Triple-Negative Breast Cancer EVs Modulate Growth and Migration of Normal Epithelial Lung Cells
Abstract
1. Introduction
2. Results and Discussion
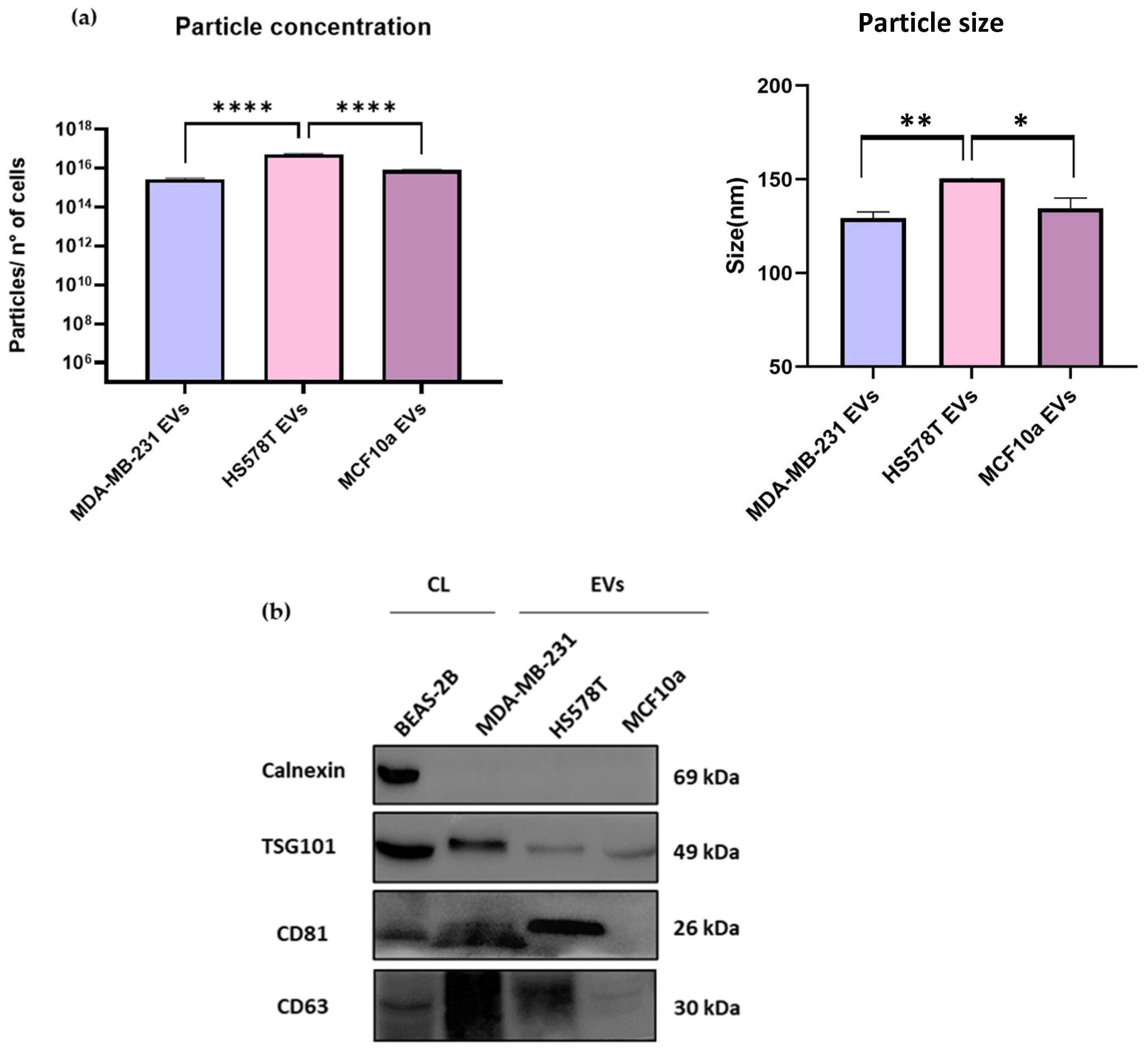
2.1. Characterization of EVs from TNBC and MCF10a Cell Lines by NTA and Immunoblotting Analysis
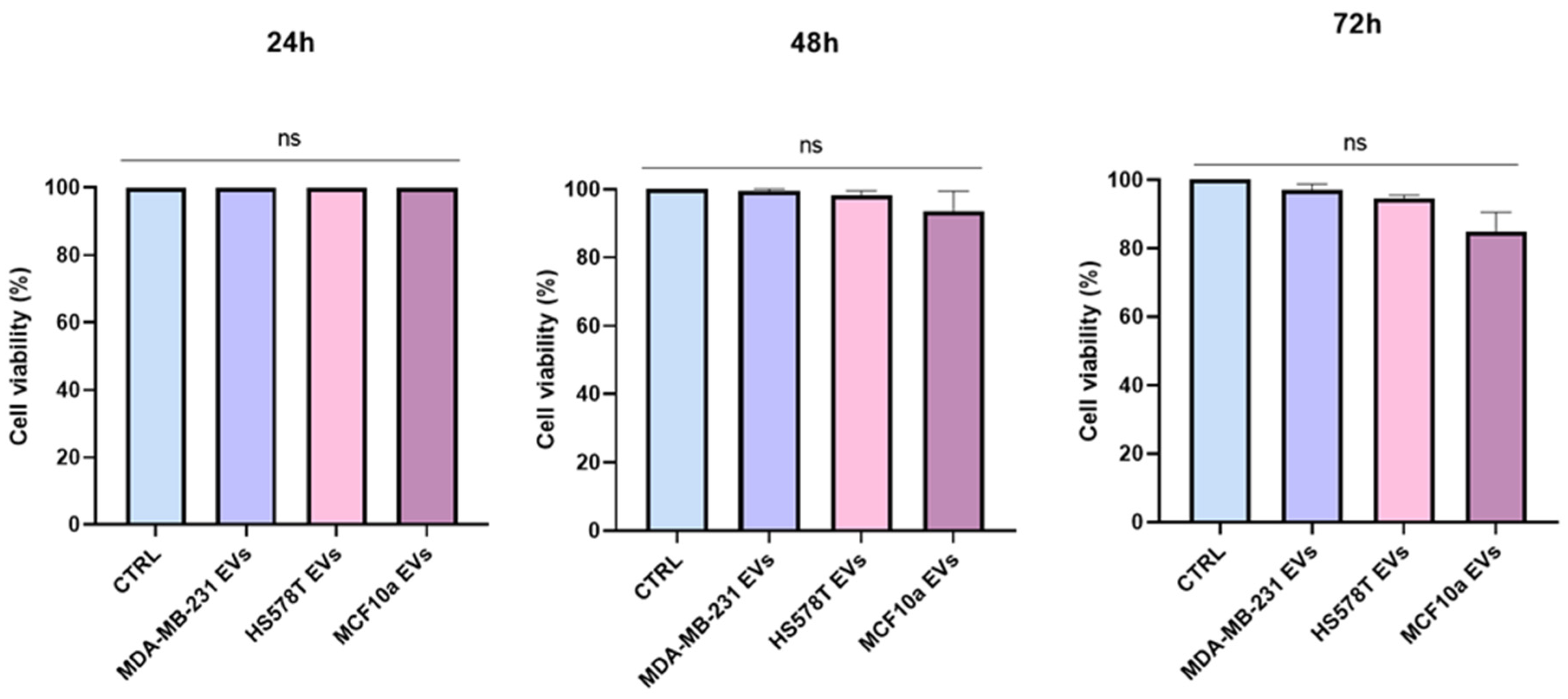
2.2. Cytoxicity Effect of EVs on Normal Human Bronchial Epithelial Cells
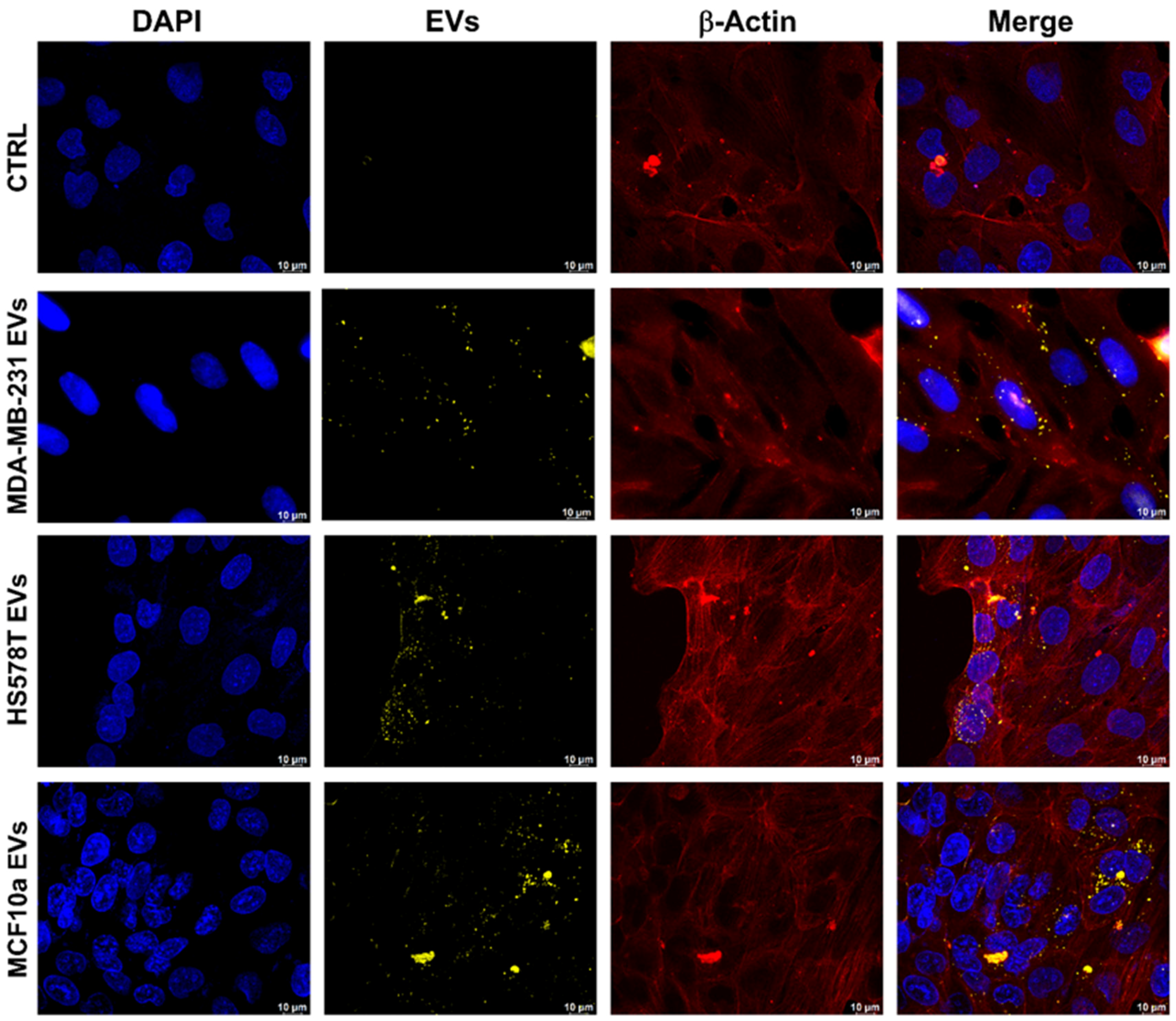
2.3. Evaluation of EVs Uptake through Confocal Microscopy
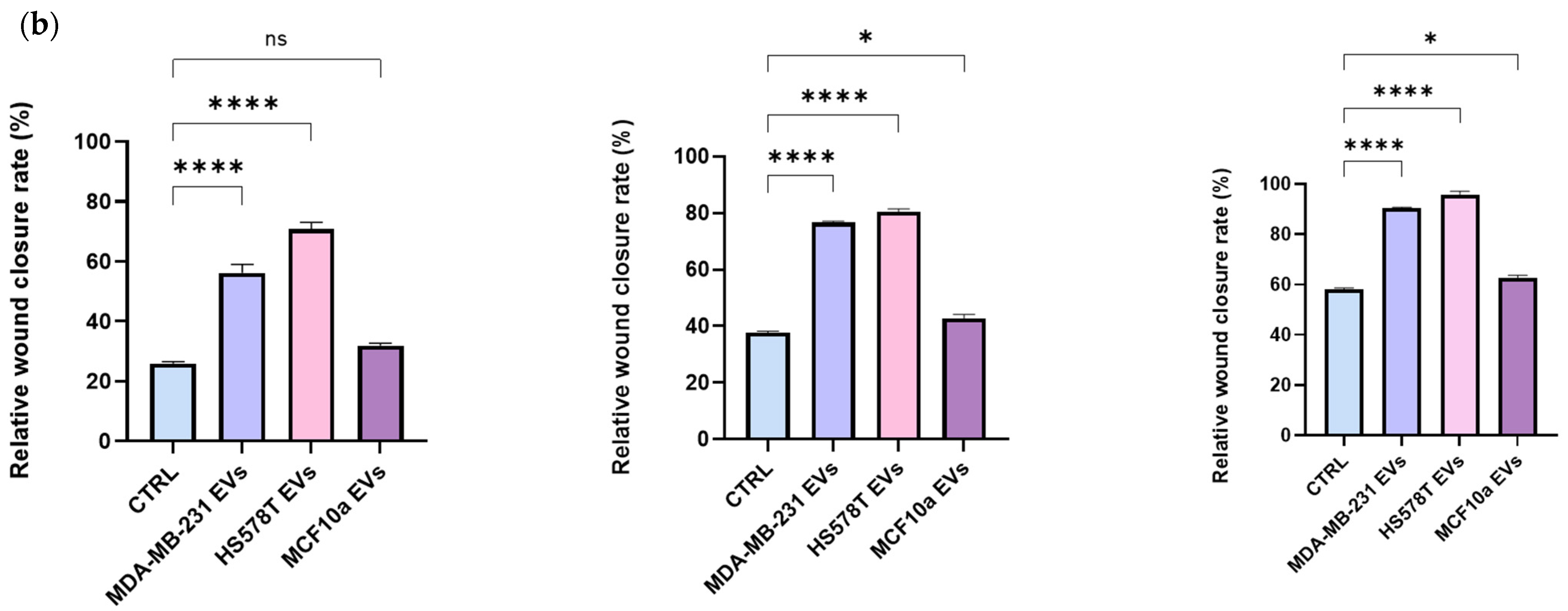
2.4. EVs Promotes Motility of Normal Human Bronchial Epithelial Cells
2.5. BEAS-2B Cells Have Higher Migration Rate in Presence of Breast Cancer-Derived EVs
3. Materials and Methods
3.1. Cell Lines
3.2. EVs Separation from Breast Cancer Cells
3.3. Particle Concentration and Size Using Nanoparticle Tracking Analysis (NTA)
3.4. Immunoblotting Analysis of EVs
3.5. Cytotoxicity Assay (MTS Assay)
3.6. Uptake of EVs Analyzed by Immunofluorescence
3.7. Scratch Assays
3.8. Transwell Migration Assay
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BSA | Bovine Serum Albumin |
| CL | cell lysate |
| CTRL | control |
| EMT | epithelial mesenchymal transition |
| ER | estrogen receptor |
| EVs | extracellular vesicles |
| EGFR2 | human epidermal growth factor receptor 2 |
| PBS | phosphate saline buffer |
| PgR | progesterone receptor |
| TEAD | TEA Domain Transcription Factor |
| TNBC | Triple-negative breast cancer |
References
- Zhu, J.W.; Charkhchi, P.; Adekunte, S.; Akbari, M.R. What Is Known about Breast Cancer in Young Women? Cancers 2023, 15, 1917. [Google Scholar] [CrossRef]
- Xu, Y.; Gong, M.; Wang, Y.; Yang, Y.; Liu, S.; Zeng, Q. Global Trends and Forecasts of Breast Cancer Incidence and Deaths. Sci. Data 2023, 10, 334. [Google Scholar] [CrossRef]
