Abstract
High-mobility group B (HMGB) proteins are a class of non-histone proteins associated with eukaryotic chromatin and are known to regulate a variety of biological processes in plants. However, the functions of HMGB genes in tomato (Solanum lycopersicum) remain largely unexplored. Here, we identified 11 members of the HMGB family in tomato using BLAST. We employed genome-wide identification, gene structure analysis, domain conservation analysis, cis-acting element analysis, collinearity analysis, and qRT-PCR-based expression analysis to study these 11 genes. These genes were categorized into four groups based on their unique protein domain structures. Despite their structural diversity, all members contain the HMG-box domain, a characteristic feature of the HMG superfamily. Syntenic analysis suggested that tomato SlHMGBs have close evolutionary relationships with their homologs in other dicots. The promoter regions of SlHMGBs are enriched with numerous cis-elements related to plant growth and development, phytohormone responsiveness, and stress responsiveness. Furthermore, SlHMGB members exhibited distinct tissue-specific expression profiles, suggesting their potential roles in regulating various aspects of plant growth and development. Most SlHMGB genes respond to a variety of abiotic stresses, including salt, drought, heat, and cold. For instance, SlHMGB2 and SlHMGB4 showed positive responses to salt, drought, and cold stresses. SlHMGB1, SlHMGB3, and SlHMGB8 were involved in responses to two types of stress: SlHMGB1 responded to drought and heat, while SlHMGB3 and SlHMGB8 responded to salt and heat. SlHMGB6 and SlHMGB11 were solely regulated by drought and heat stress, respectively. Under various treatment conditions, the number of up-regulated genes significantly outnumbered the down-regulated genes, implying that the SlHMGB family may play a crucial role in mitigating abiotic stress in tomato. These findings lay a foundation for further dissecting the precise roles of SlHMGB genes.
1. Introduction
High-mobility group (HMG) proteins were discovered in mammalian cells and named for their mobility in polyacrylamide gel electrophoresis [1]. Bustin et al. (2001) subdivided them into three superfamilies: HMGA, HMGB, and HMGN. The members containing the AT-hook domain belong to the HMGA family; those containing the HMG-box domain are assigned to the HMGB family; and those containing nucleosomal binding domain are categorized as the HMGN family [2].
In plants, HMGB proteins widely exist, and their HMG-box domain mainly consists of three α-helices arranged in an L-shape and is flanked by alkaline N-terminal and highly acidic C-terminal domains, which regulate DNA bending or binding [3,4,5]. The HMG-box can be found in HMGB, STRUCTURE-SPECIFIC RECOGNITION PROTEIN 1 (SSRP1), and AT-RICH INTERACTIVE DOMAIN (ARID) proteins, which appear to be plant specific [6,7].
At present, the HMGB genes have been cloned from a variety of plant species, including Arabidopsis [8,9], maize (Zea mays) [10,11], soybean (Glycine max) [12], wheat (Triticum aestivum) [13], pea (Pisum sativum) [14], and rice (Oryza sativa) [15,16]. The biological functions of some of these genes have been characterized. For instance, Arabidopsis AtHMGB15 controls pollen tube growth by interacting with the transcription factors responsible for pollen and pollen tube development. The athmgb15-1 mutant exhibits abnormal pollen tubes and reduced fertility [17]. AtHMGB8 and AtHMGB9 regulate cell division, such as chromosome condensation and/or segregation [7]. In mammals, extracellular HMGB1, a prototypical damage-associated molecular pattern (DAMP) molecule, functions in activating the immune response to prevent infection and promote healing after tissue injury. Interestingly, HMGB1 not only stimulates tissue repair and regeneration but also induces inflammatory response and triggers cellular apoptosis [18]. In plants, the extracellular HMGB3 of Arabidopsis exhibits a similar DAMP function. When the plants are infected by pathogens, HMGB3 is released into the extracellular space, triggering a series of typical innate immune responses. This reveals a novel function of plant HMGB proteins [19].
Furthermore, HMGB can regulate plant growth and development under stress conditions. The ectopic expression of maize HMGB1 affects primary root growth in tobacco seedlings due to the accumulation of small cells in the cytokinetic zone of the roots of transgenic plants, resulting in a reduction in primary root length. However, this phenotype is transient and gradually disappears with plant senescence [20]. In Arabidopsis, either the mutation or overexpression of AtHMGB1 results in a reduction in primary roots, while the aboveground plant remains unaffected [21]. Under salt and drought stress, Arabidopsis plants overexpressing AtHMGB2 or lacking AtHMGB5 show delayed germination and subsequent growth, but the overexpression of AtHMGB4 does not affect seed germination and plant growth [22]. Under high temperature and drought stress, ectopic expression of cucumber CsHMGB delays seed germination in Arabidopsis but does not affect postgermination growth [23]. Cold stress leads to a significant increase in the transcript levels of AtHMGB2, AtHMGB3, and AtHMGB4, and drought or salt stress down-regulates the expressions of AtHMGB2 and AtHMGB3; however, the molecular mechanism of this process is not clear [23].
In addition, OsHMGB1 in rice can interact with SWEET (Sugars Will Eventually be Exported Transporter) proteins to negatively regulate plant resistance to rice leaf blight pathogens. Meanwhile, it may also take part in the negative regulation of the immune response by interacting with the calmodulin-like protein OsCML3 to regulate the Ca2+ signaling pathway [24]. OsHMGB707 positively regulates drought resistance in rice by directly binding to the promoter of OsDREB1G, and the expression of stress-related genes is increased in OsHMGB707 overexpressing lines [25]. In cucumber, the silencing of CsHMGB reduces plant resistance to propamocarb, as evidenced by the increased malondialdehyde (MDA) content and reactive oxygen species (ROS) content, accompanied by decreased activity of antioxidant enzymes and the ascorbate–glutathione (AsA-GSH) system. In contrast, CsHMGB overexpression promotes GSH-dependent detoxification, improves antioxidant capacity, and reduces ROS accumulation [26]. The induction of BnHMGB2 isolated from oilseed rape in response to low-temperature stress potentially plays a key role in plant resistance to low-temperature stress. The 11 HMGB genes identified in Betula platyphylla respond to salt, osmotic stress, and abscisic acid (ABA) treatments, and the overexpression of BpHMG6 increases antioxidant enzyme activity along with enhanced ROS scavenging under salt stress, leading to reduced plant damage and death caused by salt stress [27].
Taken together, the HMGB genes have been studied in some plant species. However, to date, no reports have demonstrated the functions of any HMGBs in tomato. Therefore, we conducted this study and identified the HMGB members in tomato. We then analyzed the gene structures, conserved domains, phylogenetic relationships, cis-elements, expression profiles in different tissues and abiotic stress conditions, and subcellular localization in detail. These data suggest the potential roles of tomato HMGB genes in plant growth and response to abiotic stresses.
2. Results
2.1. Identification and Chromosome Localization of the HMGB Genes in Tomato
By BLAST search and conserved domain analysis, a total of 11 tomato HMGB genes were identified and subsequently named according to their location on the chromosome. According to the GFF genome annotation file, the genomic sequence length of SlHMGBs is 1556–14,596 bp, the coding sequence (CDS) length is 420–2007 bp, the encoded amino acid length is 139–668 aa, and the molecular weight is 15.52–74.46 kDa. SlHMGB3, SlHMGB4, SlHMGB6, SlHMGB8, and SlHMGB9 are acidic, and the others are basic. The subcellular prediction according to WoLF PSORT showed that all SlHMGB proteins are potentially localized in the nucleus (Table 1).

Table 1.
Profiles of the SlHMGB gene family in tomato.
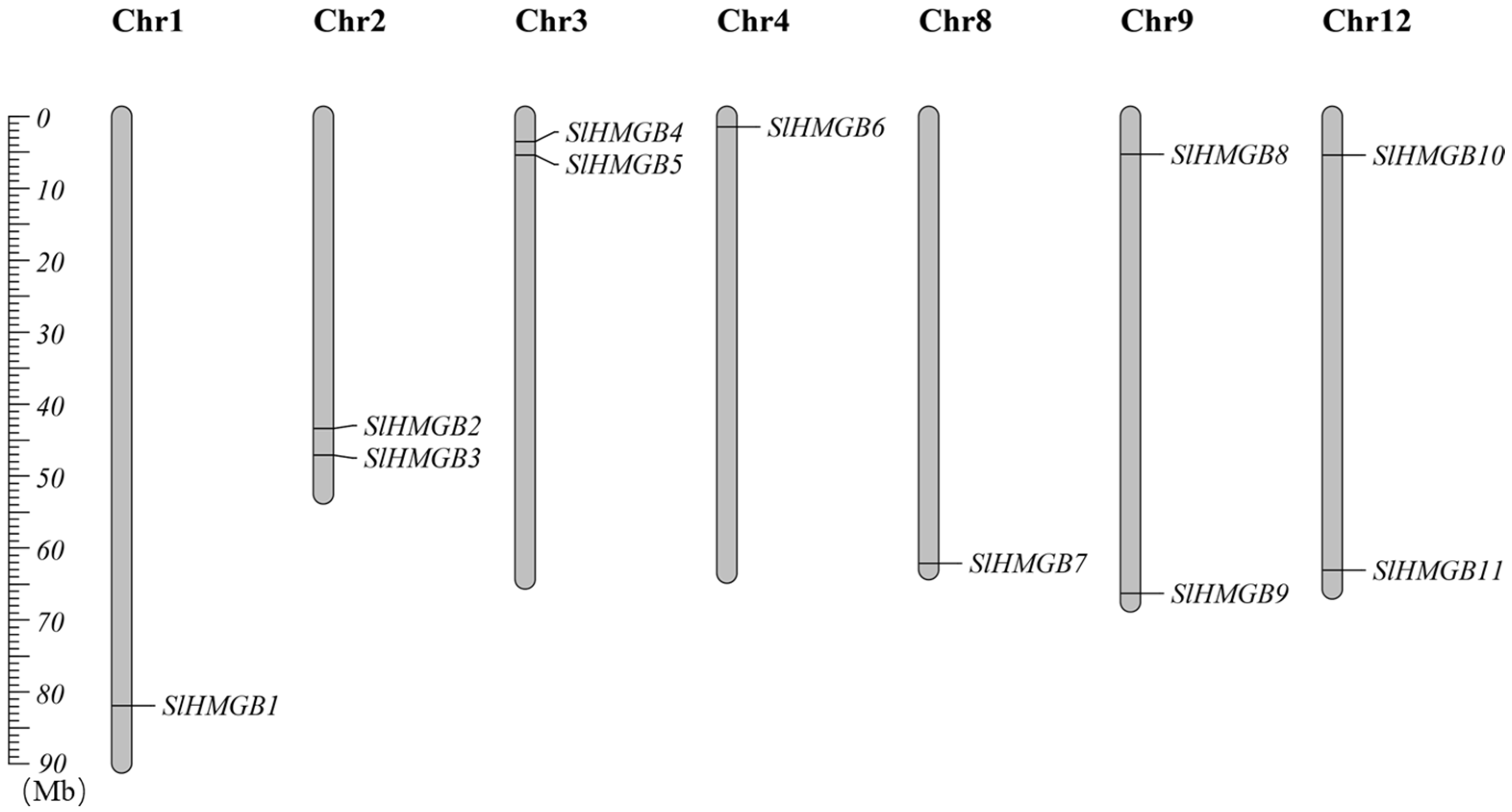
In addition, we performed chromosomal localization of SlHMGBs, which showed that they are distributed on seven chromosomes of tomato. Precisely, SlHMGB1, SlHMGB6, and SlHMGB7 are localized on chromosome 1, chromosome 4, and chromosome 8, respectively. Additionally, two genes mentioned in the parenthesis are localized on each of chromosome 2 (SlHMGB2 and SlHMGB3), chromosome 3 (SlHMGB4 and SlHMGB5), chromosome 9 (SlHMGB8 and SlHMGB9), and chromosome 12 (SlHMGB10 and SlHMGB11) (Figure 1).
