of lupus nephritis by changes of gut microbiota. Microbiome 2017, 5, 73. [Google Scholar] [CrossRef]
- Luo, X.M.; Edwards, M.R.; Mu, Q.; Yu, Y.; Vieson, M.D.; Reilly, C.M.; Ahmed, S.A.; Bankole, A.A. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl. Environ. Microbiol. 2018, 84, e02288-17. [Google Scholar] [CrossRef]
- Ma, Y.; Xu, X.; Li, M.; Cai, J.; Wei, Q.; Niu, H. Gut microbiota promote the inflammatory response in the pathogenesis of systemic lupus erythematosus. Mol. Med. 2019, 25, 35. [Google Scholar] [CrossRef]
- Hevia, A.; Milani, C.; López, P.; Cuervo, A.; Arboleya, S.; Duranti, S.; Turroni, F.; González, S.; Suárez, A.; Gueimonde, M.; et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio 2014, 5, e01548-14. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Shao, T.; Li, H.; Xie, Z.; Wen, C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 2016, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- López, P.; de Paz, B.; Rodríguez-Carrio, J.; Hevia, A.; Sánchez, B.; Margolles, A.; Suárez, A. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci. Rep. 2016, 6, 24072. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.F.; Li, X.; Li, H.X.; Zhang, Q.; Zhou, H.W.; He, Y.; Li, P.; Fu, C.; Zhang, X.H.; et al. Disordered intestinal microbes are associated with the activity of Systemic Lupus Erythematosus. Clin. Sci. 2019, 133, 821–838. [Google Scholar] [CrossRef]
- Wei, F.; Xu, H.; Yan, C.; Rong, C.; Liu, B.; Zhou, H. Changes of intestinal flora in patients with systemic lupus erythematosus in northeast China. PLoS ONE 2019, 14, e0213063. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.D.; Jia, X.M.; Xu, J.Y.; Zhao, L.D.; Ji, J.Y.; Wu, B.X.; Ma, Y.; Li, H.; Zuo, X.X.; Pan, W.Y.; et al. An autoimmunogenic and proinflammatory profile defined by the gut microbiota of patients with untreated systemic lupus erythematosus. Arthritis Rheumatol. 2021, 73, 232–243. [Google Scholar] [CrossRef]
- Zhang, S.X.; Wang, J.; Chen, J.W.; Zhang, M.X.; Zhang, Y.F.; Hu, F.Y.; Lv, Z.Q.; Gao, C.; Li, Y.F.; Li, X.F. The level of peripheral regulatory T cells is linked to changes in gut commensal microflora in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2021, 80, e177. [Google Scholar] [CrossRef]
- Toumi, E.; Goutorbe, B.; Plauzolles, A.; Bonnet, M.; Mezouar, S.; Militello, M.; Mege, J.L.; Chiche, L.; Halfon, P. Gut microbiota in systemic lupus erythematosus patients and lupus mouse model: A cross species comparative analysis for biomarker discovery. Front. Immunol. 2022, 13, 943241. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Wucherpfennig, K.W. Mechanisms for the induction of autoimmunity by infectious agents. J. Clin. Investig. 2001, 108, 1097–1104. [Google Scholar] [CrossRef]
- Damian, R.T. Molecular mimicry: Antigen sharing by parasite and host and its consequences. Am. Nat. 1964, 98, 129–149. [Google Scholar] [CrossRef]
- Rojas, M.; Restrepo-Jiménez, P.; Monsalve, D.M.; Pacheco, Y.; Acosta-Ampudia, Y.; Ramírez-Santana, C.; Leung, P.S.C.; Ansari, A.A.; Gershwin, M.E.; Anaya, J.M. Molecular mimicry and autoimmunity. J. Autoimmun. 2018, 95, 100–123. [Google Scholar] [CrossRef]
- Sundar, K.; Jacques, S.; Gottlieb, P.; Villars, R.; Benito, M.E.; Taylor, D.K.; Spatz, L.A. Expression of the Epstein-Barr virus nuclear antigen-1 (EBNA-1) in the mouse can elicit the production of anti-dsDNA and anti-Sm antibodies. J. Autoimmun. 2004, 23, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Poole, B.D.; Scofield, R.H.; Harley, J.B.; James, J.A. Epstein-Barr virus and molecular mimicry in systemic lupus erythematosus. Autoimmunity 2006, 39, 63–70. [Google Scholar] [CrossRef]
- Neo, J.Y.J.; Wee, S.Y.K.; Bonne, I.; Tay, S.H.; Raida, M.; Jovanovic, V.; Fairhurst, A.M.; Lu, J.; Hanson, B.J.; MacAry, P.A. Characterisation of a human antibody that potentially links cytomegalovirus infection with systemic lupus erythematosus. Sci. Rep. 2019, 9, 9998. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, A.H.; Kuo, C.F.; Chou, I.J.; Tseng, W.Y.; Chen, Y.F.; Yu, K.H.; Luo, S.F. Human cytomegalovirus pp65 peptide-induced autoantibodies cross-reacts with TAF9 protein and induces lupus-like autoimmunity in BALB/c mice. Sci. Rep. 2020, 10, 9662. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, Y.; Muzi, G.; Sánchez, A.; Sánchez, J.; Munera, M. Prediction of molecular mimicry between proteins from Trypanosoma sp. and human antigens associated with systemic lupus erythematosus. Microb. Pathog. 2022, 172, 105760. [Google Scholar] [CrossRef] [PubMed]