- Thomas, N.S.; Scalzo, R.L.; Wellberg, E.A. Diabetes Mellitus in Breast Cancer Survivors: Metabolic Effects of Endocrine Therapy. Nat. Rev. Endocrinol. 2024, 20, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Shafique, A.; Gonzalez, R.; Pantanowitz, L.; Tan, P.H.; Machado, A.; Cree, I.A.; Tizhoosh, H.R. A Preliminary Investigation into Search and Matching for Tumor Discrimination in World Health Organization Breast Taxonomy Using Deep Networks. Mod. Pathol. 2024, 37, 100381. [Google Scholar] [CrossRef]
- Rakha, E.A.; Green, A.R. Molecular Classification of Breast Cancer: What the Pathologist Needs to Know. Pathology 2017, 49, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Maqbool, M.; Bekele, F.; Fekadu, G. Treatment Strategies Against Triple-Negative Breast Cancer: An Updated Review. Breast Cancer Targets Ther. 2022, 14, 15–24. [Google Scholar] [CrossRef]
- Wu, Q.; Li, J.; Zhu, S.; Wu, J.; Chen, C.; Liu, Q.; Wei, W.; Zhang, Y.; Sun, S. Breast Cancer Subtypes Predict the Preferential Site of Distant Metastases: A SEER Based Study. Oncotarget 2017, 8, 27990–27996. [Google Scholar] [CrossRef] [PubMed]
- Romani, P.; Nirchio, N.; Arboit, M.; Barbieri, V.; Tosi, A.; Michielin, F.; Shibuya, S.; Benoist, T.; Wu, D.; Hindmarch, C.C.T.; et al. Mitochondrial Fission Links ECM Mechanotransduction to Metabolic Redox Homeostasis and Metastatic Chemotherapy Resistance. Nat. Cell Biol. 2022, 24, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, C.; Shao, W.; Liu, X.; Sun, L.; Yu, Z. Survival Analysis and Prognosis of Patients with Breast Cancer with Pleural Metastasis. Front. Oncol. 2023, 13, 1104246. [Google Scholar] [CrossRef]
