Elevated Temperature Effects on Protein Turnover Dynamics in Arabidopsis thaliana Seedlings Revealed by 15N-Stable Isotope Labeling and ProteinTurnover Algorithm
Abstract
1. Introduction
2. Results
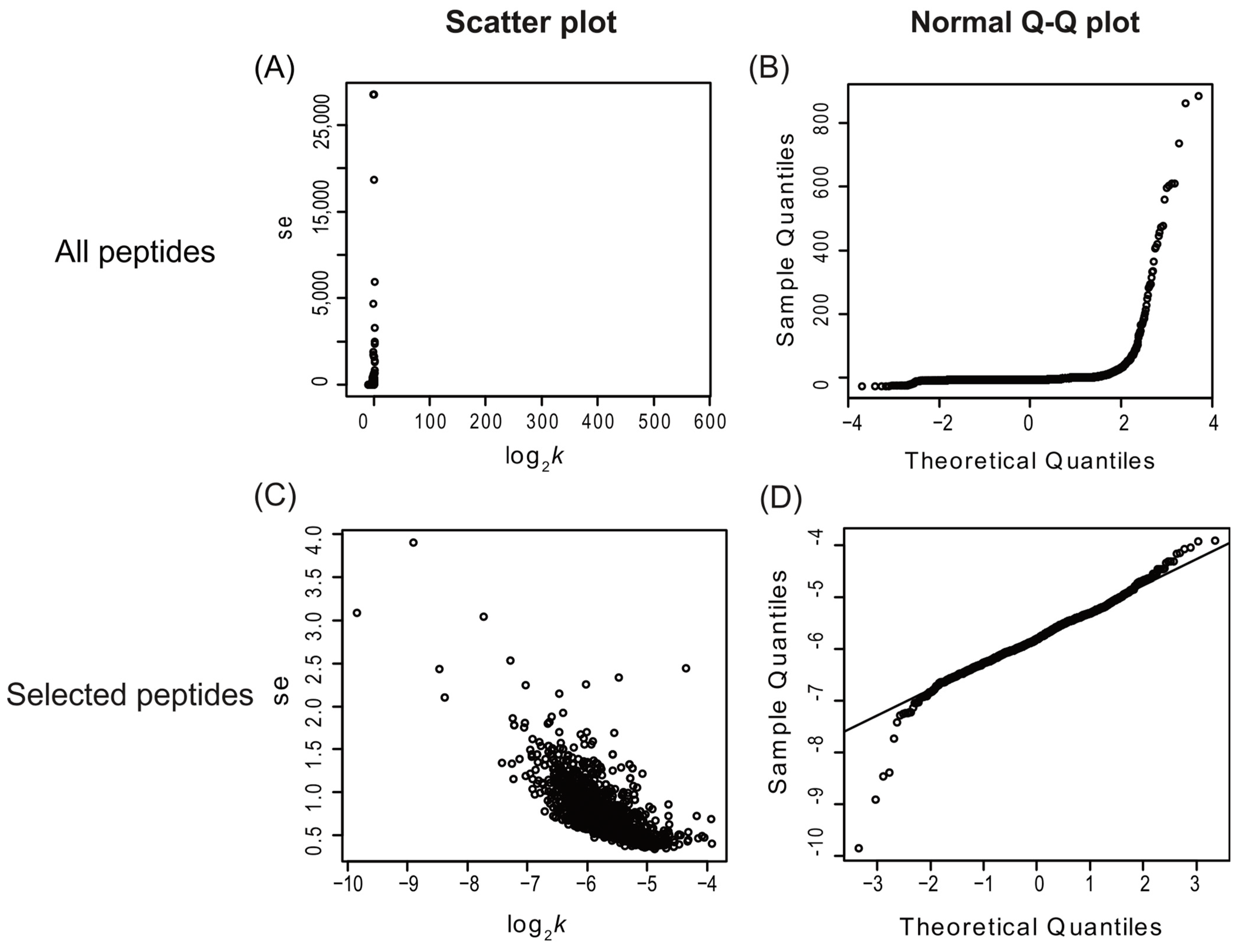
2.1. Peptide Identification and Selection Criteria for Protein Turnover Rate Measurements
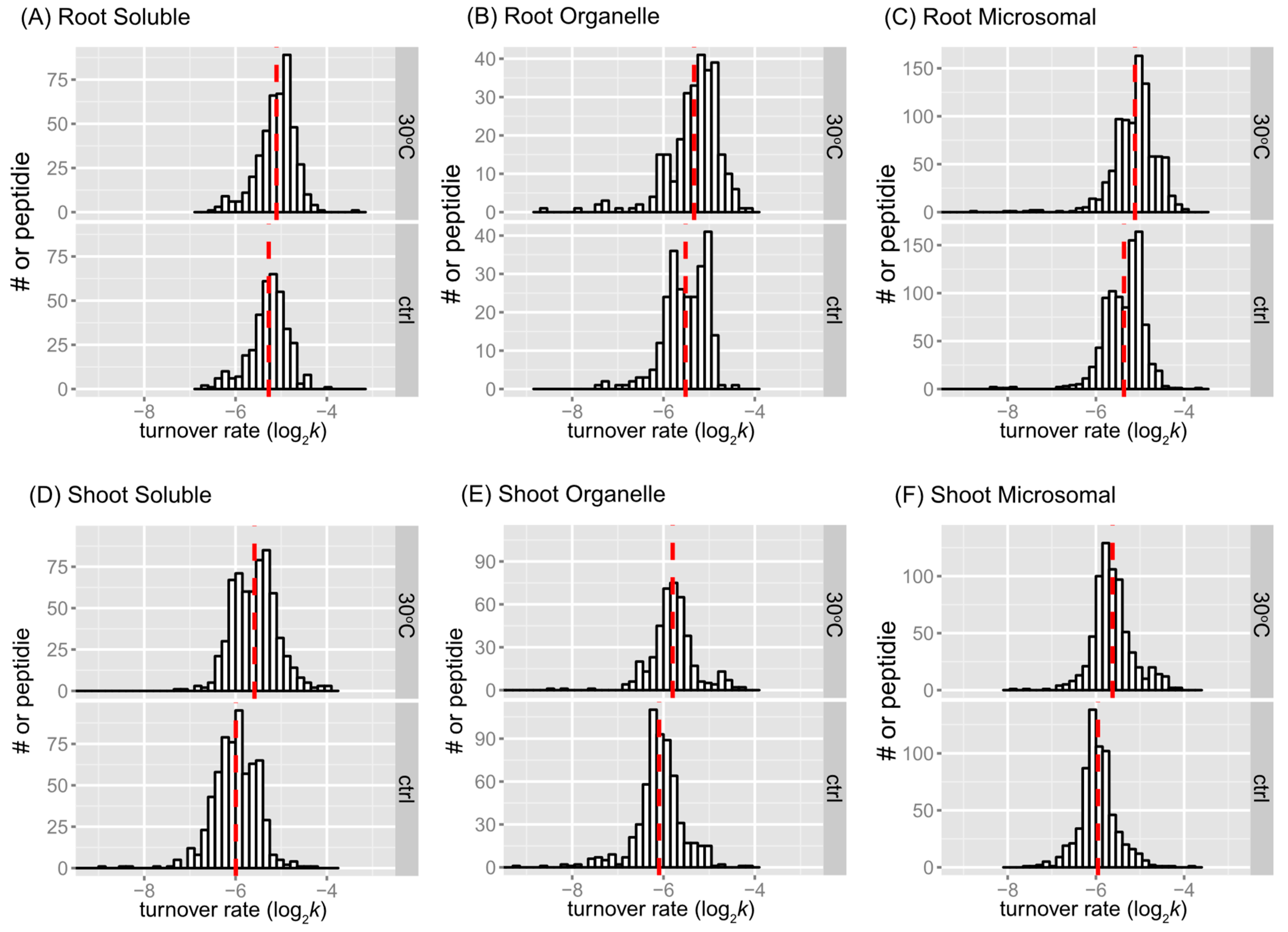
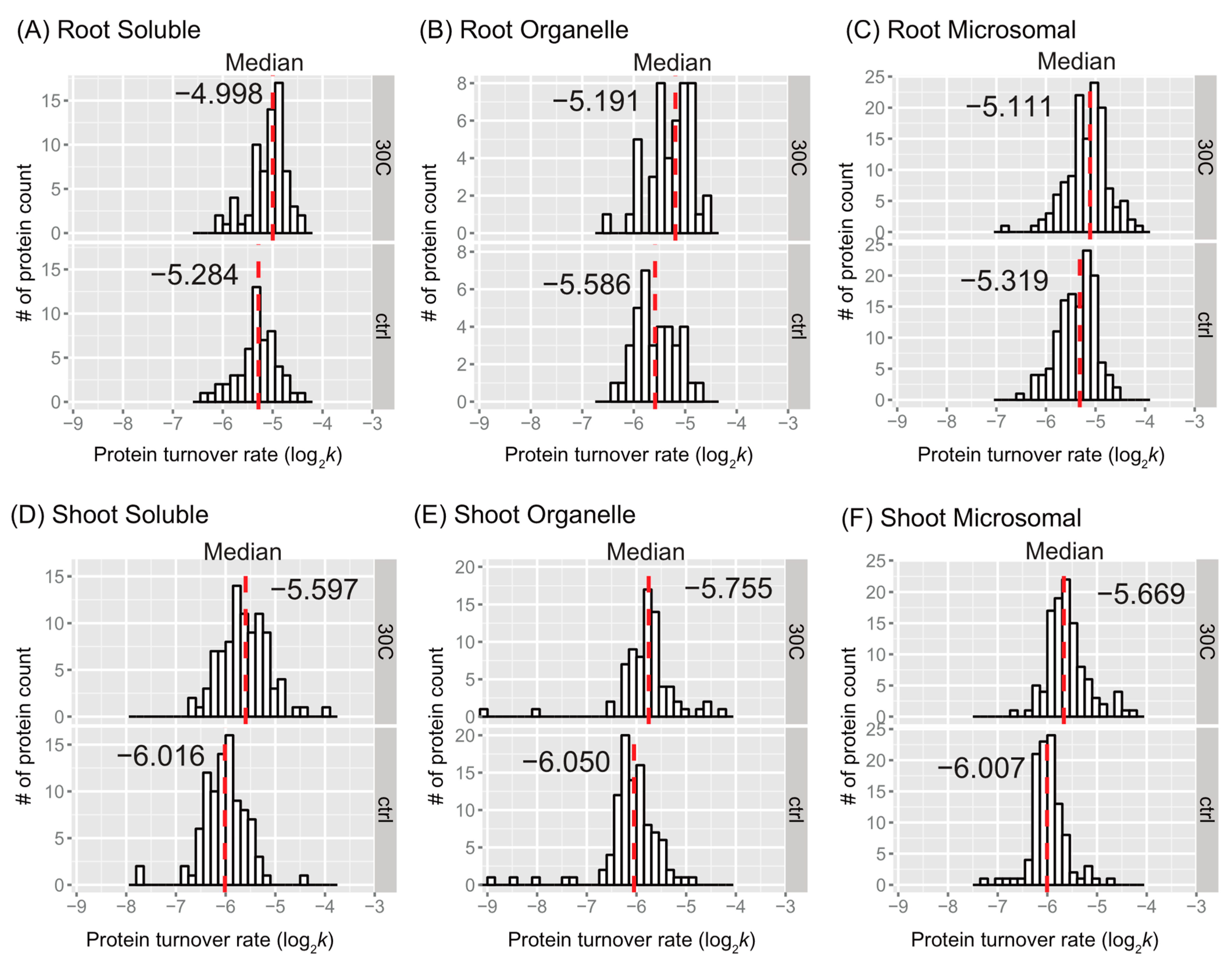
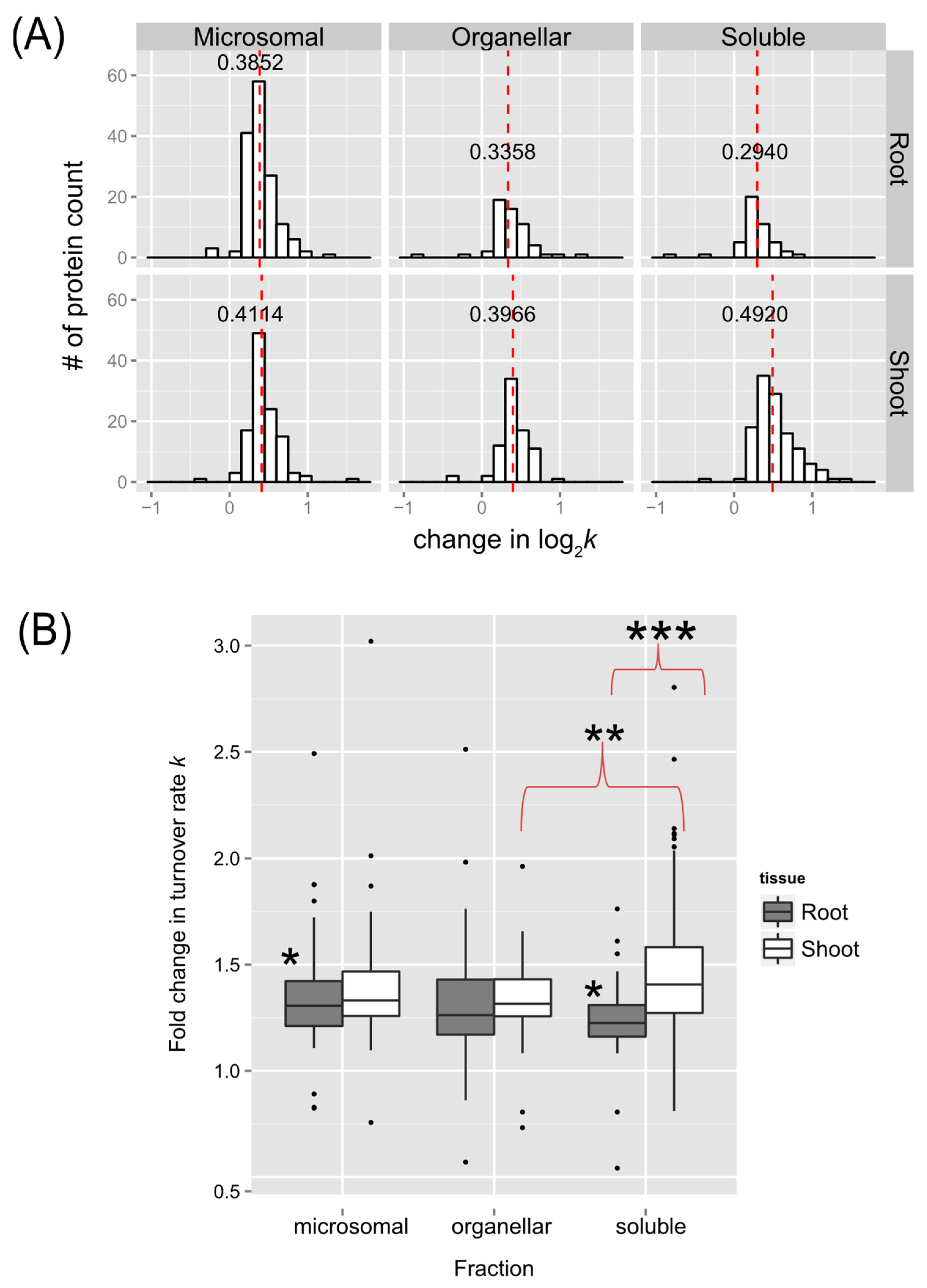
2.2. Overview of the Effects of Heat Stress on Peptide and Protein Turnover Rates
2.2.1. Trends in Peptide or Protein Turnover Rates
2.2.2. Coefficient of Variation in Protein Turnover as a Function of the Number of Peptide Observations
2.2.3. Statistical Significance of Changes in Protein Turnover Rates upon Heat Treatment
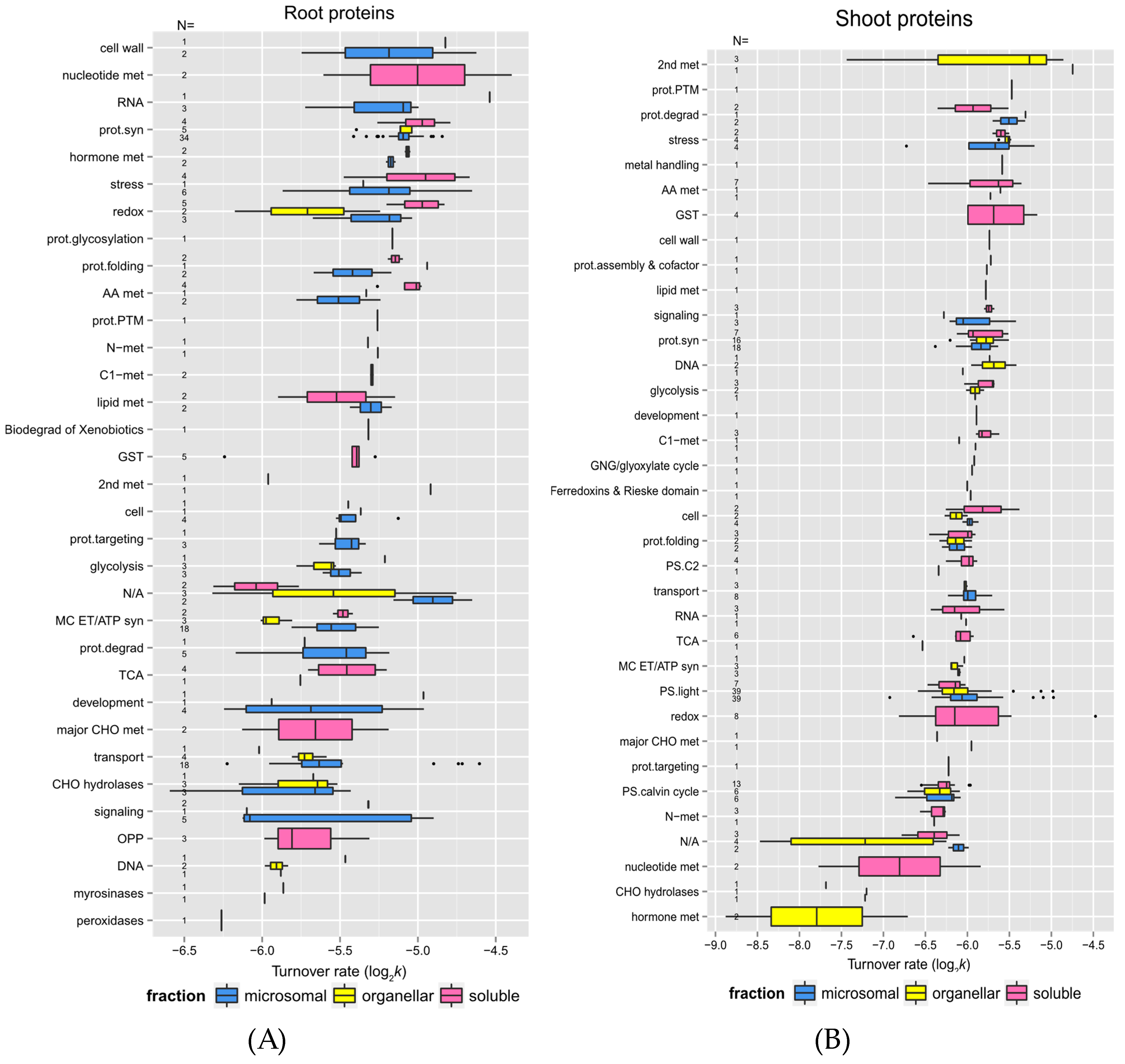
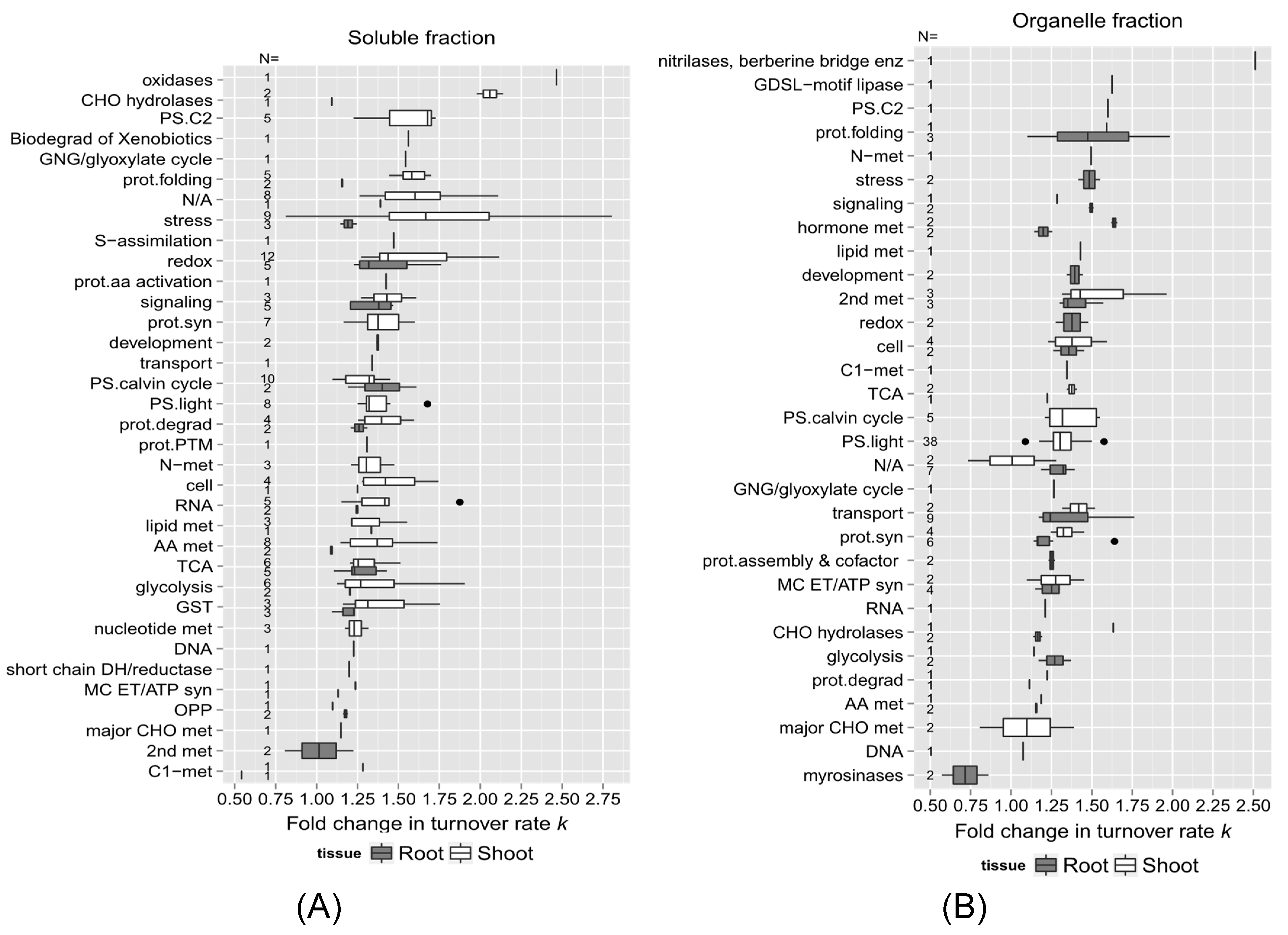
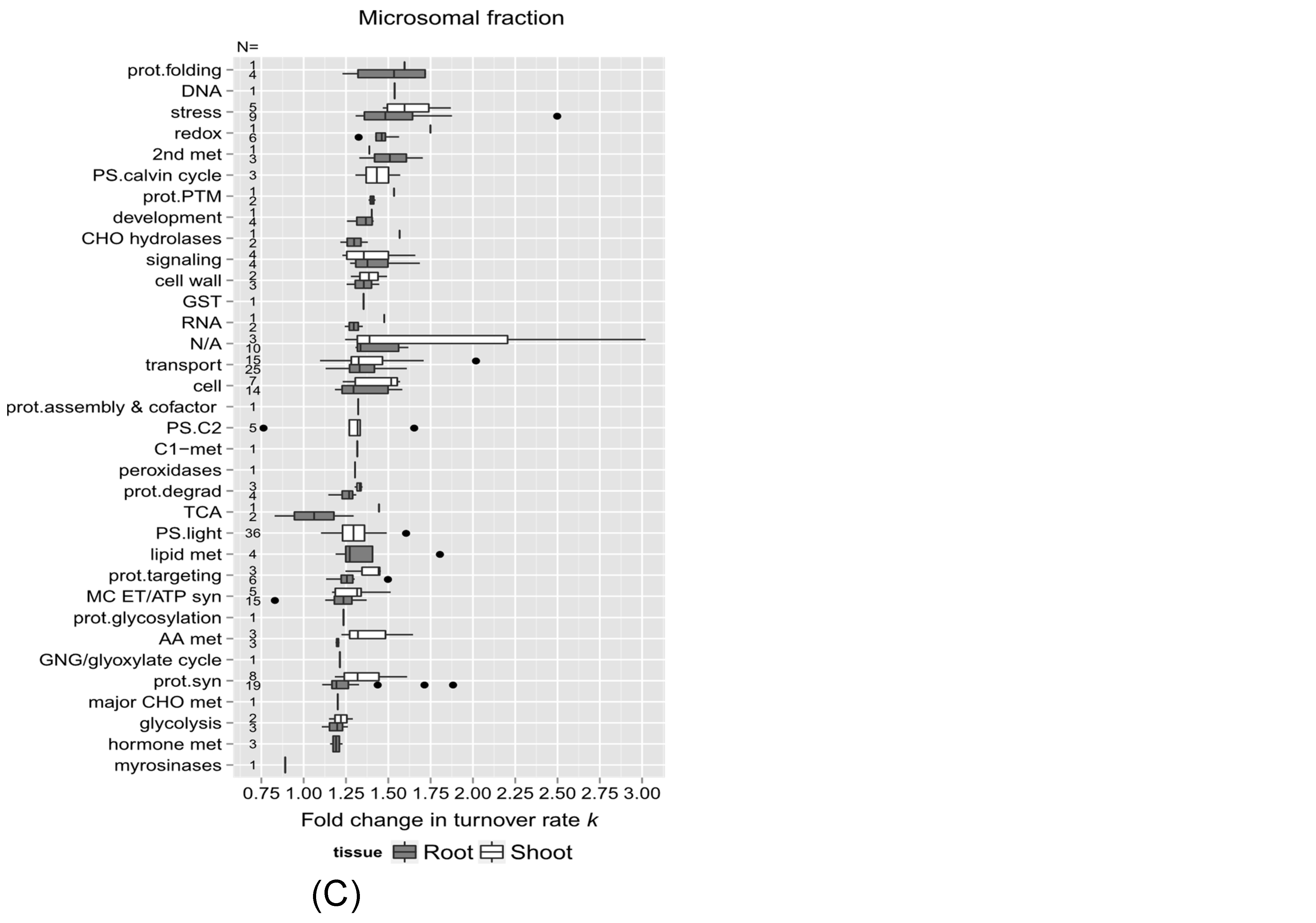
2.3. Links between Protein Functional Categories and Changes in Protein Turnover Rates upon Heat Treatment
2.3.1. Protein Function and Turnover Rates of Proteins
2.3.2. Protein Function and Change in Turnover Rates Due to High Temperature
3. Discussion
3.1. Heat Shock Proteins (HSPs) and Chaperones
3.2. Photosynthesis and Carbon Assimilation
3.3. Redox Homeostasis: HSPs, Catalases, and Peroxidases
3.4. Special Cases: Decreases or Major Increases in Protein Turnover Rates in Response to Heat Stress
3.4.1. GDSL Esterase/Lipase Family
3.4.2. 14-3-3 and V-, P-Type ATPase
3.5. Stability of Proteins Involved in Primary Metabolism and Energy Production
3.6. Differential Responses of Root and Shoot Proteomes to Heat Stress
3.7. Expanding upon Prior 15N-Labeling Studies: Progress and Limitations in the Current Study
3.8. Summary and Future Research Directions
4. Materials and Methods
4.1. Materials
4.2. Plant Growth and Labeling Conditions
4.3. Proteomic Sample Preparation
4.4. UHPLC-HRMS/MS Analysis
4.5. Protein Identification
4.6. Calculation of Protein Turnover Rates
4.7. Estimating the Difference in Log2k Due to Heat Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaloner, T.M.; Gurr, S.J.; Bebber, D.P. Plant Pathogen Infection Risk Tracks Global Crop Yields under Climate Change. Nat. Clim. Chang. 2021, 11, 710–715. [Google Scholar] [CrossRef]
- Cruz, J.-L.; Rossi-Hansberg, E. The Economic Geography of Global Warming. Rev. Econ. Stud. 2024, 91, 899–939. [Google Scholar] [CrossRef]
- Wing, I.S.; De Cian, E.; Mistry, M.N. Global Vulnerability of Crop Yields to Climate Change. J. Environ. Econ. Manag. 2021, 109, 102462. [Google Scholar] [CrossRef]
- Klueva, N.Y.; Maestri, E.; Marmiroli, N.; Nguyen, H.T. Mechanisms of Thermotolerance in Crops. In Crop Responses and Adaptations to Temperature Stress; CRC Press: Boca Raton, FL, USA, 2023; pp. 177–217. [Google Scholar]
