Genome-Wide Analysis of MADS-Box Gene Family Reveals CjSTK as a Key Regulator of Seed Abortion in Camellia japonica
Abstract
:1. Introduction
2. Results
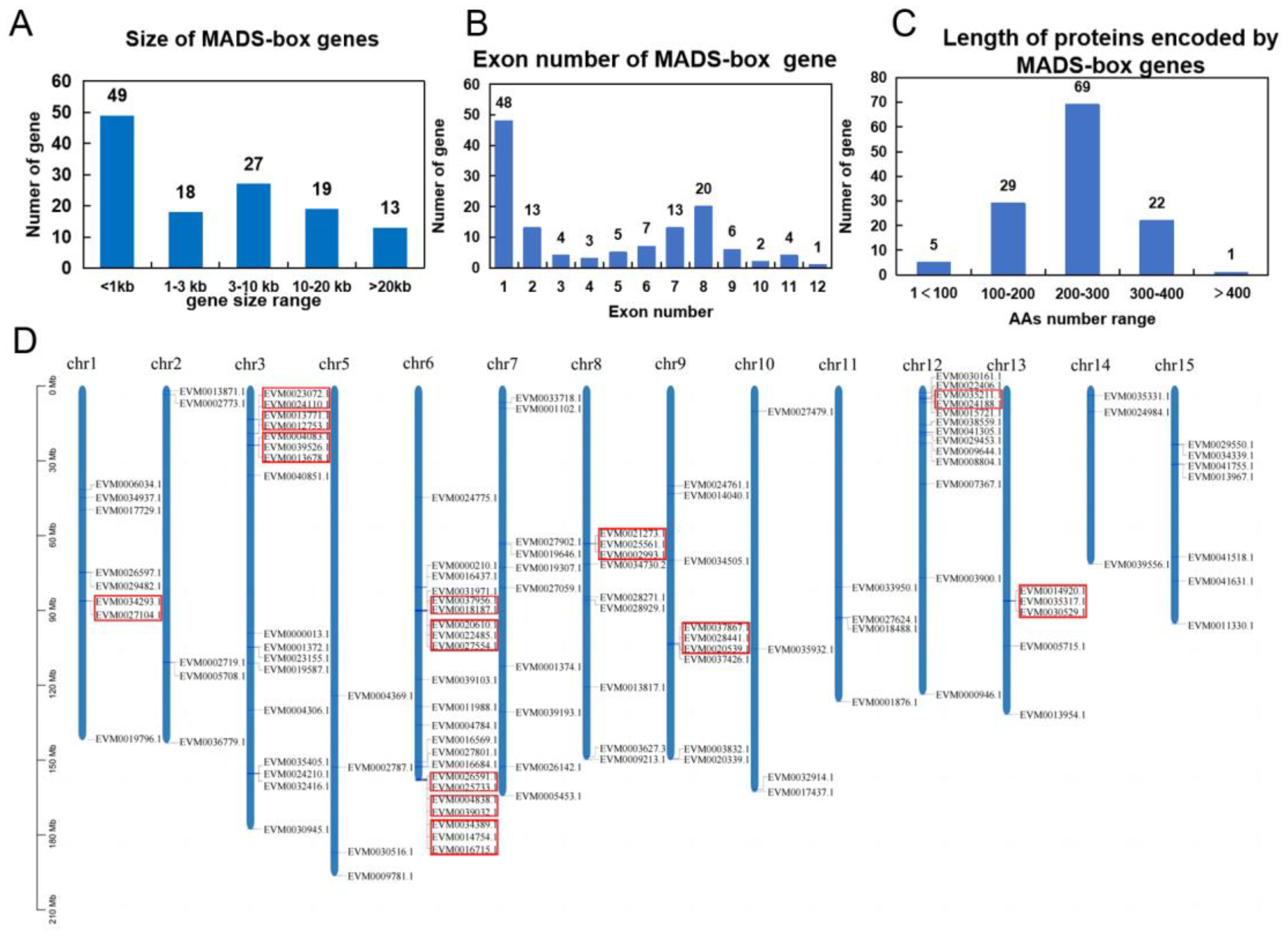
2.1. Genome-Wide Identification and Characterization of MADS-Box Genes in C. japonica (cjND) Genome
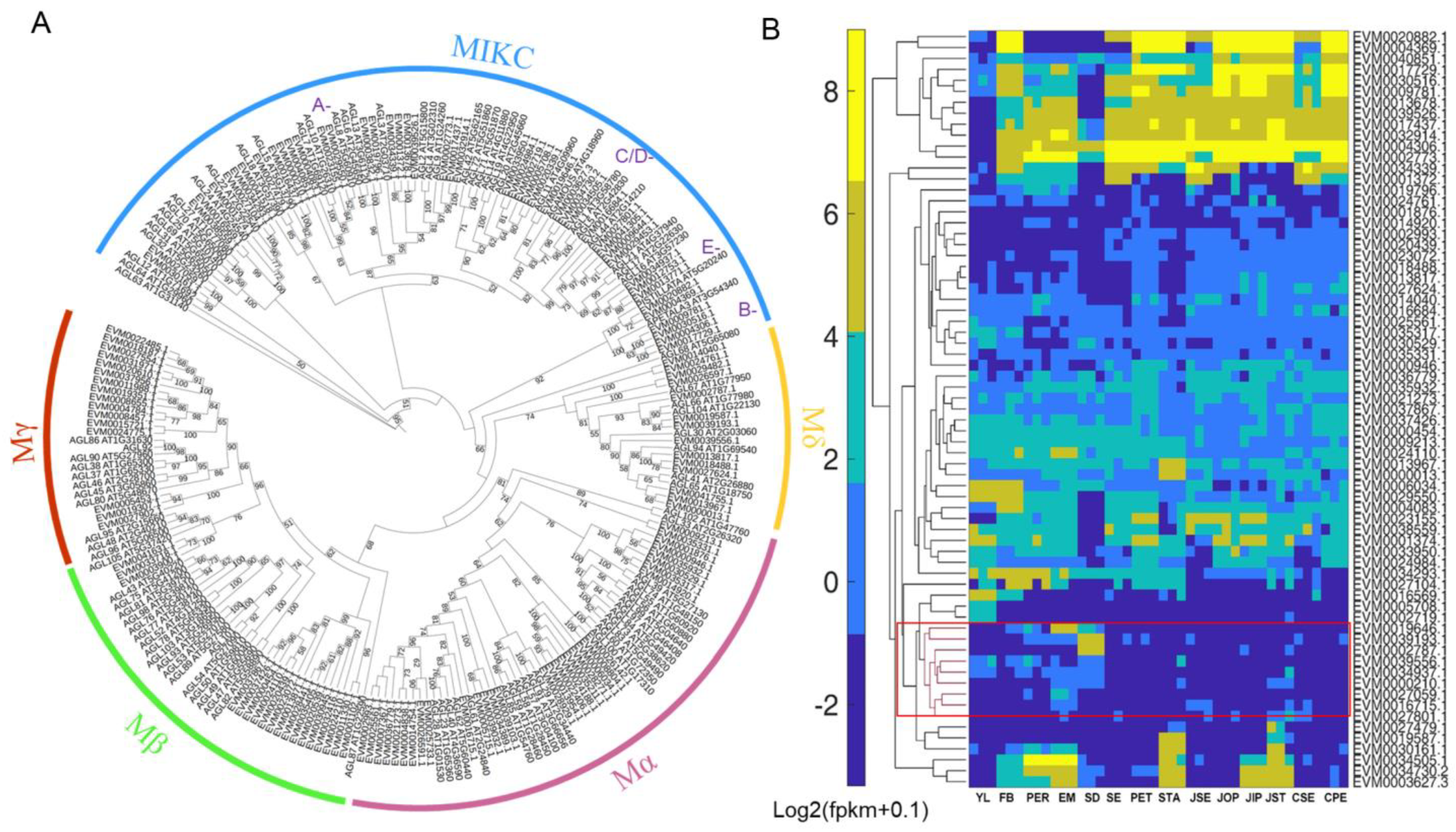
2.2. Phylogenetic Analysis of MADS-Box Genes in C. japonica
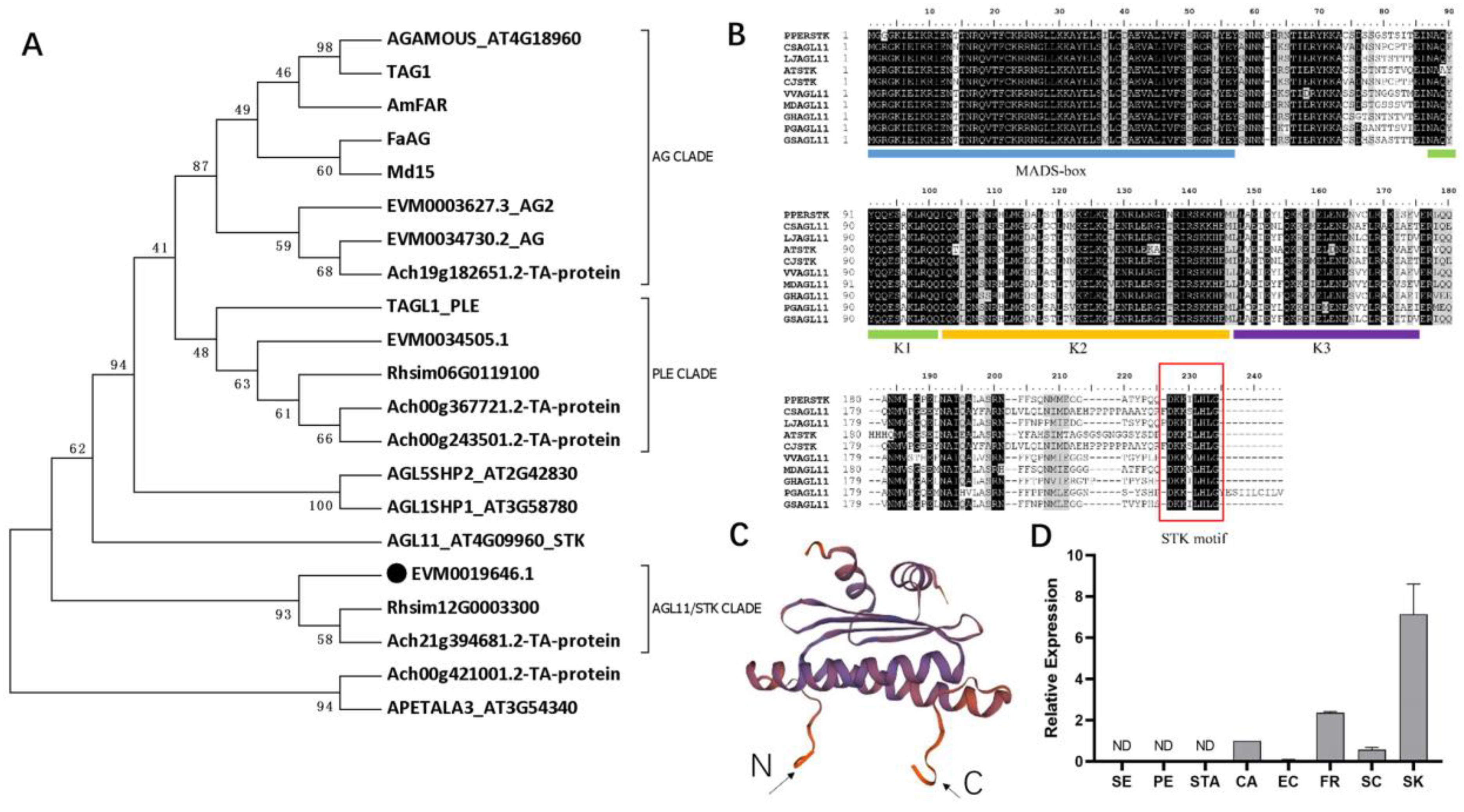
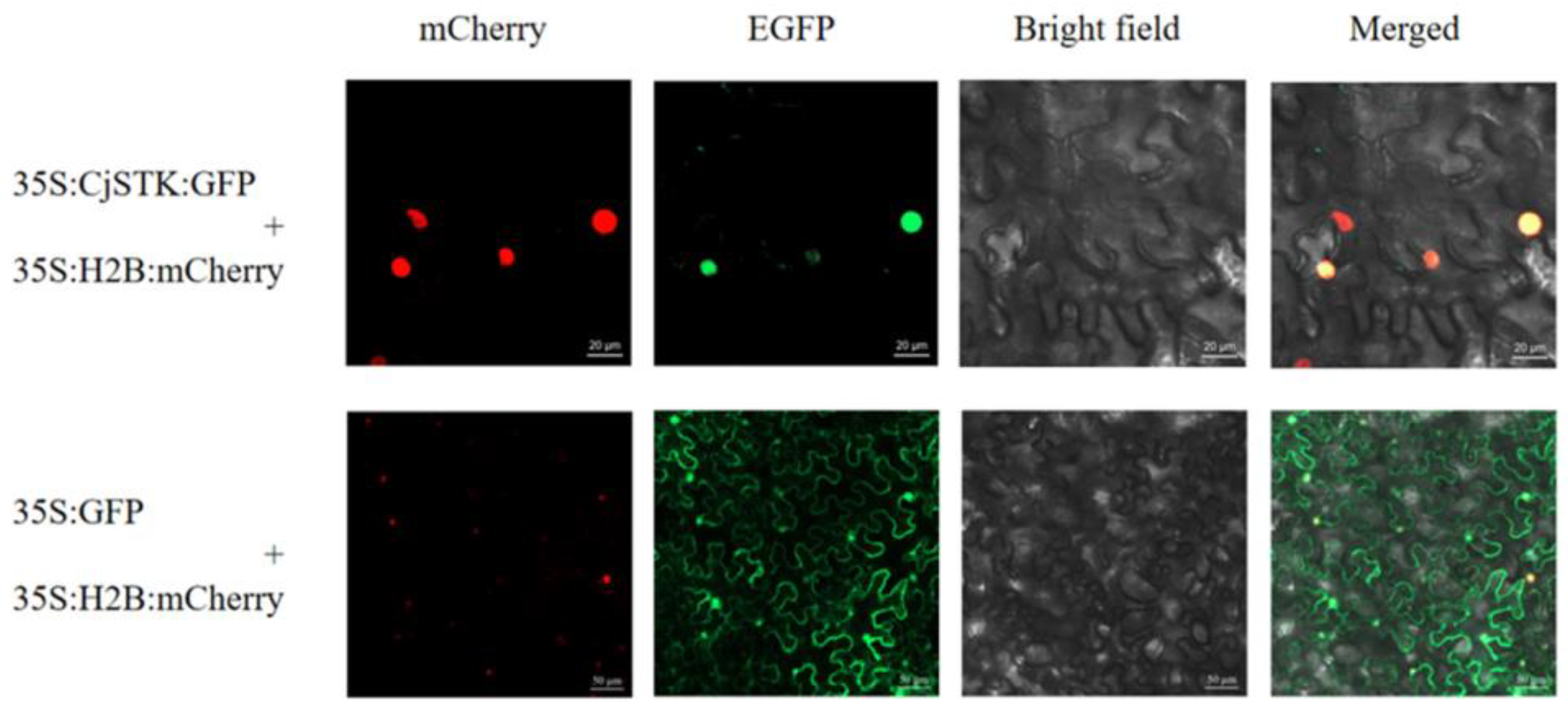
2.3. Identification and Characterization of AGL11/STK Homolog in C. japonica
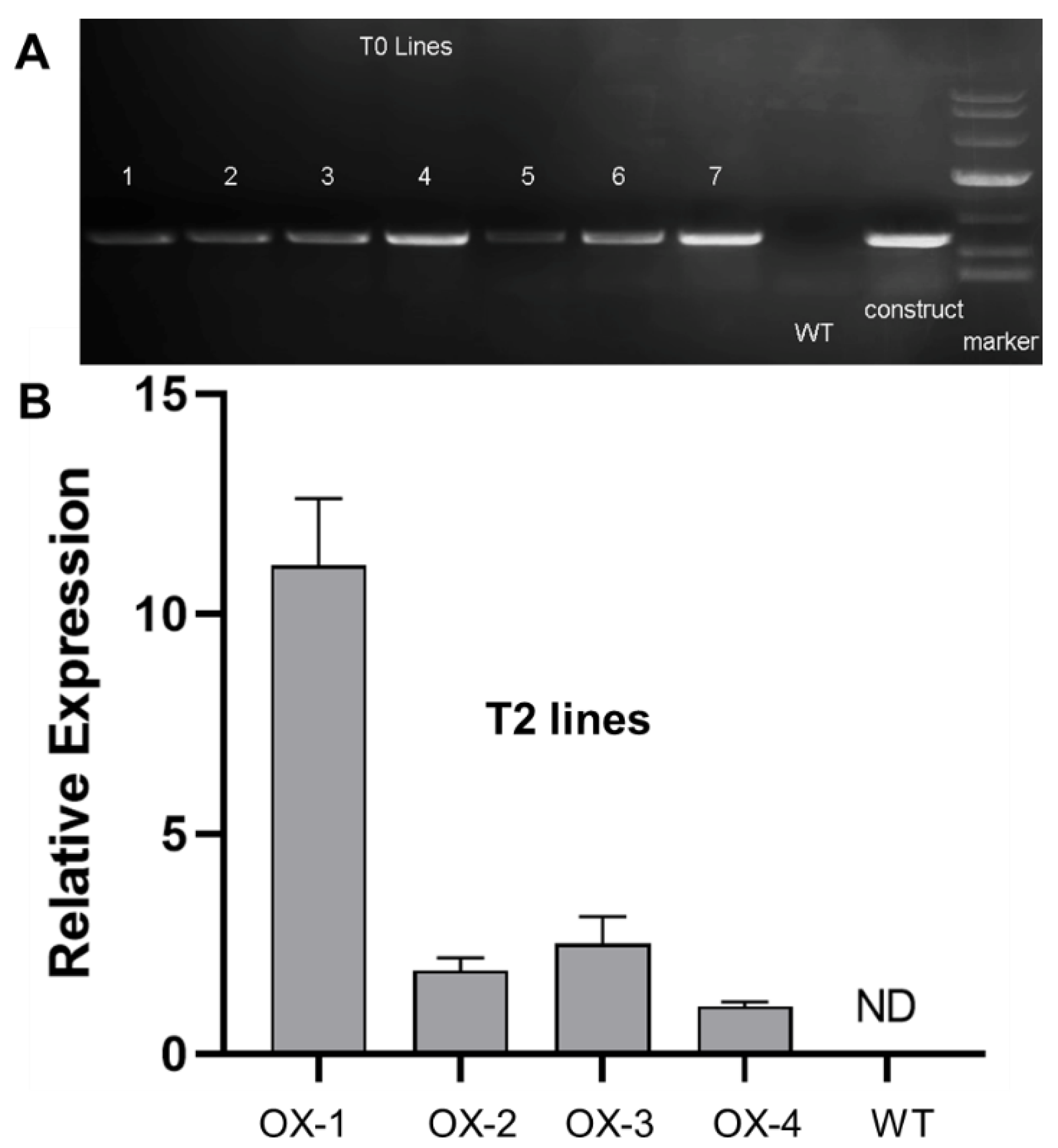
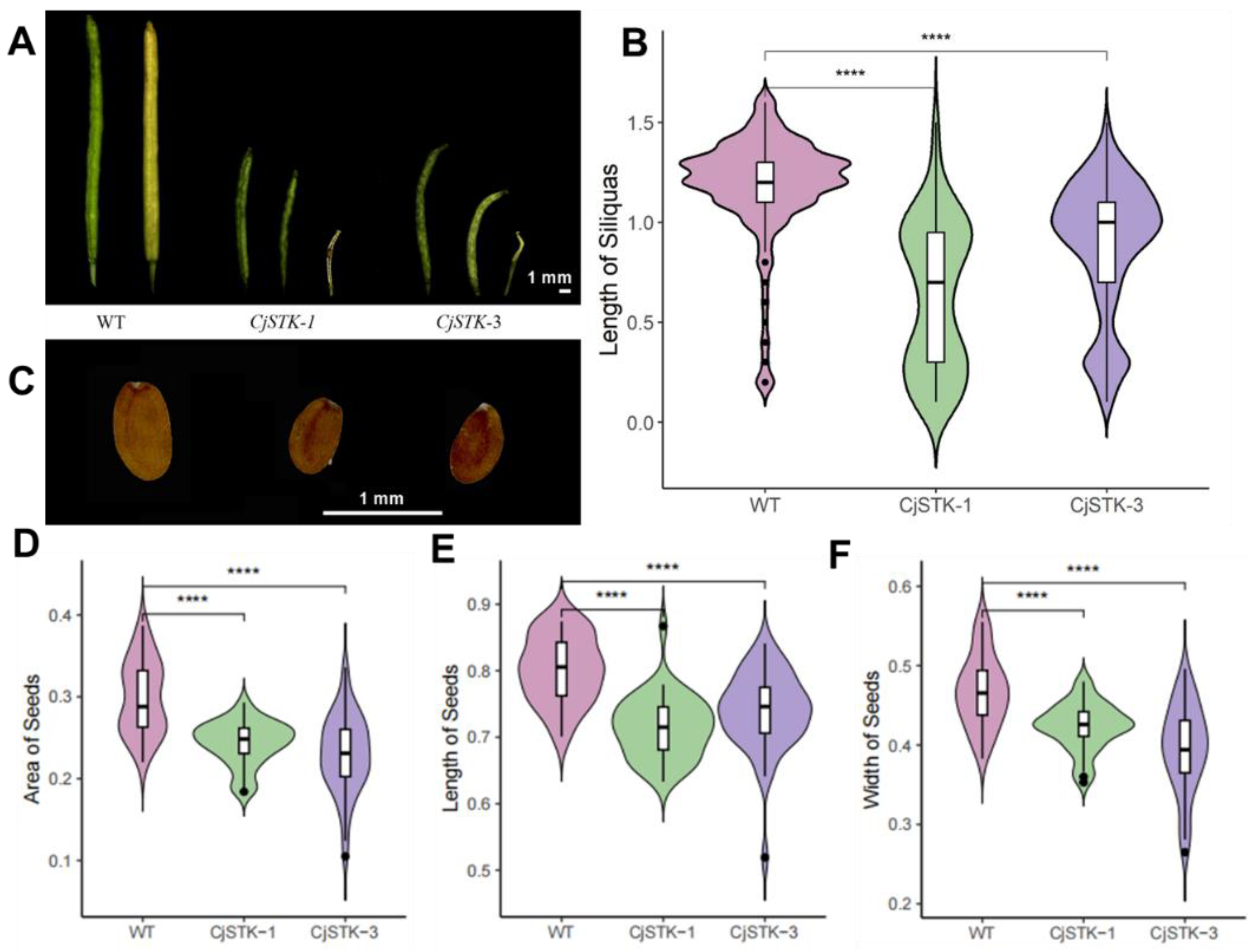
2.4. Ectopic Expression of CjSTK in Arabidopsis Causes Defects in Seed and Silique Development
3. Discussion
3.1. Conservation and Evolution of the MADS-Box Gene Family in Camellia japonica
3.2. Dual Function of the AGL11/STK Genes in Regulating Seed Development
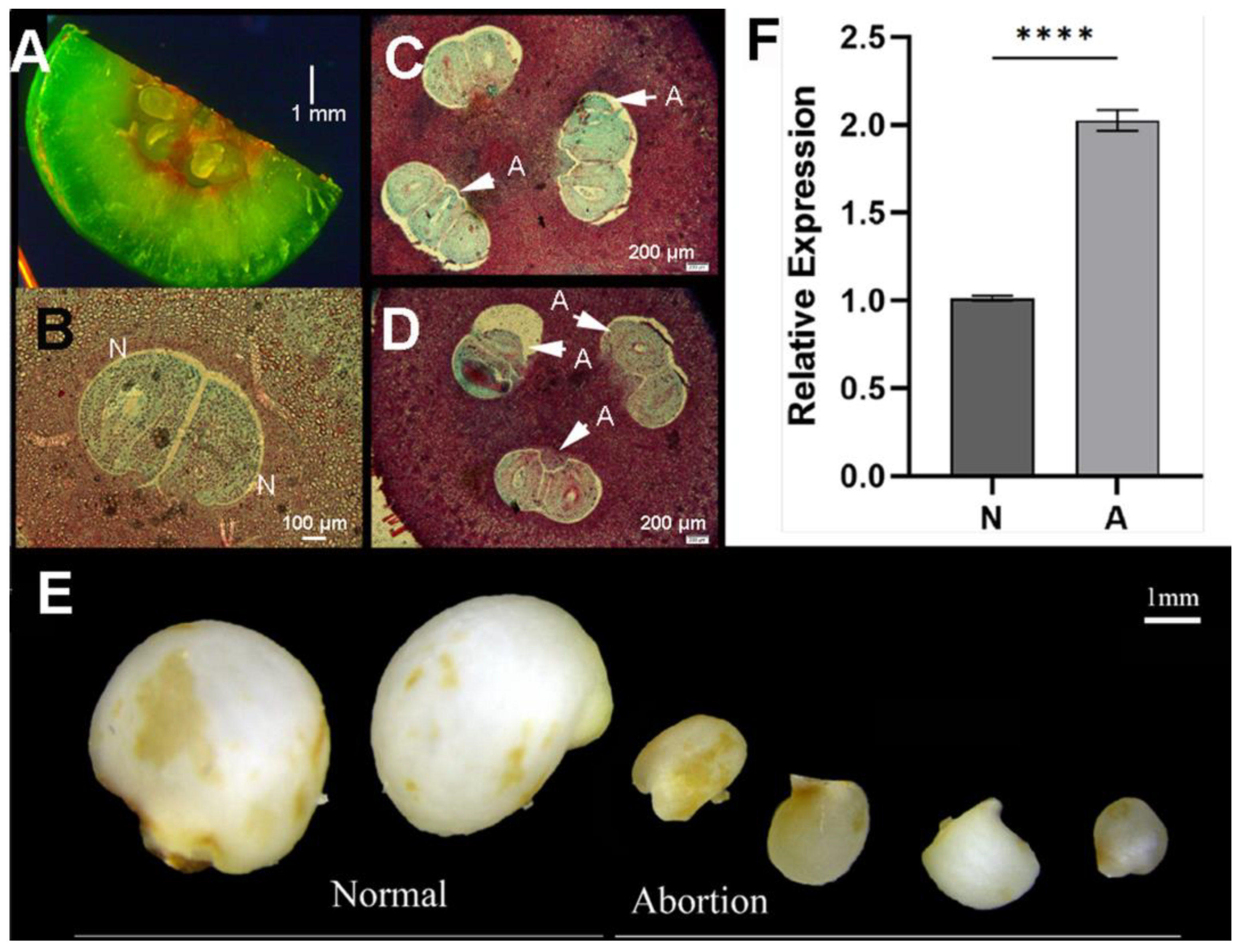
3.3. Activation and Repression of CjSTK Gene Expression Is Critical for Maintaining Seed Development
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Gene Family Analysis and Expression Profiling of MADS-Box Genes
4.3. Isolation of Total RNA and Real-Time PCR Analysis
4.4. Gene Cloning, Vector Construction and Subcellular Localization Analysis
4.5. Arabidopsis Transformation and Screening of CjSTK-OX Transgenic Lines
4.6. Statistical Analysis
4.7. Phylogenetic Tree Construction
4.8. Histological Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shore, P.; Sharrocks, A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995, 229, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenetics Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef] [PubMed]