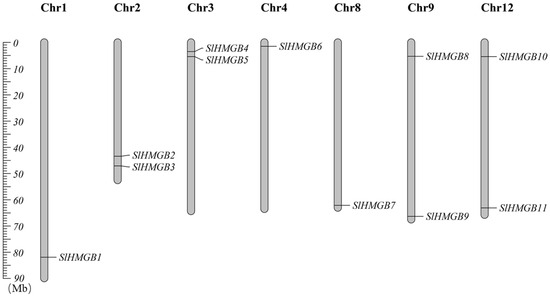
Figure 1.
Chromosomal localization of SlHMGB genes in tomato. The different chromosomes are indicated by vertical gray bars with different lengths. The positions of the genes are shown in order from top to bottom.
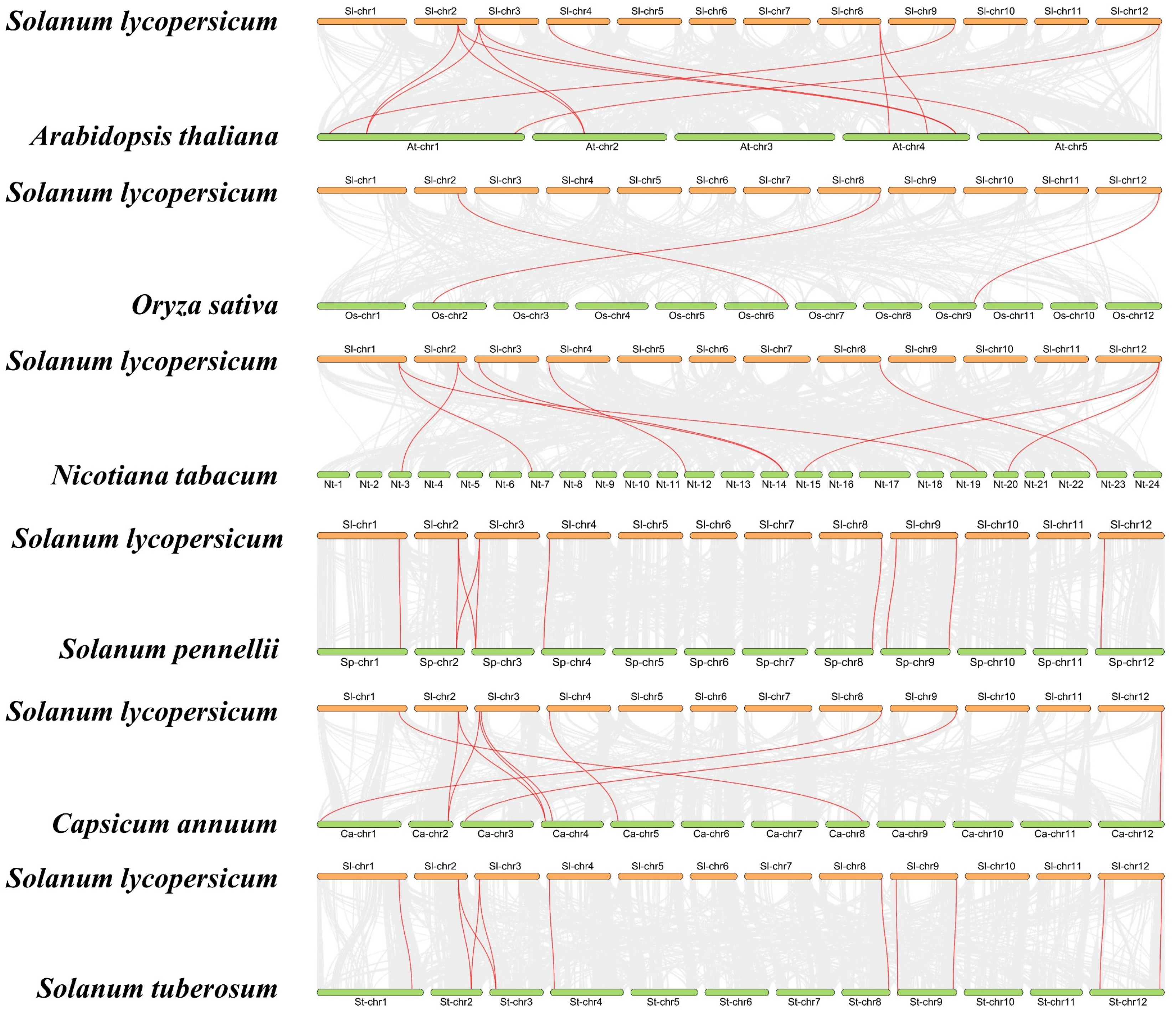
To gain a deeper understanding of the evolutionary clues about SlHMGB members, we conducted covariance analysis between tomato and other plant species, including five dicotyledonous and one monocotyledonous plant. Many genes were identified in these species that showed covariance with the tomato HMGB genes, such as eleven pairs in tomato and Arabidopsis, ten pairs in tomato and Solanum pennellii, nine pairs in tomato and tobacco (Nicotiana tabacum), ten pairs in tomato and pepper (Capsicum annuum), eleven pairs in tomato and potato (Solanum tuberosum), and three pairs in tomato and rice (Oryza sativa) (Figure 2, Table S1). After removing duplicated genes, we found that nine, eight, eight, six, and six SlHMGBs are homologous to HMGB genes of potato, Solanum pennellii, pepper, Arabidopsis, and tobacco, respectively, whereas only three SlHMGB members are homologous to rice HMGB genes, suggesting a closer evolutionary relationship of HMGBs between tomato and dicotyledonous plants. Only two members, SlHMGB2 and SlHMGB7, showed covariation of genes in all five dicotyledonous plants and rice (Table S2). In addition, SlHMGB2 and SlHMGB7 and their covariance genes might have evolved from the coevolution of dicotyledons and monocotyledons, whereas other SlHMGB members might have a common origin in dicotyledons.
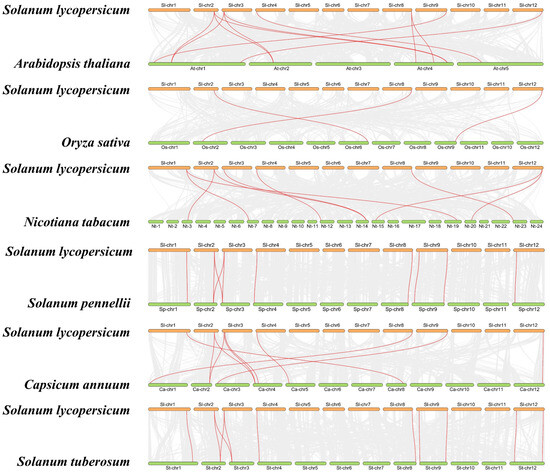
Figure 2.
Syntenic analyses of HMGB genes between tomato and other plant species. Gray lines in the background indicate all syntenic blocks between two genomes. The syntenic HMGB gene pairs are marked by red lines.
2.2. Gene Structure and Conserved Domains of SlHMGBs
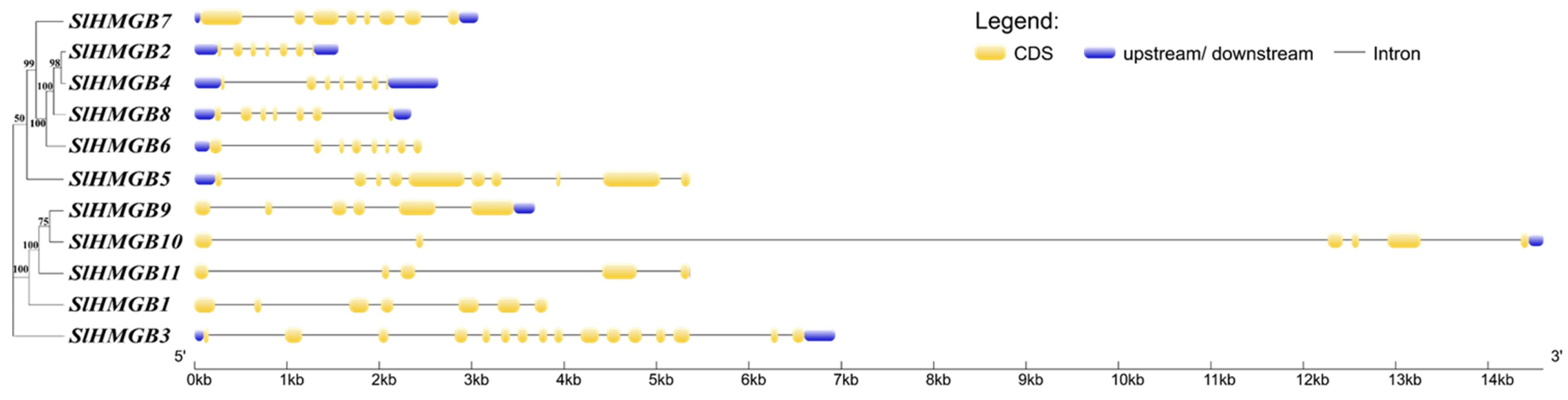
The structures of the SlHMGB genes were obtained by comparing the CDS sequence with the genomic sequence using the GSDS online website. The analysis of gene structures shows that the number of exons in the SlHMGB gene family ranges from 5 to 14, with most of the genes containing seven exons and two genes (SlHMGB3 and SlHMGB5), possessing more than ten exon numbers, and SlHMGB11 contains only five exons (Figure 3).