- Zügel, U.; Kaufmann, S.H. Immune response against heat shock proteins in infectious diseases. Immunobiology 1999, 201, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Barone, R.; Marino Gammazza, A.; Paladino, L.; Pitruzzella, A.; Spinoso, G.; Salerno, M.; Sessa, F.; Pomara, C.; Cappello, F.; Rappa, F. Morphological alterations and stress protein variations in lung biopsies obtained from autopsies of COVID-19 subjects. Cells 2021, 10, 3136. [Google Scholar] [CrossRef] [PubMed]
- Tasneem, S.; Islam, N.; Ali, R. Crossreactivity of SLE autoantibodies with 70 kDa heat shock proteins of Mycobacterium tuberculosis. Microbiol. Immunol. 2001, 45, 841–846. [Google Scholar] [CrossRef]
- Dieudé, M.; Senécal, J.L.; Raymond, Y. Induction of endothelial cell apoptosis by heat-shock protein 60-reactive antibodies from anti-endothelial cell autoantibody-positive systemic lupus erythematosus patients. Arthritis Rheum. 2004, 50, 3221–3231. [Google Scholar] [CrossRef]
- Mu, Q.; Kirby, J.; Reilly, C.M.; Luo, X.M. Leaky gut as a danger signal for autoimmune diseases. Front. Immunol. 2017, 8, 598. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Chen, Y.; Chen, D. The role of gut microbiome in autoimmune uveitis. Ophthalmic Res. 2021, 64, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Pianta, A.; Arvikar, S.L.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Two rheumatoid arthritis-specific autoantigens correlate microbial immunity with autoimmune responses in joints. J. Clin. Investig. 2017, 127, 2946–2956. [Google Scholar] [CrossRef] [PubMed]
- Harkiolaki, M.; Holmes, S.L.; Svendsen, P.; Gregersen, J.W.; Jensen, L.T.; McMahon, R.; Friese, M.A.; van Boxel, G.; Etzensperger, R.; Tzartos, J.S.; et al. T cell-mediated autoimmune disease due to low-affinity crossreactivity to common microbial peptides. Immunity 2009, 30, 348–357, Erratum in Immunity 2009, 30, 610. [Google Scholar] [CrossRef]
- Yanagisawa, N.; Haruta, I.; Shimizu, K.; Furukawa, T.; Higuchi, T.; Shibata, N.; Shiratori, K.; Yagi, J. Identification of commensal flora-associated antigen as a pathogenetic factor of autoimmune pancreatitis. Pancreatology 2014, 14, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Morel, L. Loss of gut barrier integrity in lupus. Front. Immunol. 2022, 13, 919792. [Google Scholar] [CrossRef] [PubMed]
- Greiling, T.M.; Dehner, C.; Chen, X.; Hughes, K.; Iñiguez, A.J.; Boccitto, M.; Ruiz, D.Z.; Renfroe, S.C.; Vieira, S.M.; Ruff, W.E.; et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 2018, 10, eaan2306. [Google Scholar] [CrossRef] [PubMed]
- Azzouz, D.; Omarbekova, A.; Heguy, A.; Schwudke, D.; Gisch, N.; Rovin, B.H.; Caricchio, R.; Buyon, J.P.; Alekseyenko, A.V.; Silverman, G.J.; et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 2019, 78, 947–956. [Google Scholar] [CrossRef]
- Bagavant, H.; Araszkiewicz, A.M.; Ingram, J.K.; Cizio, K.; Merrill, J.T.; Arriens, C.; Guthridge, J.M.; James, J.A.; Deshmukh, U.S. Immune response to Enterococcus gallinarum in lupus patients is associated with a subset of lupus-associated autoantibodies. Front. Immunol. 2021, 12, 635072. [Google Scholar] [CrossRef]
- Manfredo Vieira, S.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161, https://doi.org/10.1126/science.aar7201;Erratum in Science 2018, 360, eaat9922. [Google Scholar] [CrossRef]
- Stewart, L.; Edgar, J.D.M.; Blakely, G.; Patrick, S. Antigenic mimicry of ubiquitin by the gut bacterium Bacteroides fragilis: A potential link with autoimmune disease. Clin. Exp. Immunol. 2018, 194, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Handley, H.H.; Yu, J.; Yu, D.T.; Singh, B.; Gupta, R.S.; Vaughan, J.H. Autoantibodies to human heat shock protein (hsp)60 may be induced by Escherichia coli groEL. Clin. Exp. Immunol. 