- Yeh, C.H.; Kaplinsky, N.J.; Hu, C.; Charng, Y.Y. Some like It Hot, Some like It Warm: Phenotyping to Explore Thermotolerance Diversity. Plant Sci. 2012, 195, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.-D. Complexity of the Heat Stress Response in Plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat Tolerance in Plants: An Overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Crawford, A.J.; McLachlan, D.H.; Hetherington, A.M.; Franklin, K.A. High Temperature Exposure Increases Plant Cooling Capacity. Curr. Biol. 2012, 22, R396–R397. [Google Scholar] [CrossRef] [PubMed]
- Kulke, M.; Weraduwage, S.M.; Sharkey, T.D.; Vermaas, J. V Nanoscale Simulation of the Thylakoid Membrane Response to Extreme Temperatures. Plant Cell Environ. 2023, 46, 2419–2431. [Google Scholar] [CrossRef]
- Schrader, S.M.; Kleinbeck, K.R.; Sharkey, T.D. Rapid Heating of Intact Leaves Reveals Initial Effects of Stromal Oxidation on Photosynthesis. Plant Cell Environ. 2007, 30, 671–678. [Google Scholar] [CrossRef]
- Berry, J.; Bjorkman, O. Photosynthetic Response and Adaptation to Temperature in Higher Plants. Annu. Rev. Plant Physiol. 1980, 31, 491–543. [Google Scholar] [CrossRef]
- Yamane, Y.; Kashino, Y.; Koike, H.; Satoh, K. Effects of High Temperatures on the Photosynthetic Systems in Spinach: Oxygen-Evolving Activities, Fluorescence Characteristics and the Denaturation Process. Photosynth. Res. 1998, 57, 51–59. [Google Scholar] [CrossRef]
- Kurek, I.; Chang, T.K.; Bertain, S.M.; Madrigal, A.; Liu, L.; Lassner, M.W.; Zhu, G. Enhanced Thermostability of Arabidopsis Rubisco Activase Improves Photosynthesis and Growth Rates under Moderate Heat Stress. Plant Cell 2007, 19, 3230–3241. [Google Scholar] [CrossRef] [PubMed]
- Crafts-Brandner, S.J.; Salvucci, M.E. Rubisco Activase Constrains the Photosynthetic Potential of Leaves at High Temperature and CO2. Proc. Natl. Acad. Sci. USA 2000, 97, 13430–13435. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef] [PubMed]
- Kan, Y.; Mu, X.-R.; Gao, J.; Lin, H.-X.; Lin, Y. The Molecular Basis of Heat Stress Responses in Plants. Mol. Plant 2023, 16, 1612–1634. [Google Scholar] [CrossRef] [PubMed]
- Qu, A.-L.; Ding, Y.-F.; Jiang, Q.; Zhu, C. Molecular Mechanisms of the Plant Heat Stress Response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Matthew, A.; Jenks, A.J.W. (Eds.) Genes for Plant Abiotic Stress; Wiley-Blackwell: Oxford, UK, 2010; ISBN 9780813815022. [Google Scholar]
- Williams, B.; Kabbage, M.; Britt, R.; Dickman, M.B. AtBAG7, an Arabidopsis Bcl-2–Associated Athanogene, Resides in the Endoplasmic Reticulum and Is Involved in the Unfolded Protein Response. Proc. Natl. Acad. Sci. USA 2010, 107, 6088–6093. [Google Scholar] [CrossRef]
- Fields, P.A. Protein Function at Thermal Extremes: Balancing Stability and Flexibility. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2001, 129, 417–431. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When Defense Pathways Collide. The Response of Arabidopsis to a Combination of Drought and Heat Stress. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef]