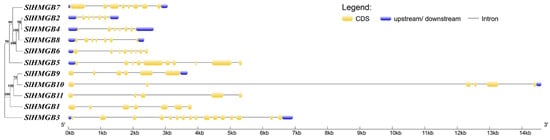
Figure 3.
Structure of SlHMGB genes. The coding sequence (CDS), upstream/downstream untranslated region (UTR), and intron are indicated by yellow boxes, blue boxes, and black lines, respectively.
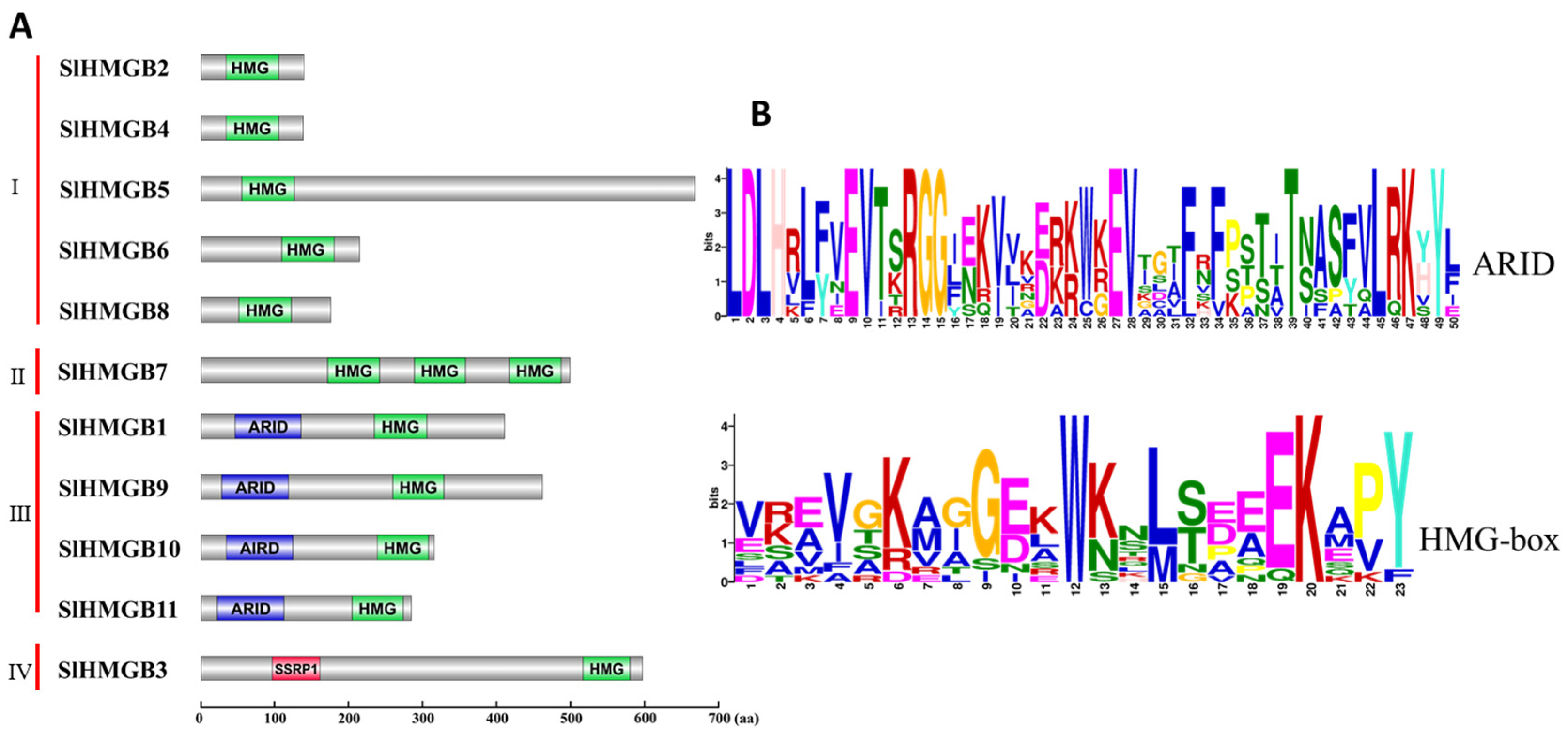
In addition, protein domain analysis of SlHMGBs and multiple sequence alignment of tomato, Arabidopsis, and rice HMGB proteins were performed. Based on the results, we categorized the SlHMGB family into four groups. Group I includes five members, SlHMGB2, 4, 5, 6, and 8, which have only one HMG-box domain; group II has one member, SlHMGB7, that contains three HMG domains; SlHMGB1, 9, 10, and 11, possessing one ARID domain and one HMG-box, are assigned to group III; and SlHMGB3 belonging to group IV has one SSRP1 domain and one HMG-box (Figure 4A and Figures S1–S4). The conserved motifs of SlHMGBs were further investigated by the MEME website (Version 5.5.5), and the sequence logos showed that the ARID-domain and HMG-box exhibit low conservation levels among SlHMGB proteins (Figure 4B).
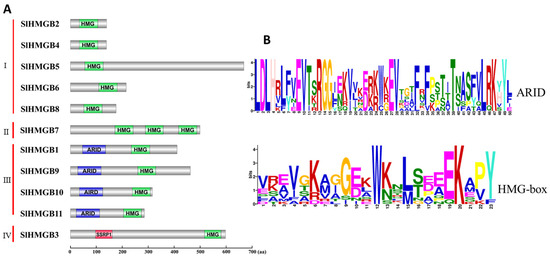
Figure 4.
Analysis of conserved domains of SlHMGB proteins. (A) Protein domain structures of SlHMGB proteins. Based on the different domain structures, the SlHMGB family was divided into four groups. HMG-box, ARID, and SSRP1 domains are marked in different colors. (B) Amino acid sequences of ARID and HMG-box domains of SlHMGB proteins. The logos of the ARID domain were generated from the alignment of four SlHMGB sequences in group III by the MEME website, and the HMG-box logos were obtained from all 11 SlHMGB members. The font size represents the conservation level of amino acids. The larger the font size, the higher the conservation.
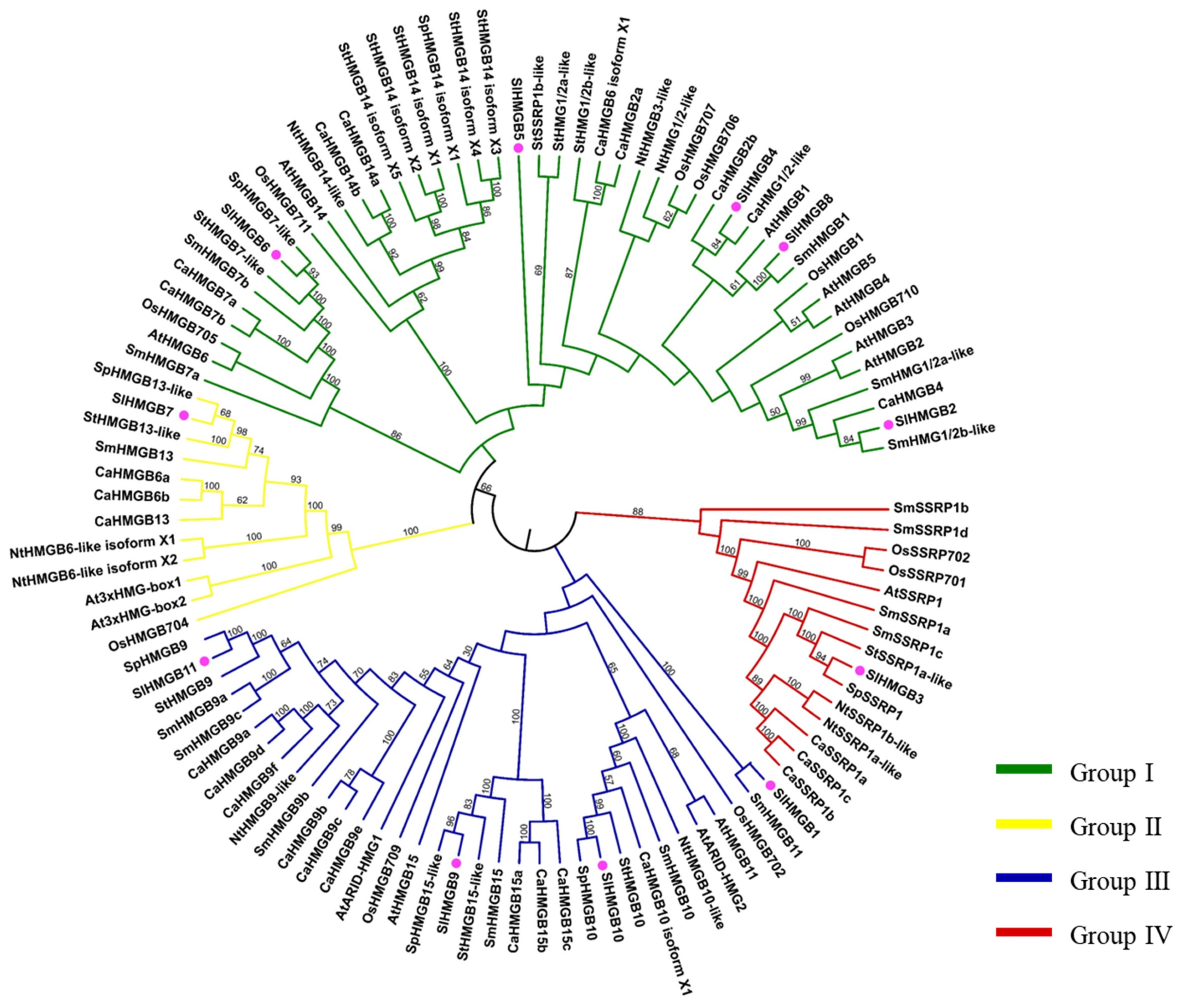
2.3. Phylogenetic Analysis of HMGB Members in Tomato
To investigate the evolutionary relationship among HMGB homologs, a total of eighty-five HMGB proteins from eight plant species, including Solanum lycopersicum (Sl), Arabidopsis thaliana (At), Oryza sativa (Os), Solanum melongena (Sm), Capsicum annuum (Ca), Solanum pennellii (Sp), Nicotiana tabacum (Nt), and Solanum tuberosum (St), were obtained and used for phylogenetic tree construction. As shown in Figure 5, these HMGB homologs were divided into four groups. In the tomato SlHMGB family, there is one protein in each of group II and group IV, four members in group III, and five proteins in group I, which are consistent with the results of protein domain analysis and sequence alignment (Figure 4A and Figures S1–S4).