- Passmore, S.; Elble, R.; Tye, B.K. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 1989, 3, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Sommer, H.; Beltrán, J.P.; Huijser, P.; Pape, H.; Lönnig, W.E.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Purugganan, M.D. The MADS-box floral homeotic gene lineages predate the origin of seed plants: Phylogenetic and molecular clock estimates. J. Mol. Evol. 1997, 45, 392–396. [Google Scholar] [CrossRef]
- Busi, M.V.; Bustamante, C.; D’angelo, C.; Hidalgo-Cuevas, M.; Boggio, S.B.; Valle, E.M.; Zabaleta, E. MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 2003, 52, 801–815. [Google Scholar]
- Callens, C.; Tucker, M.R.; Zhang, D.; Wilson, Z.A. Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 2018, 69, 2435–2459. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; García-Ponce, B.; Sánchez, M.d.l.P.; Espinosa-Soto, C.; García-Gómez, M.L.; Piñeyro-Nelson, A.; Garay-Arroyo, A. MADS-box genes underground becoming mainstream: Plant root developmental mechanisms. New Phytol. 2019, 223, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Castelán-Muñoz, N.; Herrera, J.; Cajero-Sánchez, W.; Arrizubieta, M.; Trejo, C.; García-Ponce, B.; Sánchez, M.d.l.P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. MADS-box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plants. Front. Plant Sci. 2019, 10, 853. [Google Scholar] [CrossRef] [PubMed]
- De Bodt, S.; Raes, J.; Van de Peer, Y.; Theissen, G. And then there were many: MADS goes genomic. Trends Plant Sci. 2003, 8, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Liu, N.; Li, C.; Wang, S.; Jia, L.; Zhang, R.; Li, H.; Tan, J.; Xue, H.; Zheng, W. TaMADS2-3D, a MADS transcription factor gene, regulates phosphate starvation responses in plants. Crop J. 2022, 10, 243–253. [Google Scholar] [CrossRef]
- De Bodt, S.; Raes, J.; Florquin, K.; Rombauts, S.; Rouzé, P.; Theissen, G.; Van de Peer, Y. Genomewide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. J. Mol. Evol. 2003, 56, 573–586. [Google Scholar] [CrossRef] [PubMed]