- Kappel, C.; Friedrich, T.; Oberkofler, V.; Jiang, L.; Crawford, T.; Lenhard, M.; Bäurle, I. Genomic and Epigenomic Determinants of Heat Stress-Induced Transcriptional Memory in Arabidopsis. Genome Biol. 2023, 24, 129. [Google Scholar] [CrossRef] [PubMed]
- Frank, G.; Pressman, E.; Ophir, R.; Althan, L.; Shaked, R.; Freedman, M.; Shen, S.; Firon, N. Transcriptional Profiling of Maturing Tomato (Solanum lycopersicum L.) Microspores Reveals the Involvement of Heat Shock Proteins, ROS Scavengers, Hormones, and Sugars in the Heat Stress Response. J. Exp. Bot. 2009, 60, 3891–3908. [Google Scholar] [CrossRef] [PubMed]
- Naik, B.; Kumar, V.; Rizwanuddin, S.; Chauhan, M.; Choudhary, M.; Gupta, A.K.; Kumar, P.; Kumar, V.; Saris, P.E.J.; Rather, M.A. Genomics, Proteomics, and Metabolomics Approaches to Improve Abiotic Stress Tolerance in Tomato Plant. Int. J. Mol. Sci. 2023, 24, 3025. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, H.; Hirose, T.; Kuroda, M.; Yamaguchi, T. Comprehensive Expression Profiling of Rice Grain Filling-Related Genes under High Temperature Using DNA Microarray. Plant Physiol. 2007, 144, 258–277. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Zhang, Q.; Ji, C.; Hu, G.; Zhang, P.; Wang, Y.; Yang, L.; Gu, X. Reorganization of the 3D Chromatin Architecture of Rice Genomes during Heat Stress. BMC Biol. 2021, 19, 53. [Google Scholar] [CrossRef]
- Oshino, T.; Abiko, M.; Saito, R.; Ichiishi, E.; Endo, M.; Kawagishi-Kobayashi, M.; Higashitani, A. Premature Progression of Anther Early Developmental Programs Accompanied by Comprehensive Alterations in Transcription during High-Temperature Injury in Barley Plants. Mol. Genet. Genomics 2007, 278, 31–42. [Google Scholar] [CrossRef]
- Zhang, W.; Tan, C.; Hu, H.; Pan, R.; Xiao, Y.; Ouyang, K.; Zhou, G.; Jia, Y.; Zhang, X.; Hill, C.B. Genome Architecture and Diverged Selection Shaping Pattern of Genomic Differentiation in Wild Barley. Plant Biotechnol. J. 2023, 21, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Wu, H.; Peng, H.; Yao, Y.; Ni, Z.; Li, Z.; Zhou, C.; Sun, Q. Heat Stress-Responsive Transcriptome Analysis in Heat Susceptible and Tolerant Wheat (Triticum aestivum L.) by Using Wheat Genome Array. BMC Genomics 2008, 9, 432. [Google Scholar] [CrossRef]
- Sihag, P.; Kumar, U.; Sagwal, V.; Kapoor, P.; Singh, Y.; Mehla, S.; Balyan, P.; Mir, R.R.; Varshney, R.K.; Singh, K.P. Effect of Terminal Heat Stress on Osmolyte Accumulation and Gene Expression during Grain Filling in Bread Wheat (Triticum aestivum L.). Plant Genome 2024, 17, e20307. [Google Scholar] [CrossRef]
- Fernandes, J.; Morrow, D.J.; Casati, P.; Walbot, V. Distinctive Transcriptome Responses to Adverse Environmental Conditions in Zea mays L. Plant Biotechnol. J. 2008, 6, 782–798. [Google Scholar] [CrossRef]
- Chen, Y.; Du, T.; Zhang, J.; Chen, S.; Fu, J.; Li, H.; Yang, Q. Genes and Pathways Correlated with Heat Stress Responses and Heat Tolerance in Maize Kernels. Front. Plant Sci. 2023, 14, 1228213. [Google Scholar] [CrossRef]