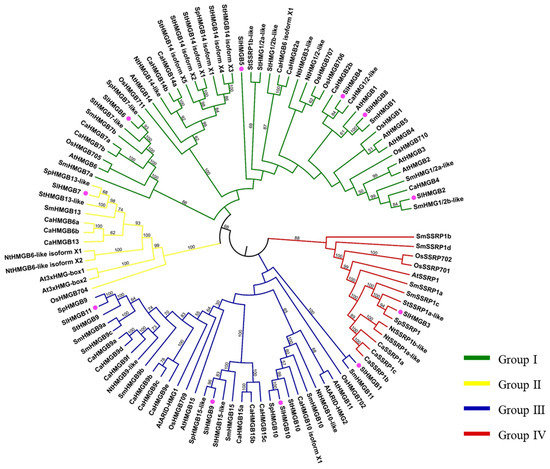
Figure 5.
Phylogenetic analysis of HMGB homologs in different species. The clades of groups I, II, III, and IV are marked in green, yellow, blue, and red, respectively. Tomato SlHMGBs are indicated by pink balls. At, Arabidopsis thaliana; Ca, Capsicum annuum; Nt, Nicotiana tabacum; Os, Oryza sativa; Sl, Solanum lycopersicum; Sp, Solanum pennellii; St, Solanum tuberosum.
2.4. Cis-Acting Element Analysis of SlHMGB Promoters
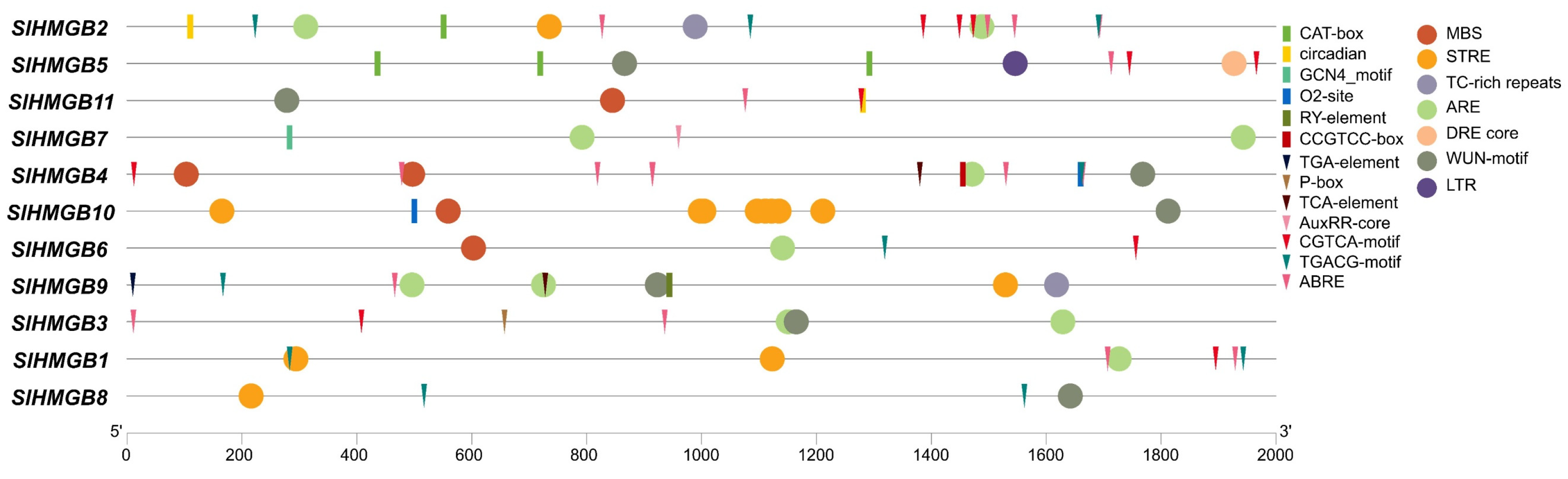
We extracted the 2 kb promoter sequences of SlHMGB genes using Tbtools (Version 2.096) and submitted them to PlantCARE (Version 1) for the identification of cis-acting elements. In addition to the common promoter elements, we obtained a total of twenty different types of cis-acting elements, which can be categorized into three major groups: plant growth and development, phytohormone responsiveness, and stress responsiveness. The CAT-box, circadian, O2-site, CCGTCC-box, GCN4_motif, and RY-element, which are involved in meristem expression, circadian control, zein metabolism, meristem activation, endosperm expression, and seed development, respectively, belong to the plant growth and development category. The phytohormone responsiveness group includes the elements of ABRE responsible for abscisic acid response, TGACG-motif and CGTCA-motif for methyl jasmonate response, P-box for gibberellin response, TCA-element for salicylic acid response, and AuxRR-core and TGA-element for auxin response. In the stress responsiveness category, the elements of ARE important for anaerobic induction response, STRE for stress response, TC-rich repeats for defense response, WUN-motif for wound response, MBS and DRE core for dehydration response, and LTR for low-temperature response are detected. Additionally, each SlHMGB gene contains different types and numbers of cis-acting elements; however, the elements related to phytohormone responsiveness or stress responsiveness are more than those related to plant growth and development in most SlHMGB genes (Figure 6).
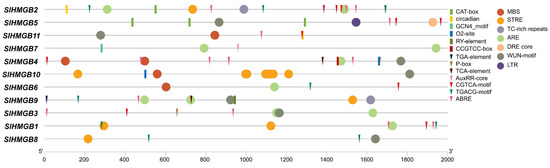
Figure 6.
Cis-element analysis in the promoters of SlHMGB genes. The elements belonging to plant growth and development, phytohormone responsiveness, and stress responsiveness categories are represented by rectangles, triangles, and circles with different colors, respectively.
2.5. Analysis of Tissue-Specific Expression Patterns of SlHMGB Genes
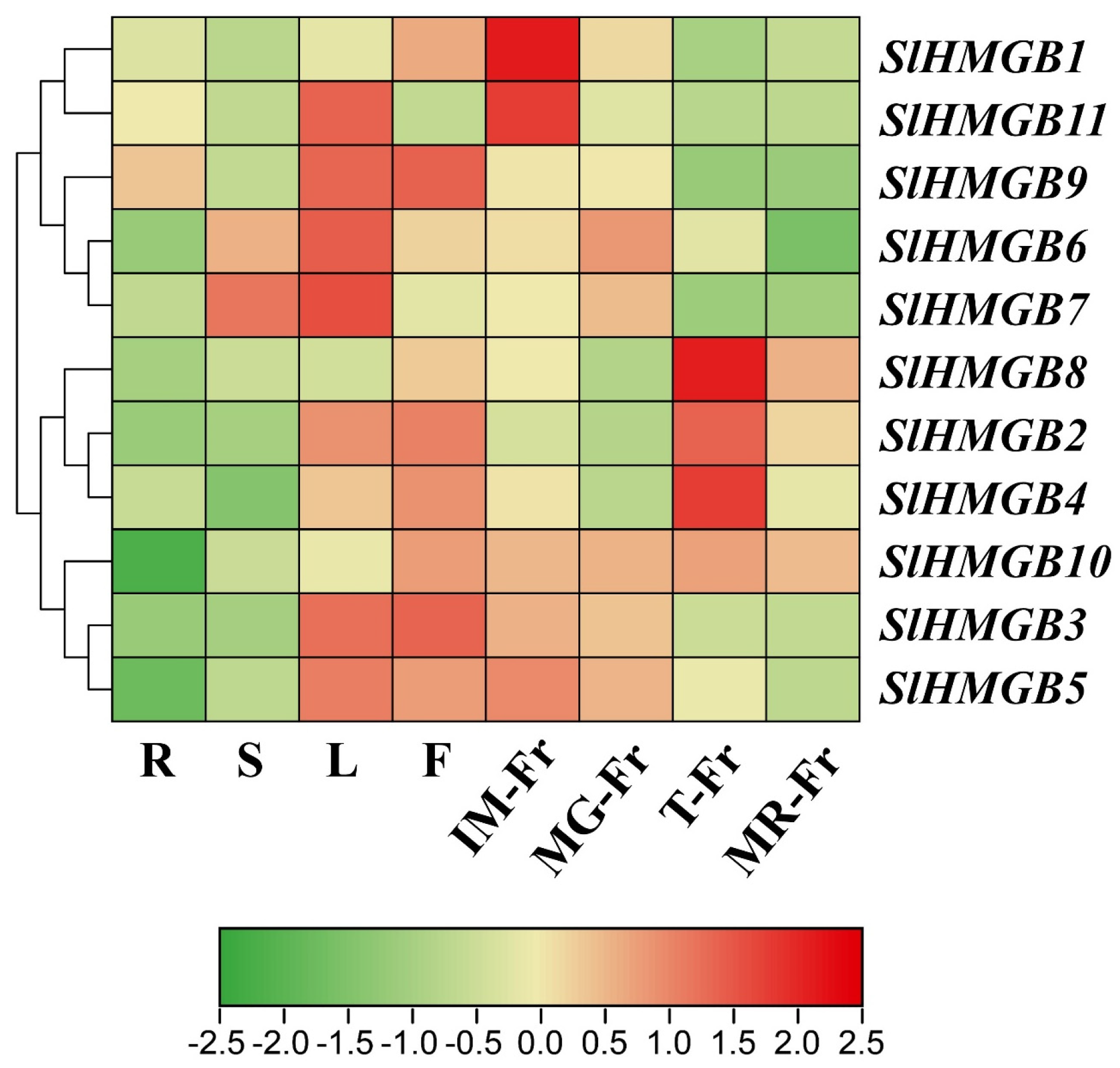
To predict the potential functions of the SlHMGB genes, we performed qRT-PCR analysis on different tomato tissues, including roots, stems, leaves, flowers, and fruits at different developmental stages. Then, a heatmap was constructed based on the results of qRT-PCR using TBtools software (Version 2.096). As shown in Figure 7, both SlHMGB1 and SlHMGB11 had the highest expression in the fruits at the mature green stage (MG-Fr). SlHMGB6, SlHMGB7, and SlHMGB9 were mainly expressed in the leaves, whereas SlHMGB6 and SlHMGB7 also showed higher expressions in the stems and immature fruits (IM-Fr), and SlHMGB9 had a higher transcript level in the flowers. SlHMGB2, SlHMGB4, and SlHMGB8 displayed the highest expression in the fruits at the turning stage (T-Fr) and the lowest expression in the roots and stems. SlHMGB3 and SlHMGB5 were principally distributed in the leaves, flowers, and fruits at the immature and mature green stages. Moreover, a higher expression of SlHMGB10 was detected in the flowers and fruits at all stages of maturity. Taken together, SlHMGB members show diverse tissue-specific expression profiles, indicating their potential roles in different aspects of plant growth and development, and there may be functional redundancy and divergence among these genes.