- de Folter, S.; Immink, R.G.; Kieffer, M.; Parenicová, L.; Henz, S.R.; Weigel, D.; Busscher, M.; Kooiker, M.; Colombo, L.; Kater, M.M.; et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 2005, 17, 1424–1433. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Vega-Léon, R.; Hugouvieux, V.; Blanc-Mathieu, R.; van der Wal, F.; Lucas, J.; Silva, C.S.; Jourdain, A.; Muino, J.M.; Nanao, M.H.; et al. The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors. Nat. Commun. 2021, 12, 4760. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hou, S.; Sun, Z.; Chen, J.; Meng, J.; Liang, D.; Wu, R.; Guo, Y. Genome-Wide Identification and Analysis of the MADS-Box Gene Family in Theobroma cacao. Genes 2021, 12, 1799. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Irish, V. The ABC model of floral development. Curr. Biol. 2017, 27, R887–R890. [Google Scholar] [CrossRef] [PubMed]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Immink, R.G.; Kaufmann, K.; Angenent, G.C. The ‘ABC’ of MADS domain protein behaviour and interactions. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2010; pp. 87–93. [Google Scholar]
- Kramer, E.M.; Jaramillo, M.A.; Di Stilio, V.S. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 2004, 166, 1011–1023. [Google Scholar] [CrossRef]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Colombo, L.; Battaglia, R.; Kater, M.M. Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 2008, 13, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.-F.; Wittich, P.; Kieft, H.; Angenent, G.; XuHan, X.; Van Lammeren, A. Temporal and spatial expression of MADS box genes, FBP7 and FBP11, during initiation and early development of ovules in wild type and mutant Petunia hybrida. Plant Biol. 2000, 2, 693–702. [Google Scholar] [CrossRef]