- Yángüez, E.; Castro-Sanz, A.B.; Fernández-Bautista, N.; Oliveros, J.C.; Castellano, M.M.; Ferna, N.; Ya, E. Analysis of Genome-Wide Changes in the Translatome of Arabidopsis Seedlings Subjected to Heat Stress. PLoS ONE 2013, 8, e71425. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Ahsan, N.; Lee, S.H.; Kyu, Y.K.; Jeong, D.B.; Lee, I.J.; Lee, B.H. A Proteomic Approach in Analyzing Heat-Responsive Proteins in Rice Leaves. Proteomics 2007, 7, 3369–3383. [Google Scholar] [CrossRef]
- Fan, K.T.; Rendahl, A.K.; Chen, W.P.; Freund, D.M.; Gray, W.M.; Cohen, J.D.; Hegeman, A.D. Proteome Scale-Protein Turnover Analysis Using High Resolution Mass Spectrometric Data from Stable-Isotope Labeled Plants. J. Proteome Res. 2016, 15, 851–867. [Google Scholar] [CrossRef]
- Evans, E.M.; Freund, D.M.; Sondervan, V.M.; Cohen, J.D.; Hegeman, A.D. Metabolic Patterns in Spirodela Polyrhiza Revealed by 15N Stable Isotope Labeling of Amino Acids in Photoautotrophic, Heterotrophic, and Mixotrophic Growth Conditions. Front. Chem. 2018, 6, 191. [Google Scholar] [CrossRef]
- Hartigan, J.A.; Hartigan, P.M. The Dip Test of Unimodality. Ann. Statis 1985, 13, 70–84. [Google Scholar] [CrossRef]
- West, B.T.; Welch, K.B.; Galecki, A.T. Linear Mixed Models: A Practical Guide Using Statistical Software; Chapman and Hall–CRC: Boca Raton, FL, USA, 2007; ISBN 1-584-88480-0. [Google Scholar]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A Guide to Using MapMan to Visualize and Compare Omics Data in Plants: A Case Study in the Crop Species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Craig, E. A Heat-Shock Proteins as Molecular Chaperones. Eur. J. Biochem. 1994, 219, 11–23. [Google Scholar] [CrossRef]
- Nelson, C.J.; Li, L.; Jacoby, R.P.; Millar, A.H. Degradation Rate of Mitochondrial Proteins in Arabidopsis thaliana Cells. J. Proteome Res. 2013, 12, 3449–3459. [Google Scholar] [CrossRef]
- Oh, S.E.; Yeung, C.; Babaei-Rad, R.; Zhao, R. Cosuppression of the Chloroplast Localized Molecular Chaperone HSP90. 5 Impairs Plant Development and Chloroplast Biogenesis in Arabidopsis. BMC Res. Notes 2014, 7, 643. [Google Scholar] [CrossRef]
- Mattoo, R.U.H.; Goloubinoff, P. Molecular Chaperones Are Nanomachines That Catalytically Unfold Misfolded and Alternatively Folded Proteins. Cell. Mol. Life Sci. 2014, 71, 3311–3325. [Google Scholar] [CrossRef]
- Suzuki, K.; Nakanishi, H.; Bower, J.; Yoder, D.W.; Osteryoung, K.W.; Miyagishima, S. Plastid Chaperonin Proteins Cpn60 Alpha and Cpn60 Beta Are Required for Plastid Division in Arabidopsis thaliana. BMC Plant Biol. 2009, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Nash, D.; Miyao, M.; Murata, N. Heat Inactivation of Oxygen Evolution in Photosystem II Particles and Its Acceleration by Chloride Depletion and Exogenous Manganese. Biochim. Biophys. Acta Bioenerg. 1985, 807, 127–133. [Google Scholar] [CrossRef]