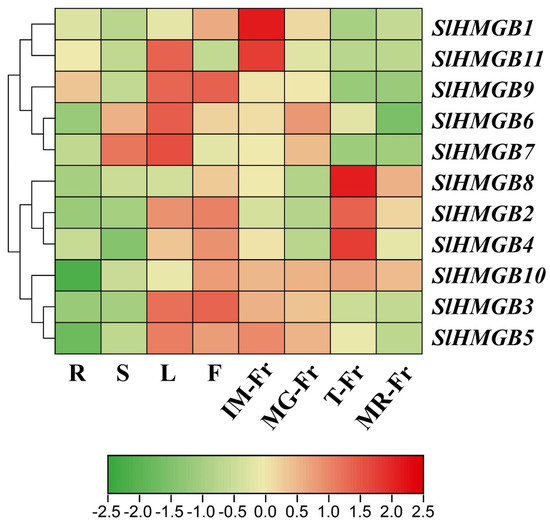
Figure 7.
The tissue-specific expression patterns of SlHMGB genes. Log2 normalization of qRT-PCR data was performed by TBtools software (Version 2.096) and used to construct the heatmap. R, roots; S, stems; L, leaves; F, flowers; IM-Fr, immature fruits; MG-Fr, mature green fruits; T-Fr, fruits at the turning stage; MR-Fr, mature red fruits. The numerical values in the box represent the values of log2 normalization of qRT-PCR data. The larger the numerical value, the higher the gene expression level.
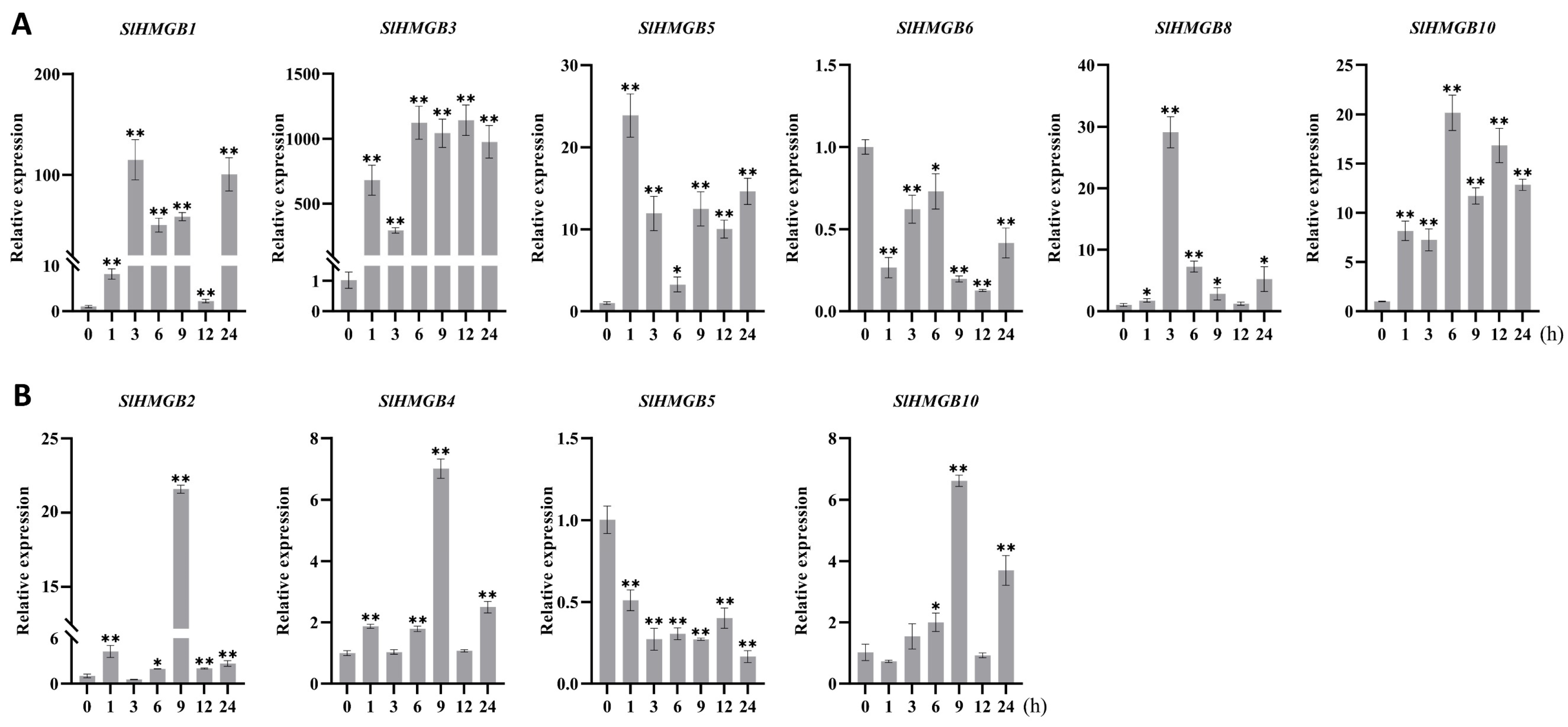
2.6. Expression Analysis of SlHMGB Genes under Abiotic Stress
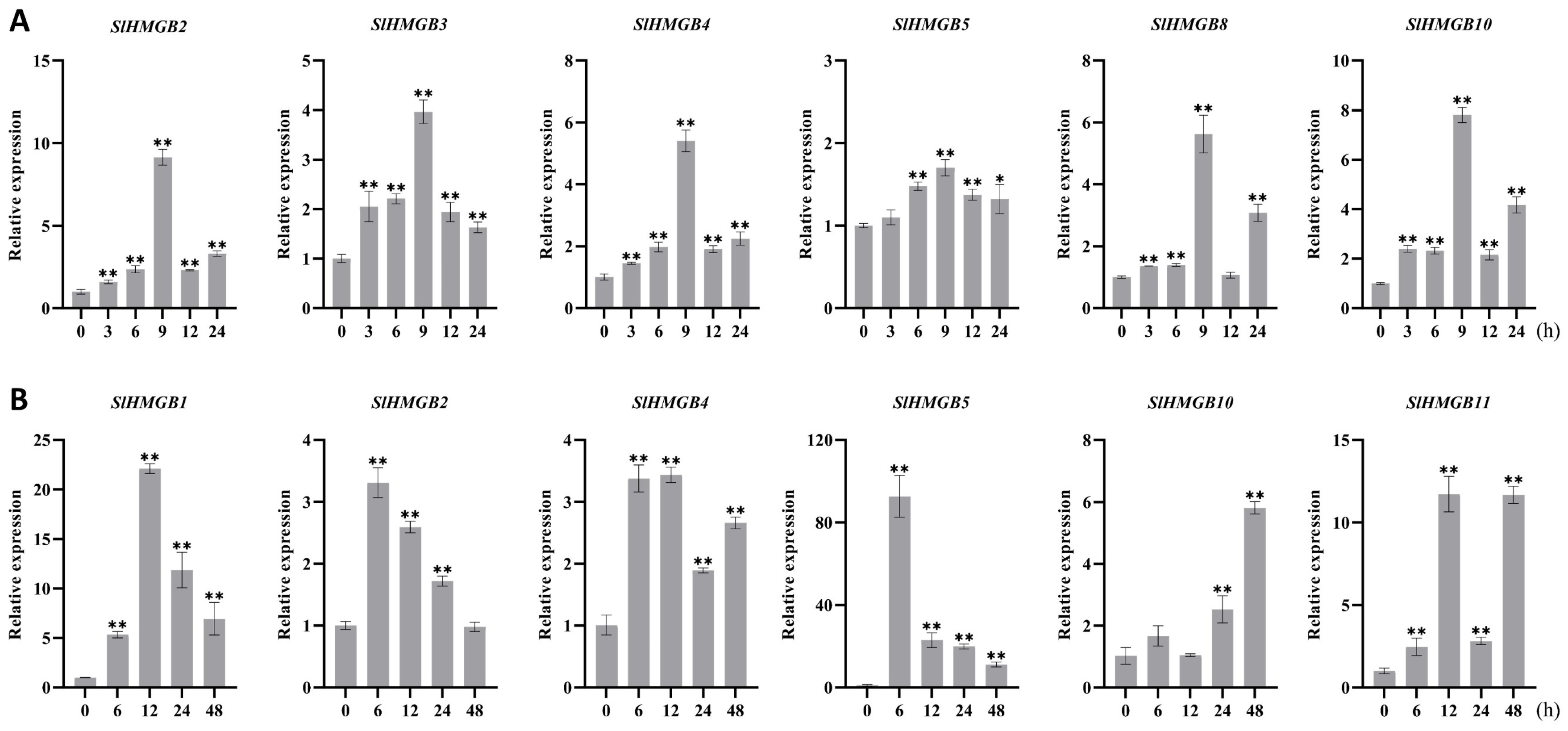
The cis-acting elements of SlHMGB promoters were detected to be related to stress responsiveness, suggesting that SlHMGB genes may play important roles in plant response to environmental stress (Figure 6). Therefore, we further analyzed the expressions of SlHMGB genes under salt, drought, heat, and cold treatments. Overall, the SlHMGB members showed different expression patterns under different abiotic stresses. For example, under salt treatment, six genes, SlHMGB2, SlHMGB3, SlHMGB4, SlHMGB5, SlHMGB8, and SlHMGB10, were up-regulated, and all the peaks occurred at 9 h after treatment (Figure 8A). In addition, the other five genes showed disordered expression patterns after salt treatment (Figure S5A). Upon imposition of drought stress by PEG-6000, six SlHMGB genes showed up-regulation in their expression patterns. With a few exceptions, the expression levels of SlHMGB1, SlHMGB2, SlHMGB4, SlHMGB5, SlHMGB10, and SlHMGB11 were higher than the control during the whole treatment period, but their peaks occurred at different time points (Figure 8B). Moreover, the additional five SlHMGB members displayed inconsistent expression trends at various time points under PEG treatment (Figure S5B).
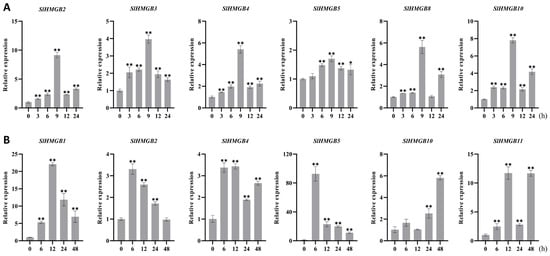
Figure 8.
qRT-PCR analyses of SlHMGB genes under salt and drought stresses. (A,B) Relative expression levels of SlHMGBs under salt (A) and drought (B) treatments. Each value is the mean ± SD of three biological replicates. The asterisks indicate significant differences between abiotic stress treatments (different time points) and the control (0 h) by Student’s t-tests (* p < 0.05, ** p < 0.01).