- Schlappack, O.K.; Baur, M.; Steger, G.; Dittrich, C.; Moser, K. The Clinical Course of Lung Metastases from Breast Cancer. Klin. Wochenschr. 1988, 66, 790–795. [Google Scholar] [CrossRef]
- Liu, Y.; Mao, D.; Wang, H.; Che, X.; Chen, Y. Formation of Pre-Metastatic Niches Induced by Tumor Extracellular Vesicles in Lung Metastasis. Pharmacol. Res. 2023, 188, 106669. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Shelke, G.V.; Lässer, C.; Brismar, H.; Lötvall, J. Extracellular Vesicles from Mast Cells Induce Mesenchymal Transition in Airway Epithelial Cells. Respir. Res. 2020, 21, 101. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Ma, X.; Wei, H. Multiple Biological Roles of Extracellular Vesicles in Lung Injury and Inflammation Microenvironment. BioMed Res. Int. 2020, 2020, 5608382. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal Information for Studies of Extracellular Vesicles (MISEV2023): From Basic to Advanced Approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.; Goh, C.Y.; Gorzel, K.W.; Higgins, M.; McCann, A. The Potential Role of Cofilin-1 in Promoting Triple Negative Breast Cancer (TNBC) Metastasis via the Extracellular Vesicles (EVs). Transl. Oncol. 2022, 15, 101247. [Google Scholar] [CrossRef] [PubMed]
- Patel, U.; Susman, D.; Allan, A.L. Influence of Extracellular Vesicles on Lung Stromal Cells during Breast Cancer Metastasis. Int. J. Mol. Sci. 2023, 24, 11801. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Su, X.; Xu, M.; Xiao, X.; Li, X.; Li, H.; Keating, A.; Zhao, R.C. Exosomes Secreted by Mesenchymal Stromal/Stem Cell-Derived Adipocytes Promote Breast Cancer Cell Growth via Activation of Hippo Signaling Pathway. Stem. Cell Res. Ther. 2019, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Zhou, M.; Xu, Y.; Peng, B.; Gao, Y.; Mo, Y. Knockdown of CCM3 Promotes Angiogenesis through Activation and Nuclear Translocation of YAP/TAZ. Biochem. Biophys. Res. Commun. 2024, 701, 149525. [Google Scholar] [CrossRef]