- Armond, P.A.; Schreiber, U.; Björkman, O. Photosynthetic Acclimation to Temperature in the Desert Shrub, Larrea divaricata: II. Light-Harvesting Efficiency and Electron Transport. Plant Physiol. 1978, 61, 411–415. [Google Scholar] [CrossRef]
- Yamashita, T.; Butler, W.L. Inhibition of Chloroplasts by UV-Irradiation and Heat-Treatment. Plant Physiol. 1968, 43, 2037–2040. [Google Scholar] [CrossRef]
- Betterle, N.; Ballottari, M.; Hienerwadel, R.; Dall’Osto, L.; Bassi, R. Dynamics of Zeaxanthin Binding to the Photosystem II Monomeric Antenna Protein Lhcb6 (CP24) and Modulation of Its Photoprotection Properties. Arch. Biochem. Biophys. 2010, 504, 67–77. [Google Scholar] [CrossRef]
- Marutani, Y.; Yamauchi, Y.; Miyoshi, A.; Inoue, K.; Ikeda, K.; Mizutani, M.; Sugimoto, Y. Regulation of Photochemical Energy Transfer Accompanied by Structural Changes in Thylakoid Membranes of Heat-Stressed Wheat. Int. J. Mol. Sci. 2014, 15, 23042–23058. [Google Scholar] [CrossRef] [PubMed]
- Volkov, R.A.; Panchuk, I.I.; Mullineaux, P.M.; Schöffl, F. Heat Stress-Induced H2O2 Is Required for Effective Expression of Heat Shock Genes in Arabidopsis. Plant Mol. Biol. 2006, 61, 733–746. [Google Scholar] [CrossRef]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef]
- Pnueli, L.; Liang, H.; Rozenberg, M.; Mittler, R. Growth Suppression, Altered Stomatal Responses, and Augmented Induction of Heat Shock Proteins in Cytosolic Ascorbate Peroxidase (Apx1)-deficient Arabidopsis Plants. Plant J. 2003, 34, 187–203. [Google Scholar] [CrossRef]
- Correa-Aragunde, N.; Foresi, N.; Delledonne, M.; Lamattina, L. Auxin Induces Redox Regulation of Ascorbate Peroxidase 1 Activity by S-Nitrosylation/Denitrosylation Balance Resulting in Changes of Root Growth Pattern in Arabidopsis. J. Exp. Bot. 2013, 64, 3339–3349. [Google Scholar] [CrossRef] [PubMed]
- Nagano, A.J.; Fukao, Y.; Fujiwara, M.; Nishimura, M.; Hara-Nishimura, I. Antagonistic Jacalin-Related Lectins Regulate the Size of ER Body-Type β-Glucosidase Complexes in Arabidopsis Thaliana. Plant Cell Physiol. 2008, 49, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, J.D.; Folta, K.M.; Paul, A.-L.; Ferl, R.J. The 14-3-3 Proteins Mu and Upsilon Influence Transition to Flowering and Early Phytochrome Response. Plant Physiol. 2007, 145, 1692–1702. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, J.D.; Paul, A.-L.; Ferl, R.J. The 14-3-3 Proteins of Arabidopsis Regulate Root Growth and Chloroplast Development as Components of the Photosensory System. J. Exp. Bot. 2012, 63, 3061–3070. [Google Scholar] [CrossRef] [PubMed]
- Gampala, S.S.; Kim, T.W.; He, J.X.; Tang, W.; Deng, Z.; Bai, M.Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An Essential Role for 14-3-3 Proteins in Brassinosteroid Signal Transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef]