Furthermore, six SlHMGB genes responded to heat stress (42 °C); among them, five members were up-regulated and one member was down-regulated. In detail, the increased expressions of SlHMGB1, SlHMGB3, SlHMGB5, and SlHMGB10 were discovered at each time point after heat treatment, while SlHMGB8 expression was induced at most time points but was unchanged at 12 h. The transcript level of SlHMGB6 was reduced during the whole treatment period (Figure 9A). The remaining five SlHMGB members showed confused expression profiles under drought stress (Figure S6A). Under cold stress (4 °C), three and one SlHMGB genes were positively and negatively regulated, respectively. The expressions of SlHMGB2, SlHMGB4, and SlHMGB10 were increased at partial time points and unchanged at other points compared with the control, while the SlHMGB5 level was decreased throughout the treatment period (Figure 9B). The rest of the SlHMGB members exhibited disorganized expression patterns (Figure S6B).
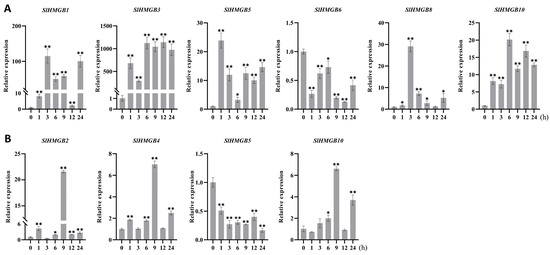
Figure 9.
qRT-PCR analyses of SlHMGBs under heat and cold stresses. (A,B) Relative transcript levels of SlHMGBs under heat (A) and cold (B) treatments. Values are the means ± SD of three biological replicates. The asterisks indicate significant differences between abiotic stress treatments (different time points) and the control (0 h) by Student’s t-tests (* p < 0.05, ** p < 0.01).
Based on the SlHMGB members involved in plant response to the four abiotic stresses, we produced Venn plots (Figure S7). Two genes out of eleven SlHMGB family members, SlHMGB5 and SlHMGB10, responded to all four abiotic stresses, wherein SlHMGB10 was up-regulated by different treatments, suggesting that this gene might play key roles in tomato stress tolerance. Furthermore, both SlHMGB2 and SlHMGB4 positively responded to salt, drought, and cold stresses. SlHMGB1, SlHMGB3, and SlHMGB8 were positively involved in response to two stresses (SlHMGB1 for drought and heat stresses, SlHMGB3 and SlHMGB8 for salt and heat stresses). SlHMGB6 and SlHMGB11 were only regulated by drought or heat stress, respectively. In conclusion, most SlHMGB genes probably played important roles in mitigating abiotic stresses in tomato, despite showing different roles under different stresses.
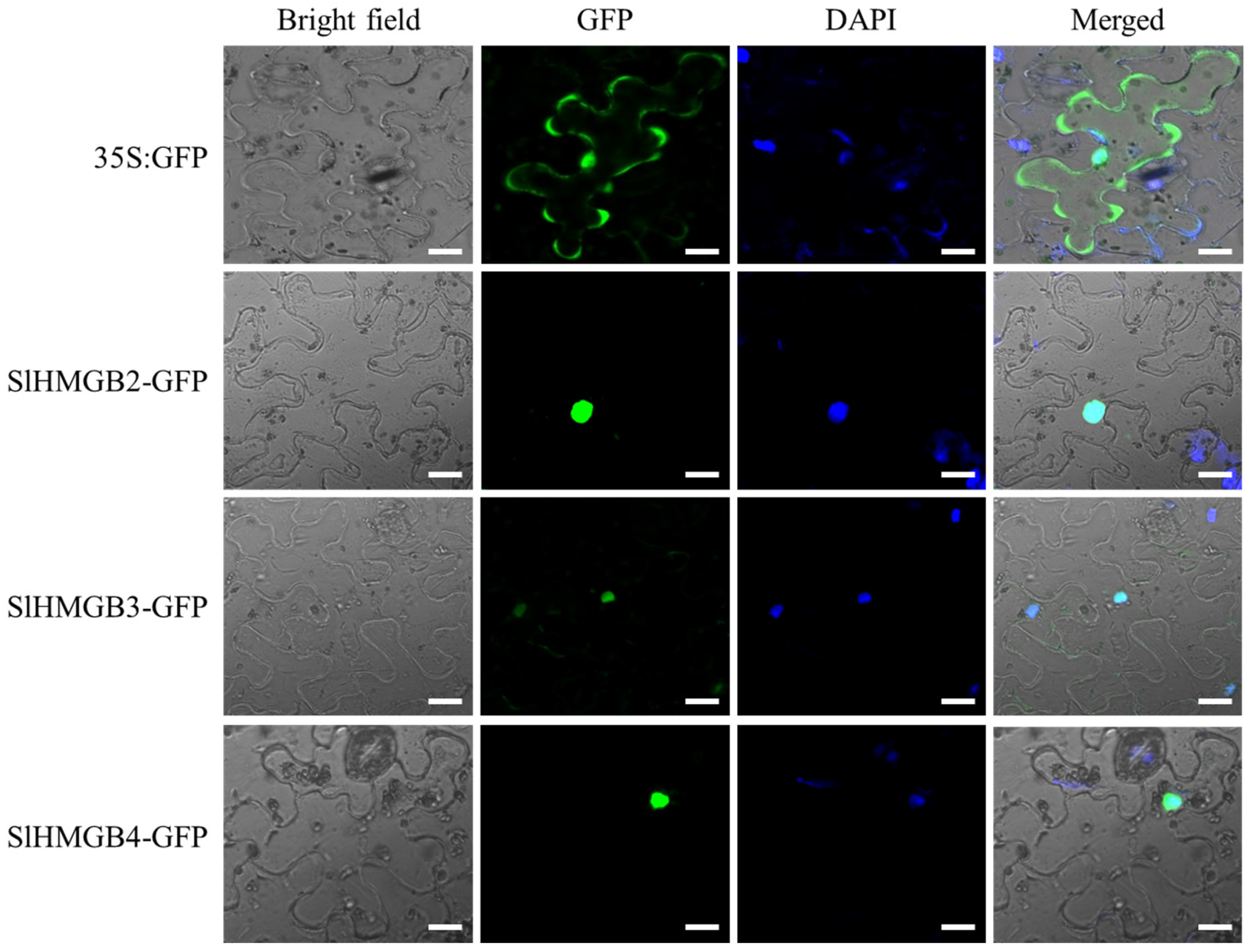
2.7. Subcellular Localization of SlHMGB Proteins
According to the subcellular localization prediction results, all SlHMGB proteins are potentially localized in the nucleus (Table 1). To verify these results, we selected three SlHMGB proteins, SlHMGB2, SlHMGB3, and SlHMGB4, for subcellular localization in tobacco leaves. As shown in Figure 10, SlHMGB2-GFP, SlHMGB3-GFP, and SlHMGB4-GFP fusion proteins were merged with DAPI (a nucleus marker), respectively, indicating that all three proteins were localized to the nucleus, which is consistent with the predicted results. As the control, the 35S:GFP signal was distributed throughout the cell.
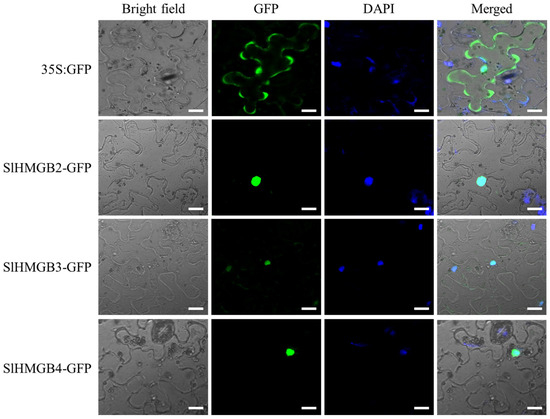
Figure 10.
Subcellular localization of SlHMGB proteins. The constructs of 35S:GFP, SlHMGB2-GFP, SlHMGB3-GFP, SlHMGB4-GFP, and DAPI were co-transformed into the tobacco leaves. 35S:GFP served as a control. The fluorescence signals were detected by a laser-scanning confocal microscope. Bars = 20 μm.
3. Discussion
Until now, the HMGB genes have been extensively identified in many plant species, such as Arabidopsis [8,9], rice [15,16], maize [10], Vicia faba [28], Canavalia gladiata [29], Pharbitis nil [30], soybean [12], and pea [14]. However, SlHMGBs in tomato have received little attention to date. In this study, we identified a total of 11 tomato HMGB genes (Table 1), which were named SlHMGB1-SlHMGB11 based on their locations on the chromosome. Through protein structural domain analysis and amino acid multiple sequence alignment, we found that SlHMGB proteins possess a conserved HMG-box domain (Figure 4), a typical structure of the HMGB family. This domain can mediate non-sequence-specific or sequence-specific DNA binding [6]. However, multiple sequence alignment and motif analyses revealed that the HMG-box domains exhibit low similarity among HMGB homologs of different plant species, as well as among different SlHMGB members in tomato (Figures S1–S4 and Figure 4B). This suggests that HMGBs display low conservation during evolution.
Furthermore, the phylogenetic tree categorizes the HMGB members into four groups based on the structural domains. Group I includes the members containing the HMG-box, group II has the members containing the 3xHMG-box, group III possesses the members containing the ARID domain, and group IV has the members containing the SSRP1 box (Figure 5). The HMGB gene pairings were compared between tomato and other species, such as Arabidopsis, rice, tobacco, Solanum pennellii, pepper, and potato (Figure 2, Tables S2 and S3). The results showed that SlHMGB exhibits a close evolutionary relationship with its homologs in dicotyledonous plants. In addition, SlHMGB2 and SlHMGB7 have syntenic pairs in both dicotyledons and monocotyledons (Table S2), suggesting that these homologs may have existed prior to divergence.
HMGB genes have previously been proved to modulate plant growth and development, such as seed germination [23], leaf growth, flowering induction [31], pollen maturation, pollen tube development [17], reduction in seed dormancy [32], and anthocyanin accumulation [33]. Herein, our results showed that the SlHMGB genes exhibit diverse expression patterns in different tomato tissues (Figure 7). Expressions of SlHMGB genes are generally low in roots, except SlHMGB9. Two members are highly expressed in the stems, eight members in the leaves, nine members in the flowers, five members in the IM-fruits, five members in the MG-fruits, four members in the T-fruits, and two members in the MR-fruits. This implied the potentially important roles of the SlHMGB family in the growth and development of different organs. Moreover, some SlHMGB members show similar expression patterns, indicating they may have redundant functions in plant growth and development of tomato.