- Mui, C.W.; Chan, W.N.; Chen, B.; Cheung, A.H.-K.; Yu, J.; Lo, K.W.; Ke, H.; Kang, W.; To, K.F. Targeting YAP1/TAZ in Nonsmall-Cell Lung Carcinoma: From Molecular Mechanisms to Precision Medicine. Int. J. Cancer 2023, 152, 558–571. [Google Scholar] [CrossRef]
- Ramírez-Ricardo, J.; Leal-Orta, E.; Martínez-Baeza, E.; Ortiz-Mendoza, C.; Breton-Mora, F.; Herrera-Torres, A.; Elizalde-Acosta, I.; Cortes-Reynosa, P.; Thompson-Bonilla, R.; Salazar, E.P. Circulating Extracellular Vesicles from Patients with Breast Cancer Enhance Migration and Invasion via a Src-Dependent Pathway in MDA-MB-231 Breast Cancer Cells. Mol. Med. Rep. 2020, 22, 1932–1948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, L.; Fang, Y.; Zheng, D.; Lin, T.; Wang, H. Label-Free Exosomal Detection and Classification in Rapid Discriminating Different Cancer Types Based on Specific Raman Phenotypes and Multivariate Statistical Analysis. Molecules 2019, 24, 2947. [Google Scholar] [CrossRef]
- Ozawa, P.M.M.; Alkhilaiwi, F.; Cavalli, I.J.; Malheiros, D.; de Souza Fonseca Ribeiro, E.M.; Cavalli, L.R. Extracellular Vesicles from Triple-Negative Breast Cancer Cells Promote Proliferation and Drug Resistance in Non-Tumorigenic Breast Cells. Breast Cancer Res. Treat. 2018, 172, 713–723. [Google Scholar] [CrossRef]
- Maji, S.; Yan, I.K.; Parasramka, M.; Mohankumar, S.; Matsuda, A.; Patel, T. In Vitro Toxicology Studies of Extracellular Vesicles. J. Appl. Toxicol. 2017, 37, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and Mechanisms of Extracellular Vesicle Uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed]