- Plaxton, W.C. The Organization and Regulation of Plant Glycolysis. Annu. Rev. Plant Biol. 1996, 47, 185–214. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Whelan, J.; Soole, K.L.; Day, D.A. Organization and Regulation of Mitochondrial Respiration in Plants. Annu. Rev. Plant Biol. 2011, 62, 79–104. [Google Scholar] [CrossRef]
- Baena-González, E.; Sheen, J. Convergent Energy and Stress Signaling. Trends Plant Sci. 2008, 13, 474–482. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, X. Reprogramming of Plant Central Metabolism in Response to Abiotic Stresses: A Metabolomics View. Int. J. Mol. Sci. 2022, 23, 5716. [Google Scholar] [CrossRef]
- Howell, N.; Akagi, J.M.; Himes, R.H. Thermostability of Glycolytic Enzymes from Thermophilic Clostridia. Can. J. Microbiol. 1969, 15, 461–464. [Google Scholar] [CrossRef]
- Jones, M.V.; Spencer, W.N. Thermostability of Enzymes of the Tricarboxylic Acid Cycle of Bacillus Coagulans. Antonie Van Leeuwenhoek 1985, 51, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Imanaka, T. Molecular Bases of Thermophily in Hyperthermophiles. Proc. Japan Acad. Ser. B 2011, 87, 587–602. [Google Scholar] [CrossRef] [PubMed]
- Almalki, A.F.Y.; Arabdin, M.; Khan, A. The Role of Heat Shock Proteins in Cellular Homeostasis and Cell Survival. Cureus 2021, 13, e18316. [Google Scholar]
- Wang, X.; Chen, S.; Zhang, H.; Shi, L.; Cao, F.; Guo, L.; Xie, Y.; Wang, T.; Yan, X.; Dai, S. Desiccation Tolerance Mechanism in Resurrection Fern-Ally Selaginella Tamariscina Revealed by Physiological and Proteomic Analysis. J. Proteome Res. 2010, 9, 6561–6577. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, N.; Wu, L.; Li, C.; Li, C.; Zhang, L.; Liu, T.; Wang, W. Quantitative ITRAQ-Based Proteomic Analysis of Phosphoproteins and ABA-Regulated Phosphoproteins in Maize Leaves under Osmotic Stress. Sci. Rep. 2015, 5, 15626. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Huang, B. Growth and Physiological Responses of Creeping Bentgrass to Changes in Air and Soil Temperatures. Crop Sci. 2000, 40, 1363–1368. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mechanisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Xu, C. Identification and Characterization of Proteins Associated with Plant Tolerance to Heat Stress. J. Integr. Plant Biol. 2008, 50, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Agrawal, D.; Jajoo, A. Photosynthesis: Response to High Temperature Stress. J. Photochem. Photobiol. B Biol. 2014, 137, 116–126. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T. Proteomics of Stress Responses in Wheat and Barley—Search for Potential Protein Markers of Stress Tolerance. Front. Plant Sci. 2014, 5, 122688. [Google Scholar] [CrossRef]