1996, 103, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.I.; Hirata, D.; Minota, S.; Higashiyama, T.; Kurimoto, M.; Yanagi, H.; Yura, T.; Kubota, H. Autoantibodies against chaperonin CCT in human sera with rheumatic autoimmune diseases: Comparison with antibodies against other Hsp60 family proteins. Cell Stress Chaperones 2000, 5, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Yamashiro, Y.; Ohtsuka, Y.; Shimizu, T.; Sakurai, Y.; Misawa, S.; Ito, T. Heat shock proteins and superantigenic properties of bacteria from the gastrointestinal tract of patients with Kawasaki disease. Immunology 2009, 128, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cho, K.S.; Vu, T.H.K.; Shen, C.H.; Kaur, M.; Chen, G.; Mathew, R.; McHam, M.L.; Fazelat, A.; Lashkari, K.; et al. Commensal microflora-induced T cell responses mediate progressive neurodegeneration in glaucoma. Nat. Commun. 2018, 9, 3209, Erratum in Nat. Commun. 2018, 9, 3914. [Google Scholar] [CrossRef] [PubMed]
- Genest, O.; Wickner, S.; Doyle, S.M. Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J. Biol. Chem. 2019, 294, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Tukaj, S.; Sitko, K. Heat Shock Protein 90 (Hsp90) and Hsp70 as potential therapeutic targets in autoimmune skin diseases. Biomolecules 2022, 12, 1153. [Google Scholar] [CrossRef] [PubMed]
- Bruce, I.N.; Buie, J.; Bloch, L.; Bae, S.C.; Costenbader, K.; Levy, R.A.; Werth, V.P.; Marion, A.; Sangodkar, S.; Manzi, S. Lupus spectrum ambiguity has long-term negative implications for patients. Lupus Sci. Med. 2023, 10, e000856. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, A.; Bonsmann, G.; Anders, H.J.; Herzer, P.; Tenbrock, K.; Schneider, M. The diagnosis and treatment of systemic lupus erythematosus. Dtsch. Ärzteblatt Int. 2015, 112, 423–432. [Google Scholar] [CrossRef]
- Kernder, A.; Richter, J.G.; Fischer-Betz, R.; Winkler-Rohlfing, B.; Brinks, R.; Aringer, M.; Schneider, M.; Chehab, G. Delayed diagnosis adversely affects outcome in systemic lupus erythematosus: Cross sectional analysis of the LuLa cohort. Lupus 2021, 30, 431–438. [Google Scholar] [CrossRef]
- Yu, H.; Nagafuchi, Y.; Fujio, K. Clinical and Immunological Biomarkers for Systemic Lupus Erythematosus. Biomolecules 2021, 11, 928. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Bañuelos, E.; Fava, A.; Andrade, F. An update on autoantibodies in systemic lupus erythematosus. Curr. Opin. Rheumatol. 2023, 35, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Papini, A.M. The use of post-translationally modified peptides for detection of biomarkers of immune-mediated diseases. J. Pept. Sci. 2009, 15, 621–628. [Google Scholar] [CrossRef] [PubMed]
- García-Moreno, C.; Gómara, M.J.; Castellanos-Moreira, R.; Sanmartí, R.; Haro, I. Peptides bearing multiple post-translational modifications as antigenic targets for severe rheumatoid arthritis patients. Int. J. Mol. Sci. 2021, 22, 13290. [Google Scholar] [CrossRef]
- Vitale, A.M.; Conway de Macario, E.; Alessandro, R.; Cappello, F.; Macario, A.J.L.; Marino Gammazza, A. Missense mutations of human Hsp60: A computational analysis to unveil their pathological significance. Front. Genet. 2020, 11, 969. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitale, A.M.; Paladino, L.; Caruso Bavisotto, C.; Barone, R.; Rappa, F.; Conway de Macario, E.; Cappello, F.; Macario, A.J.L.; Marino Gammazza, A. Interplay between the Chaperone System and Gut Microbiota Dysbiosis in Systemic Lupus Erythematosus Pathogenesis: Is Molecular Mimicry the Missing Link between Those Two Factors? Int. J. Mol. Sci. 2024, 25, 5608. https://doi.org/10.3390/ijms25115608
Vitale AM, Paladino L, Caruso Bavisotto C, Barone R, Rappa F, Conway de Macario E, Cappello F, Macario AJL, Marino Gammazza A. Interplay between the Chaperone System and Gut Microbiota Dysbiosis in Systemic Lupus Erythematosus Pathogenesis: Is Molecular Mimicry the Missing Link between Those Two Factors? International Journal of Molecular Sciences. 2024; 25(11):5608. https://doi.org/10.3390/ijms25115608
Chicago/Turabian StyleVitale, Alessandra Maria, Letizia Paladino, Celeste Caruso Bavisotto, Rosario Barone, Francesca Rappa, Everly Conway de Macario, Francesco Cappello, Alberto J. L. Macario, and Antonella Marino Gammazza. 2024. "Interplay between the Chaperone System and Gut Microbiota Dysbiosis in Systemic Lupus Erythematosus Pathogenesis: Is Molecular Mimicry the Missing Link between Those Two Factors?" International Journal of Molecular Sciences 25, no. 11: 5608. https://doi.org/10.3390/ijms25115608