- Leal-Orta, E.; Ramirez-Ricardo, J.; Garcia-Hernandez, A.; Cortes-Reynosa, P.; Salazar, E.P. Extracellular Vesicles from MDA-MB-231 Breast Cancer Cells Stimulated with Insulin-like Growth Factor 1 Mediate an Epithelial–Mesenchymal Transition Process in MCF10A Mammary Epithelial Cells. J. Cell Commun. Signal 2022, 16, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Somiya, M.; Yoshioka, Y.; Ochiya, T. Biocompatibility of Highly Purified Bovine Milk-Derived Extracellular Vesicles. J. Extracell. Vesicles 2018, 7, 1440132. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Zhao, Y.; Cao, M.; Wang, J.; Tran, S.V.; Song, Z.; Hsueh, B.W.; Wang, S.E. Lung Fibroblasts Take up Breast Cancer Cell-Derived Extracellular Vesicles Partially Through MEK2-Dependent Macropinocytosis. Cancer Res. Commun. 2024, 4, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, J.; Chen, Y.; Li, T.; Yu, Q.; Sun, X. Breast Cancer Cell-Derived EVs Promote HUVECs Proliferation and Migration by Regulting JAK2/STAT3 Pathway. 2022; preprint. [Google Scholar] [CrossRef]
- O’Brien, K.; Rani, S.; Corcoran, C.; Wallace, R.; Hughes, L.; Friel, A.M.; McDonnell, S.; Crown, J.; Radomski, M.W.; O’Driscoll, L. Exosomes from Triple-Negative Breast Cancer Cells Can Transfer Phenotypic Traits Representing Their Cells of Origin to Secondary Cells. Eur. J. Cancer 2013, 49, 1845–1859. [Google Scholar] [CrossRef]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xiao, Z.; Zhu, W. The Roles of Small Extracellular Vesicles as Prognostic Biomarkers and Treatment Approaches in Triple-Negative Breast Cancer. Front. Oncol. 2022, 12, 998964. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, I.; Cocca, L.; Puoti, I.; Palma, F.; Ingenito, F.; Quintavalle, C.; Affinito, A.; Roscigno, G.; Nuzzo, S.; Chianese, R.V.; et al. Exosomal microRNAs Synergistically Trigger Stromal Fibroblasts in Breast Cancer. Mol. Ther.-Nucleic Acids 2022, 28, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, P.; Incoronato, M.; Coppola, L.; Infante, T.; Parente, C.A.; Nicolai, E.; Soricelli, A.; Salvatore, M. SDN Biobank: Bioresource of Human Samples Associated with Functional and/or Morphological Bioimaging Results for the Study of Oncological, Cardiological, Neurological, and Metabolic Diseases. Open J. Bioresour. 2017, 4, 2. [Google Scholar] [CrossRef]
- Shelke, G.V.; Lässer, C.; Gho, Y.S.; Lötvall, J. Importance of Exosome Depletion Protocols to Eliminate Functional and RNA-Containing Extracellular Vesicles from Fetal Bovine Serum. J. Extracell. Vesicles 2014, 3, 24783. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Pacheco, S.; O’Driscoll, L. Evidence for the Need to Evaluate More Than One Source of Extracellular Vesicles, Rather Than Single or Pooled Samples Only, When Comparing Extracellular Vesicles Separation Methods. Cancers 2021, 13, 4021. [Google Scholar] [CrossRef]
- Santoro, J.; Mukhopadhya, A.; Oliver, C.; Brodkorb, A.; Giblin, L.; O’Driscoll, L. An Investigation of Extracellular Vesicles in Bovine Colostrum, First Milk and Milk over the Lactation Curve. Food Chem. 2023, 401, 134029. [Google Scholar] [CrossRef] [PubMed]
- Santoro, J.; Carrese, B.; Peluso, M.S.; Coppola, L.; D’Aiuto, M.; Mossetti, G.; Salvatore, M.; Smaldone, G. Influence of Breast Cancer Extracellular Vesicles on Immune Cell Activation: A Pilot Study. Biology 2023, 12, 1531. [Google Scholar] [CrossRef] [PubMed]