- Gao, H.; Ge, W.; Bai, L.; Zhang, T.; Zhao, L.; Li, J.; Shen, J.; Xu, N.; Zhang, H.; Wang, G. Proteomic Analysis of Leaves and Roots during Drought Stress and Recovery in Setaria italica L. Front. Plant Sci. 2023, 14, 1240164. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A Glutathione Responsive Rice Glyoxalase II, Os GLYII-2, Functions in Salinity Adaptation by Maintaining Better Photosynthesis Efficiency and Anti-oxidant Pool. Plant J. 2014, 80, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Hossain, Z. Organ-Specific Proteome Analysis for Identification of Abiotic Stress Response Mechanism in Crop. Front. Plant Sci. 2013, 4, 44308. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Renaut, J. Plant Proteome Changes under Abiotic Stress—Contribution of Proteomics Studies to Understanding Plant Stress Response. J. Proteomics 2011, 74, 1301–1322. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Nelson, C.J.; Solheim, C.; Whelan, J.; Millar, A.H. Determining Degradation and Synthesis Rates of Arabidopsis Proteins Using the Kinetics of Progressive 15N Labeling of Two-Dimensional Gel-Separated Protein Spots. Mol. Cell. Proteomics 2012, 11, M111.010025. [Google Scholar] [CrossRef] [PubMed]
- Sperling, E.; Bunner, A.E.; Sykes, M.T.; Williamson, J.R. Quantitative Analysis of Isotope Distributions in Proteomic Mass Spectrometry Using Least-Squares Fourier Transform Convolution. Anal. Chem. 2008, 80, 4906–4917. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.J.; Alexova, R.; Jacoby, R.P.; Harvey Millar, A. Proteins with High Turnover Rate in Barley Leaves Estimated by Proteome Analysis Combined with in Planta Isotope Labeling. Plant Physiol. 2014, 166, 91–108. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, H.; Obata, T.; Sulpice, R.; Fernie, A.R.; Stitt, M. Quantifying Protein Synthesis and Degradation in Arabidopsis by Dynamic 13CO2 Labeling and Analysis of Enrichment in Individual Amino Acids in Their Free Pools and in Protein. Plant Physiol. 2015, 168, 74–93. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Nelson, C.J.; Trösch, J.; Castleden, I.; Huang, S.; Millar, A.H. Protein Degradation Rate in Arabidopsis thaliana Leaf Growth and Development. Plant Cell 2017, 29, 207–228. [Google Scholar] [CrossRef]
- Liebler, D.C.; Zimmerman, L.J. Targeted Quantitation of Proteins by Mass Spectrometry. Biochemistry 2013, 52, 3797–3806. [Google Scholar] [CrossRef]
- Hinkson, I.V.; Elias, J.E. The Dynamic State of Protein Turnover: It’s about Time. Trends Cell Biol. 2011, 21, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Wigge, P.A. H2A. Z-Containing Nucleosomes Mediate the Thermosensory Response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Hu, A.; Noble, W.S.; Wolf-Yadlin, A. Technical Advances in Proteomics: New Developments in Data-Independent Acquisition. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Budnik, B.; Levy, E.; Harmange, G.; Slavov, N. SCoPE-MS: Mass Spectrometry of Single Mammalian Cells Quantifies Proteome Heterogeneity during Cell Differentiation. Genome Biol. 2018, 19, 161. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Assmann, S.M.; Albert, R. Predicting Essential Components of Signal Transduction Networks: A Dynamic Model of Guard Cell Abscisic Acid Signaling. PLoS Biol. 2006, 4, e312. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front. Plant Sci. 2018, 9, 325823. [Google Scholar] [CrossRef]