Moreover, HMGB genes have been functionally characterized in some plant species, especially Arabidopsis [13,14,15,16,29,34,35,36]. Here, we deduced the functional similarity and discrepancy between tomato and Arabidopsis HMGB homologs based on the tissue-specific expression patterns (Figure 7), syntenic analyses (Figure 2, Table S2), and previous studies. For example, AtHMGB2 is syntenic with SlHMGB2 and SlHMGB4 (Figure 2), and SlHMGB2 and SlHMGB4 are up-regulated under salt, drought, and cold stresses (Figure 8 and Figure 9), suggesting that they may play important roles in mitigating abiotic stress in tomato, whereas AtHMGB2 expression is induced by cold stress and inhibited by drought and salt stresses [23], indicating that AtHMGB2 also responds to abiotic stress, but the effects are different from SlHMGB2 and SlHMGB4 under diverse stresses. SlHMGB9 is principally expressed in the flowers (Figure 7), revealing its potential involvement in flower development. Correspondingly, its syntenic gene, AtHMGB15, has been demonstrated to play crucial roles in pollen development and pollen tube growth in Arabidopsis [17]. This suggests that there may be similar functions between SlHMGB9 and AtHMGB15. Based on these findings, we speculated that tomato SlHMGB genes may exhibit conserved and distinct functions with their homologs of other plant species.
In Arabidopsis, cold stress results in a significant increase in the transcript levels of AtHMGB2, AtHMGB3, and AtHMGB4, while drought or salt stress suppresses the expression of AtHMGB2 and AtHMGB3 [23]. By analyzing the qPCR results in this study, we found that tomato SlHMGB genes also respond to abiotic stresses, for example, salt treatment causes differential expressions of six genes, all of which are up-regulated (Figure 8A), while the drought and heat treatments lead to increased expressions of five genes and decreased expression of one gene (Figure 8B and Figure 9A). Three genes are positively regulated, and one gene is negatively regulated by cold treatment (Figure 9B). Overall, the number of up-regulated SlHMGB genes significantly outnumbers the down-regulated genes under various abiotic stresses, suggesting that the SlHMGB gene family may play a crucial role in mitigating abiotic stress in tomato. In addition, Venn diagrams of the qPCR results showed that two SlHMGB genes are involved in response to four abiotic stresses (Figure S7), of which SlHMGB10 is significantly up-regulated under all treatments (Figure 8 and Figure 9). This indicates that SlHMGB10 may play a critical role in enhancing stress tolerance in tomato. In conclusion, the tomato SlHMGB family genes are extensively involved in response to multiple abiotic stresses.
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatments
The tomato cultivar Micro-Tom was used in this study. The plants were cultivated in a greenhouse at Shandong Agricultural University (Tai’an, China; 34° N, 117° E) with a daytime temperature of 25 °C for 16 h and a nighttime temperature of 18 °C for 8 h. Field management was carried out following the standard practices.
To analyze the tissue-specific expression, the roots, stems, and leaves from the plants at the four true-leaf stage, fully opened flowers, and fruits at the immature (IM, 15 days post anthesis), mature green (MG), turning (T), and mature red (MR) stages were collected for RNA extraction. Three biological replicates were prepared for each tissue.
For salt stress, the tomato seedlings were cultivated in Hoagland’s nutrient solution and then subjected to 150 mM salt treatment at the four true-leaf stage. At 0 h, 3 h, 6 h, 9 h, 12 h, and 24 h after treatment, the leaves were collected, respectively. Additionally, the plants cultivated in the pots containing the matrix were prepared for drought, heat, and cold treatments, and the matrix conditions (including volume and composition) are the same in every pot under each treatment. For drought stress, the plants at the four-week-old stage were watered with a solution containing 5% polyethylene glycol (PEG)-6000, and a 50 mL solution was used for each plant. After treatment, the leaf samples were collected at 0 h, 6 h, 12 h, 24 h, and 48 h, respectively. With heat and cold treatments, the plants at the four-week-old stage were exposed to 42 °C and 4 °C, respectively. The leaf samples were then collected at various time points after treatments, including 0 h, 1 h, 3 h, 6 h, 9 h, 12 h, and 24 h. Three biological replicates were performed for each treatment.
4.2. Identification of HMGB Genes in Tomato
To identify the HMGB members in tomato, all tomato protein sequences were downloaded from the Solanaceae genome, and the BLAST searches were performed by TBtools software (Version 2.096) using the full-length amino acid sequences of Arabidopsis and rice HMGBs with a blast E-value of 10−5 [37]. Then, the retrieved HMGB candidate sequences were submitted to the InterPro website (https://www.ebi.ac.uk/interpro/ accessed on 21 May 2023) to annotate the structural domains [38], and only the candidates containing the HMG-boxes were referred to as HMGB homologs.
TBtools software (Version 2.096) was used to analyze the physicochemical properties of tomato SlHMGB proteins, including molecular weight and isoelectric point [39]. Subcellular localization prediction was performed using the WoLF PSORT (https://www.genscript.com/wolf-psort.html, accessed on 11 November 2022) [40].
4.3. Gene Structure, Chromosome Mapping, and Syntenic Analysis
Tomato genome annotation files were used to obtain CDS, exon, and UTR information, which were then submitted to the Gene Structure Display Server online software (https://gsds.gao-lab.org/, accessed on 8 February 2023) to map the gene structure schematically [41].
The chromosome location of the SlHMGB genes was detected by TBtools software using the tomato genome annotation file. Subsequently, a map was generated through Mapchart software (Version 2.32) to visualize the distribution of SlHMGB genes on the chromosomes [42].
Genome sequence files and annotation files of different species (Solanum lycopersicum, Solanum pennellii, Solanum tuberosum, Nicotiana tabacum, Capsicum annuum, Arabidopsis thaliana, and Oryza sativa) were downloaded from SGN (https://solgenomics.net/, accessed on 20 June 2023), TAIR (https://www.arabidopsis.org/, accessed on 20 June 2023), and MSU-RGAP (http://rice.uga.edu/, accessed on 20 June 2023) databases. Then the syntenic analysis was performed using the One Step MCScanX-Super fast program within the TBtools software (Version 2.096) package, and the syntenic plots between tomato and other species were visualized by the Dual Synteny Plot program with in the TBtools (Version 2.096). The co-linear SlHMGB gene pairs were highlighted [43].
4.4. Protein Structural Domain and Motif Analysis
The sites of protein structural domains were obtained according to the NCBI online software (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 11 March 2023) [44], and then the DOG software (Version 2.0) was used to draw the schematic diagram of protein structural domains [45].
A comparison of major structural domains of tomato HMGB protein, including HMG-box, ARID, and SSRP elements, was performed using the MEME online program (https://meme-suite.org/meme/tools/meme, v5.1.1, accessed on 11 March 2023) [46].
4.5. Phylogenetic Analysis
To further understand the evolutionary relationship of SlHMGB with HMGB in other species, HMGB protein sequences of Arabidopsis thaliana (At), Oryza sativa (Os), Solanum melongena (Sm), Capsicum annuum (Ca), Solanum Pennellii (Sp), Nicotiana tabacum (Nt), and Solanum tuberosum (St) were obtained from the Solanaceae Genomics Network and NCBI databases. The phylogenetic tree was constructed using the neighbor-joining method in MEGA-X software (Version 10.2.6) [47]. The bootstrap method was selected for the test of phylogeny, the number of repetitions was set to 1000, and the Poisson model was selected for the Model/Method [48]. Subsequently, the phylogenetic tree was annotated in detail by the iTOL tool (https://itol.embl.de/, accessed on 15 April 2023) [49].
4.6. Cis-Acting Elements Prediction
The 2000 bp sequence upstream of the promoter of the SlHMGB gene was extracted by TBtools and then submitted to the online database PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 20 April 2023)to predict the cis-acting elements [50].
4.7. Expression Analyses by Quantitative Real-Time PCR (qRT-PCR)
Total RNA from different samples was extracted by the Trizol method and then reverse transcribed into cDNA using the TransGen EasyScript One-Step cDNA Synthesis SuperMix kit (TransGen Biotech, Beijing, China). Primers were designed for the target genes by DNAMAN software (Version 6.0.3.99)and then amplified by fluorescence qRT-PCR using the PerfectStar Green qPCR SuperMix kit with cDNA as a template on an ABI7500 machine. Finally, the relative expression of target genes was calculated by 2−ΔΔCt relative quantitative analysis, and the experiment was performed with three biological replicates. The primers are listed in Supplementary Table S1 [40].
4.8. Subcellular Localization
Four SlHMGB genes were cloned and fused to the pPZP211-GFP vector. Then, the constructs were introduced into Agrobacterium (GV3101). Then, the recombinant Agrobacterium cells were injected into tobacco (Nicotiana benthamiana) leaves. After transformation, the tobacco plants were incubated at 25 °C in darkness for 48 h. The fluorescence signals were observed using an LSM880 laser scanning confocal fluorescence microscope (Carl Zeiss, Oberkochen, Germany) [51].
4.9. Statistical Analysis
Data were analyzed by Microsoft Excel 2021 using three biological replicates, and error bars represent the standard deviation (SD). The significance of differences was assayed using a two-tailed Student’s t-test (* p < 0.05; ** p < 0.01) with SPSS 26.0.
5. Conclusions
In this study, we identified a total of eleven SlHMGB genes, which can be divided into four categories based on the phylogenetic analysis. All SlHMGB proteins contain the conserved HMG-box domain, a characteristic structure of the HMGB family. Syntenic analysis suggested that the HMGB homologs between tomato and other dicots share close evolutionary relationships. Analyses of cis-elements and gene expression patterns imply that SlHMGB genes may play a role in regulating organ growth, development, and response to multiple abiotic stresses in tomato. Furthermore, subcellular localization detected nuclear localization of SlHMGB2, SlHMGB3, or SlHMGB4. Thus, our data provide valuable insights into the potential biological functions of SlHMGB genes in tomato.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25115850/s1.