- Pužar Dominkuš, P.; Stenovec, M.; Sitar, S.; Lasič, E.; Zorec, R.; Plemenitaš, A.; Žagar, E.; Kreft, M.; Lenassi, M. PKH26 Labeling of Extracellular Vesicles: Characterization and Cellular Internalization of Contaminating PKH26 Nanoparticles. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1350–1361. [Google Scholar] [CrossRef]
- Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; Bertier, L.; et al. EV-TRACK: Transparent Reporting and Centralizing Knowledge in Extracellular Vesicle Research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leone, I.; Santoro, J.; Soricelli, A.; Febbraro, A.; Santoriello, A.; Carrese, B. Triple-Negative Breast Cancer EVs Modulate Growth and Migration of Normal Epithelial Lung Cells. Int. J. Mol. Sci. 2024, 25, 5864. https://doi.org/10.3390/ijms25115864
Leone I, Santoro J, Soricelli A, Febbraro A, Santoriello A, Carrese B. Triple-Negative Breast Cancer EVs Modulate Growth and Migration of Normal Epithelial Lung Cells. International Journal of Molecular Sciences. 2024; 25(11):5864. https://doi.org/10.3390/ijms25115864
Chicago/Turabian StyleLeone, Ilaria, Jessie Santoro, Andrea Soricelli, Antonio Febbraro, Antonio Santoriello, and Barbara Carrese. 2024. "Triple-Negative Breast Cancer EVs Modulate Growth and Migration of Normal Epithelial Lung Cells" International Journal of Molecular Sciences 25, no. 11: 5864. https://doi.org/10.3390/ijms25115864
APA StyleLeone, I., Santoro, J., Soricelli, A., Febbraro, A., Santoriello, A., & Carrese, B. (2024). Triple-Negative Breast Cancer EVs Modulate Growth and Migration of Normal Epithelial Lung Cells. International Journal of Molecular Sciences, 25(11), 5864. https://doi.org/10.3390/ijms25115864