- Colombo, L.; Franken, J.; Koetje, E.; van Went, J.; Dons, H.; Angenent, G.C.; van Tunen, A.J. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 1995, 7, 1859–1868. [Google Scholar] [PubMed]
- Angenent, G.C.; Franken, J.; Busscher, M.; van Dijken, A.; van Went, J.L.; Dons, H.J.; van Tunen, A.J. A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 1995, 7, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.Y.; Li, X.G.; Li, Q.Z.; Bai, S.N.; Lu, W.L.; Zhang, X.S. Characterization of HoMADS 1 and its induction by plant hormones during in vitro ovule development in Hyacinthus orientalis L. Plant Mol. Biol. 2004, 55, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Dreni, L.; Jacchia, S.; Fornara, F.; Fornari, M.; Ouwerkerk, P.B.; An, G.; Colombo, L.; Kater, M.M. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 2007, 52, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Balasubramani, S.P.; Basha, S.M. Molecular Characterization and Phylogenetic Analysis of MADS-Box Gene VroAGL11 Associated with Stenospermocarpic Seedlessness in Muscadine Grapes. Genes 2021, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Malabarba, J.; Buffon, V.; Mariath, J.E.A.; Maraschin, F.S.; Margis-Pinheiro, M.; Pasquali, G.; Revers, L.F. Manipulation of VviAGL11 expression changes the seed content in grapevine (Vitis vinifera L.). Plant Sci. 2018, 269, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, M.; Herrera-Ubaldo, H.; Caporali, E.; Novák, O.; Strnad, M.; Balanzà, V.; Ezquer, I.; Mendes, M.A.; de Folter, S.; Colombo, L. SEEDSTICK Controls Arabidopsis Fruit Size by Regulating Cytokinin Levels and FRUITFULL. Cell Rep. 2020, 30, 2846–2857.e2843. [Google Scholar] [CrossRef] [PubMed]