Author Contributions
Investigation, methodology, data curation, formal analysis, and writing—original draft, J.Z.; methodology, data curation, and formal analysis, H.T.; methodology and formal analysis, J.W.; methodology and formal analysis, Y.L.; data curation and formal analysis, L.G.; data curation and formal analysis, G.L.; methodology and data curation, Q.S.; conceptualization, methodology, supervision, project administration, and writing—review and editing, Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32372798, 32372723) and the Key Research and Development Program of Shandong Province (2023CXPT018, 2022TZXD0025).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data supporting the findings of this study are available within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Goodwin, G.H.; Sanders, C.; Johns, E.W. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur. J. Biochem. 1973, 38, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Bustin, M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 2001, 26, 152–153. [Google Scholar] [CrossRef] [PubMed]
- Grasser, K.D.; Grill, S.; Duroux, M.; Launholt, D.; Thomsen, M.S.; Nielsen, B.V.; Nielsen, H.K.; Merkle, T. HMGB6 from Arabidopsis thaliana specifies a novel type of plant chromosomal HMGB protein. Biochemistry 2004, 43, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
- Grasser, K.D.; Launholt, D.; Grasser, M. High mobility group proteins of the plant HMGB family: Dynamic chromatin modulators. Biochim. Biophys. Acta Gene Struct. Expr. 2007, 1769, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Štros, M. HMGB proteins: Interactions with DNA and chromatin. Biochim. Biophys. Acta 2010, 1799, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Štros, M.; Launholt, D.; Grasser, K.D. The HMG-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 2007, 64, 2590–2606. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, D.S.; Coppens, F.; Ma, L.; Antosch, M.; Marktl, B.; Merkle, T.; Beemster, G.T.; Houben, A.; Grasser, K.D. The plant-specific family of DNA-binding proteins containing three HMG-box domains interacts with mitotic and meiotic chromosomes. New Phytol. 2011, 192, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.K.; Williams, L.A.; Brady-Passerini, K.; Brown, R.H.; Gasser, C.S. Positive- and negative-acting regulatory elements contribute to the tissue-specific expression of INNER NO OUTER, a YABBY-type transcription factor gene in Arabidopsis. BMC Plant Biol. 2012, 12, 214. [Google Scholar] [CrossRef] [PubMed]
- Pfab, A.; Grønlund, J.T.; Holzinger, P.; Längst, G.; Grasser, K.D. The Arabidopsis histone chaperone FACT: Role of the HMG-Box domain of SSRP1. J. Mol. Biol. 2018, 430, 2747–2759. [Google Scholar] [CrossRef]
- Grasser, K.D.; Wurz, A.; Feix, G. Isolation and characterization of high-mobility-group proteins from maize. Planta 1991, 185, 350–355. [Google Scholar] [CrossRef]
- Grasser, K.D.; Krech, A.B.; Feix, G. The maize chromosomal HMGa protein recognizes structural features of DNA and increases DNA flexibility. Plant J. 1994, 6, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Laux, T.; Goldberg, R.B. A plant DNA binding protein shares highly conserved sequence motifs with HMG-box proteins. Nucleic Acids Res. 1991, 19, 4769. [Google Scholar] [CrossRef] [PubMed]
- Spiker, S. High-mobility group chromosomal proteins of wheat. J. Biol. Chem. 1984, 259, 12007–12013. [Google Scholar] [CrossRef]
- Webster, C.I.; Packman, L.C.; Pwee, K.H.; Gray, J.C. High mobility group proteins HMG-1 and HMG-I/Y bind to a positive regulatory region of the pea plastocyanin gene promoter. Plant J. 1997, 11, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhang, W.; Pwee, K.-H.; Kumar, P.P. Cloning and characterization of rice HMGB1 gene. Gene 2003, 312, 103–109. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, W.; Pwee, K.-H.; Kumar, P.P. Rice HMGB1 protein recognizes DNA structures and bends DNA efficiently. Arch. Biochem. Biophys. 2003, 411, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Wang, Y.J.; Liang, Y.; Niu, Q.K.; Tan, X.Y.; Chu, L.C.; Chen, L.Q.; Zhang, X.Q.; Ye, D. The ARID-HMG DNA-binding protein AtHMGB15 is required for pollen tube growth in Arabidopsis thaliana. Plant J. 2014, 79, 741–756. [Google Scholar] [CrossRef]
- Bianchi, M.E.; Manfredi, A.A. Dangers in and out. Science 2009, 323, 1683–1684. [Google Scholar] [CrossRef]
- Dinesh-Kumar, S.P.; Choi, H.W.; Manohar, M.; Manosalva, P.; Tian, M.; Moreau, M.; Klessig, D.F. Activation of plant innate immunity by extracellular high mobility group box 3 and its inhibition by salicylic acid. PLoS Pathog. 2016, 12, e1005518. [Google Scholar]
- Lichota, J.; Ritt, C.; Grasser, K.D. Ectopic expression of the maize chromosomal HMGB1 protein causes defects in root development of tobacco seedlings. Biochem. Biophys. Res. Commun. 2004, 318, 317–322. [Google Scholar] [CrossRef]
- Lildballe, D.L.; Pedersen, D.S.; Kalamajka, R.; Emmersen, J.; Houben, A.; Grasser, K.D. The expression level of the chromatin-associated HMGB1 protein influences growth, stress tolerance, and transcriptome in Arabidopsis. J. Mol. Biol. 2008, 384, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Kwak, K.J.; Kang, H. Expression of a high mobility group protein isolated from Cucumis sativus affects the germination of Arabidopsis thaliana under abiotic stress conditions. J. Integr. Plant Biol. 2008, 50, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Kwak, K.J.; Kim, J.Y.; Kim, Y.O.; Kang, H. Characterization of transgenic Arabidopsis plants overexpressing high mobility group B proteins under high salinity, drought or cold stress. Plant Cell Physiol. 2007, 48, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Chinpongpanich, A.; Phean-O-Pas, S.; Thongchuang, M.; Qu, L.-J.; Buaboocha, T. C-terminal extension of calmodulin-like 3 protein from Oryza sativa L.: Interaction with a high mobility group target protein. Acta Biochim. Biophys. Sin. 2015, 47, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Chen, S.; Li, T.; Yu, S.; Zhao, H.; Liu, H.; Luo, L. Overexpression of OsHMGB707, a high mobility group protein, enhances rice drought tolerance by promoting stress-related gene expression. Front. Plant Sci. 2021, 12, 711271. [Google Scholar] [CrossRef]
- Li, S.; Xin, M.; Luan, J.; Liu, D.; Wang, C.; Liu, C.; Zhang, W.; Zhou, X.; Qin, Z. Overexpression of CsHMGB alleviates phytotoxicity and propamocarb residues in cucumber. Front. Plant Sci. 2020, 11, 738. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Liu, Z.; Li, X.; Tan, B.; Wu, J.; Gao, C. Screening and functional identification of salt tolerance HMG genes in Betula platyphylla. Environ. Exp. Bot. 2021, 181, 104235. [Google Scholar] [CrossRef]
- Grasser, K.D.; Wohlfarth, T.; Bäumlein, H.; Feix, G. Comparative analysis of chromosomal HMG proteins from monocotyledons and dicotyledons. Plant Mol. Biol. 1993, 23, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Minamikawa, T. Isolation and characterization of a cDNA encoding a high mobility group protein HMG-1 from Canavalia gladiata D.C. Biochim. Biophys. Acta. 1998, 1396, 47–50. [Google Scholar] [CrossRef]
- Zheng, C.C.; Bui, A.Q.; O’Neill, S.D. Abundance of an mRNA encoding a high mobility group DNA-binding protein is regulated by light and an endogenous rhythm. Plant Mol. Biol. 1993, 23, 813–823. [Google Scholar] [CrossRef]
- Van Lijsebettens, M.; Grasser, K.D. The role of the transcript elongation factors FACT and HUB1 in leaf growth and the induction of flowering. Plant Signal. Behav. 2010, 5, 715–717. [Google Scholar] [CrossRef]
- Michl-Holzinger, P.; Mortensen, S.A.; Grasser, K.D. The SSRP1 subunit of the histone chaperone FACT is required for seed dormancy in Arabidopsis. J. Plant Physiol. 2019, 236, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Pfab, A.; Breindl, M.; Grasser, K.D. The Arabidopsis histone chaperone FACT is required for stress-induced expression of anthocyanin biosynthetic genes. Plant Mol. Biol. 2018, 96, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, T.J.; Arwood, L.J.; Spiker, S.; Guiltinan, M.J.; Thompson, W.F. High mobility group chromosomal proteins bind to AT-rich tracts flanking plant genes. Plant Mol. Biol. 1991, 16, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Ritt, C.; Grimm, R.; Fernandez, S.; Alonso, J.C.; Grasser, K.D. Basic and acidic regions flanking the HMG domain of maize HMGa modulate the interactions with DNA and the self-association of the protein. Biochemistry 1998, 37, 2673–2681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, Q.; Pwee, K.H.; Manjunatha Kini, R. Interaction of wheat high-mobility-group proteins with four-way-junction DNA and characterization of the structure and expression of HMGA gene. Arch. Biochem. Biophys. 2003, 409, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Antosch, M.; Mortensen, S.A.; Grasser, K.D. Plant Proteins Containing High Mobility Group Box DNA-Binding Domains Modulate Different Nuclear Processes. Plant Physiol. 2012, 159, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Yang, T.; He, Y.; Niu, S.; Yan, S.; Zhang, Y. Identification and characterization of the CONSTANS (CO)/CONSTANS-like (COL) genes related to photoperiodic signaling and flowering in tomato. Plant Sci. 2020, 301, 110653. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef]
- Han, W.; Zhang, Q.; Suo, Y.; Li, H.; Diao, S.; Sun, P.; Huang, L.; Fu, J. Identification and Expression Analysis of the bHLH Gene Family Members in Diospyros kaki. Horticulturae 2023, 9, 380. [Google Scholar] [CrossRef]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, L.; Yun, L.; Ji, L.; Li, G.; Ji, M.; Shi, Y.; Zheng, X. Catalase (CAT) gene family in wheat (Triticum aestivum L.): Evolution, expression pattern and function analysis. Int. J. Mol. Sci. 2022, 23, 542. [Google Scholar] [CrossRef]
- He, Y.; Yang, T.; Yan, S.; Niu, S.; Zhang, Y. Identification and characterization of the BEL1-like genes reveal their potential roles in plant growth and abiotic stress response in tomato. Int. J. Biol. Macromol. 2022, 200, 193–205. [Google Scholar] [CrossRef]
- Yin, L.; Sun, Y.; Chen, X.; Liu, J.; Feng, K.; Luo, D.; Sun, M.; Wang, L.; Xu, W.; Liu, L.; et al. Genome-wide analysis of the hd-zip gene family in Chinese cabbage (Brassica rapa subsp. pekinensis) and the expression pattern at high temperatures and in carotenoids regulation. Agronomy 2023, 13, 1324. [Google Scholar]
- Chu, Z.; Wang, X.; Li, Y.; Yu, H.; Li, J.; Lu, Y.; Li, H.; Ouyang, B. Genomic organization, phylogenetic and expression analysis of the B-box gene family in tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).