- Franks, R.G.; Mizzotti, C.; Ezquer, I.; Paolo, D.; Rueda-Romero, P.; Guerra, R.F.; Battaglia, R.; Rogachev, I.; Aharoni, A.; Kater, M.M.; et al. SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat. PLoS Genet. 2014, 10, e1004856. [Google Scholar] [CrossRef] [PubMed]
- Petrella, R.; Caselli, F.; Roig-Villanova, I.; Vignati, V.; Chiara, M.; Ezquer, I.; Tadini, L.; Kater, M.M.; Gregis, V. BPC transcription factors and a Polycomb Group protein confine the expression of the ovule identity gene SEEDSTICK in Arabidopsis. Plant J. 2020, 102, 582–599. [Google Scholar] [CrossRef] [PubMed]
- Kooiker, M.; Airoldi, C.A.; Losa, A.; Manzotti, P.S.; Finzi, L.; Kater, M.M.; Colombo, L. BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 2005, 17, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Ezquer, I.; Mizzotti, C.; Nguema-Ona, E.; Gotte, M.; Beauzamy, L.; Viana, V.E.; Dubrulle, N.; Costa de Oliveira, A.; Caporali, E.; Koroney, A.S.; et al. The Developmental Regulator SEEDSTICK Controls Structural and Mechanical Properties of the Arabidopsis Seed Coat. Plant Cell 2016, 28, 2478–2492. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhong, X.; Zhang, Q.; Li, H. Overexpression of the GmGAL2 Gene Accelerates Flowering in Arabidopsis. Plant Mol. Biol. Rep. 2010, 28, 704–711. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Ma, R.; Liu, Z.; Zhang, D.; Wang, S.; Guo, Y.; Chen, M. Overexpression of BnaAGL11, a MADS-Box Transcription Factor, Regulates Leaf Morphogenesis and Senescence in Brassica napus. J. Agric. Food Chem. 2022, 70, 3420–3434. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Routaboul, J.M.; Liu, M.; Deng, W.; Maza, E.; Mila, I.; Hu, G.; Zouine, M.; Frasse, P.; Vrebalov, J.T.; et al. Overexpression of the class D MADS-box gene Sl-AGL11 impacts fleshy tissue differentiation and structure in tomato fruits. J. Exp. Bot. 2017, 68, 4869–4884. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Cao, Z.; Wu, P.; Zhang, X.; Lou, J.; Liu, Y.; Wang, Q.; Hu, Y.; Si, S.; Sun, X.; et al. Genome-wide identification, interaction of the MADS-box proteins in Zanthoxylum armatum and functional characterization of ZaMADS80 in floral development. Front. Plant Sci. 2022, 13, 1038828. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Tian, X.; Qiu, Q.; Zhang, Y.; Fan, X.; Yuan, D. Dynamic cytological and transcriptomic analyses provide novel insights into the mechanisms of sex determination in Castanea henryi. Front. Plant Sci. 2023, 14, 1257541. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Fan, Z.; Li, S.; Wang, M.; Huang, M.; Ma, X.; Liu, W.; Wang, Y.; Yu, Y.; Li, Y. Author Correction: Genomics insights into flowering and floral pattern formation: Regional duplication and seasonal pattern of gene expression in Camellia. BMC Biol. 2024, 22, 57. [Google Scholar] [CrossRef] [PubMed]
- Parenicova, L.; De Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Lyu, T.; Yan, C.; Wang, Y.; Ye, N.; Fan, Z.; Li, X.; Li, J.; Yin, H. Identification of alternatively spliced gene isoforms and novel noncoding RNAs by single-molecule long-read sequencing in Camellia. RNA Biol. 2020, 17, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.; Fan, Z.; Liu, Z.; Tanaka, T.; Yin, H. Global gene expression defines faded whorl specification of double flower domestication in Camellia. Sci. Rep. 2017, 7, 3197. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Qu, Y.; Wang, Z.; Huang, B.; Wen, Q.; Xin, Y.; Ni, Z.; Xu, L.a. Gene structural specificity and expression of MADS-box gene family in camellia chekiangoleosa. Int. J. Mol. Sci. 2023, 24, 3434. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chen, Q.; Idrees, A.; Bi, W.; Lai, Z.; Sun, Y. Structural and functional analysis of the MADS-box genes reveals their functions in cold stress responses and flower development in tea plant (Camellia sinensis). Plants 2023, 12, 2929. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Nie, Z.; Huang, H.; Yan, C.; Li, S.; Hu, Z.; Wang, Y.; Yin, H. Small RNA profiling reveals that an ovule-specific microRNA, cja-miR5179, targets a B-class MADS-box gene in Camellia japonica. Ann. Bot. 2023, 132, 1007–1020. [Google Scholar] [CrossRef]
- Singh, R.; Low, E.T.; Ooi, L.C.; Ong-Abdullah, M.; Ting, N.C.; Nagappan, J.; Nookiah, R.; Amiruddin, M.D.; Rosli, R.; Manaf, M.A.; et al. The oil palm SHELL gene controls oil yield and encodes a homologue of SEEDSTICK. Nature 2013, 500, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-r.; Sun, J.-C.; Sun, Z.-l.; Yu, X.; Zhang, Q.; Fang, K.-f.; Cao, Q.-Q.; Ling, Q. The MADS-box transcription factor CmAGL11 modulates somatic embryogenesis in Chinese chestnut (Castanea mollissima Blume). J. Integr. Agric. 2020, 19, 1033–1043. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, B.-Y.; Li, M.; Li, X.-J.; Zhang, Z.-T.; Li, Y.; Li, X.-B. A MADS-box gene is specifically expressed in fibers of cotton (Gossypium hirsutum) and influences plant growth of transgenic Arabidopsis in a GA-dependent manner. Plant Physiol. Biochem. 2014, 75, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bent, A. Arabidopsis thaliana floral dip transformation method. Agrobacterium Protoc. 2006, 343, 87–104. [Google Scholar]
- Van Bel, M.; Proost, S.; Wischnitzki, E.; Movahedi, S.; Scheerlinck, C.; Van de Peer, Y.; Vandepoele, K. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 2012, 158, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Fan, Z.; Li, J.; Li, X.; Wu, B.; Wang, J.; Liu, Z.; Yin, H. Genome-wide transcriptome profiling provides insights into floral bud development of summer-flowering Camellia azalea. Sci. Rep. 2015, 5, 9729. [Google Scholar] [CrossRef]
| Primer Name | Primer Sequence (5′→3′) | Use |
|---|---|---|
| CjSTK-F | ATGGGGCGAGGAAAAATTG | Gene cloning |
| CjSTK-R | TTATCCAAGATGGAGAGAC | |
| ExCjSTK-F | ATGGGGCGAGGAAAAATTG | Vector construction |
| ExCjSTK-R | TTATCCAAGATGGAGAGAC | |
| GAPDH-F | GGGAATCCTTGGTTACACTGAG | Internal control in C. japonica |
| GAPDH-R | ACCCCATTCGTTGTCATACC | |
| qCjSTK-F | CATTCCTTCGAGCCAAGATAGC | qRT-PCR for CjSTK |
| qCjSTK-R | TCTCCTGGCACCATGTTTTG | |
| AtACTIN-F | GCACCCTGTTCTTCTTACCG | Internal control in Arabidopsis |
| AtACTIN-R | AACCCTCGTAGATTGGCACA | |
| HYG_F | TGACCTATTGCATCTCCCGC | Hygomycin selection |
| HYG_R | ATTTGTGTACGCCCGACAGT | |
| 5′RACE-F | GCTGTCAACGATACGCTACGTAAC | 5′RACE for CjSTK |
| 5′RACE-R | CTCATAGACGCGTCCTCGACTA | Gene-specific primer |
| 3′RACE-F | AGTCACATTCTGCAAGCGGAGAA | 3′RACE for CjSTK |
| 3′RACE-R | GCGAGCACAGAATTAATACGACT | Gene-specific primer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Chu, X.; Ma, X.; Hu, Z.; Wang, M.; Li, J.; Yin, H. Genome-Wide Analysis of MADS-Box Gene Family Reveals CjSTK as a Key Regulator of Seed Abortion in Camellia japonica. Int. J. Mol. Sci. 2024, 25, 5770. https://doi.org/10.3390/ijms25115770
Yu Y, Chu X, Ma X, Hu Z, Wang M, Li J, Yin H. Genome-Wide Analysis of MADS-Box Gene Family Reveals CjSTK as a Key Regulator of Seed Abortion in Camellia japonica. International Journal of Molecular Sciences. 2024; 25(11):5770. https://doi.org/10.3390/ijms25115770
Chicago/Turabian StyleYu, Yifan, Xian Chu, Xianjin Ma, Zhikang Hu, Minyan Wang, Jiyuan Li, and Hengfu Yin. 2024. "Genome-Wide Analysis of MADS-Box Gene Family Reveals CjSTK as a Key Regulator of Seed Abortion in Camellia japonica" International Journal of Molecular Sciences 25, no. 11: 5770. https://doi.org/10.3390/ijms25115770