- Antoniewicz, M.R. A Guide to Metabolic Flux Analysis in Metabolic Engineering: Methods, Tools and Applications. Metab. Eng. 2021, 63, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, R.G.; Shachar-Hill, Y. Measuring Multiple Fluxes through Plant Metabolic Networks. Plant J. 2006, 45, 490–511. [Google Scholar] [CrossRef]
- Burgess, A.J.; Degen, G.E. The Long and the Short of It: Adaptation of Carbon Uptake and Metabolic Flux to Different Daylengths. Plant Physiol. 2024, 194, 317–318. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, X.; Sharkey, T.D.; Shachar-Hill, Y.; Walker, B.J. The Metabolic Origins of Non-Photorespiratory CO2 Release during Photosynthesis: A Metabolic Flux Analysis. Plant Physiol. 2021, 186, 297–314. [Google Scholar] [CrossRef]
- Xu, Y.; Wieloch, T.; Kaste, J.A.M.; Shachar-Hill, Y.; Sharkey, T.D. Reimport of Carbon from Cytosolic and Vacuolar Sugar Pools into the Calvin–Benson Cycle Explains Photosynthesis Labeling Anomalies. Proc. Natl. Acad. Sci. USA 2022, 119, e2121531119. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Koroma, A.A.; Weise, S.E.; Fu, X.; Sharkey, T.D.; Shachar-Hill, Y. Daylength Variation Affects Growth, Photosynthesis, Leaf Metabolism, Partitioning, and Metabolic Fluxes. Plant Physiol. 2024, 194, 475–490. [Google Scholar] [CrossRef]
- Li, T.; Pang, N.; He, L.; Xu, Y.; Fu, X.; Tang, Y.; Shachar-Hill, Y.; Chen, S. Re-Programing Glucose Catabolism in the Microalga Chlorella sorokiniana under Light Condition. Biomolecules 2022, 12, 939. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Freund, D.M.; Hegeman, A.D.; Cohen, J.D. Metabolic Signatures of Arabidopsis thaliana Abiotic Stress Responses Elucidate Patterns in Stress Priming, Acclimation, and Recovery. Stress Biol. 2022, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Urano, K.; Kurihara, Y.; Seki, M.; Shinozaki, K. ‘Omics’ Analyses of Regulatory Networks in Plant Abiotic Stress Responses. Curr. Opin. Plant Biol. 2010, 13, 132–138. [Google Scholar] [CrossRef]
- Yang, X.; Chen, W.; Rendahl, A.K.; Hegeman, A.D.; Gray, W.M.; Cohen, J.D. Measuring the Turnover Rates of Arabidopsis Proteins Using Deuterium Oxide: An Auxin Signaling Case Study. Plant J. 2010, 63, 680–695. [Google Scholar] [CrossRef]


| ID b | Protein | AGI c | Fraction d | Turnover Rate e | SD f | Functional Category g | |
|---|---|---|---|---|---|---|---|
| Fastest | Q9M0A7 | Putative uncharacterized protein (Gamma-glutamyl peptidase 1) | At4g30530 | S | −4.397 | 0.0238 | nucleotide met |
| A8MRQ4_A8MSB9_F4JTU2_Q9SVM8 | Glycine-rich RNA-binding protein 2, mitochondrial | At4g13850 | S | −4.539 | 0.1128 | RNA | |
| P20649 | ATPase 1, plasma membrane-type | At2g18960 | M | −4.607 | 0.0528 | transport | |
| Q9SYM5 | Trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 | At1g78570 | M | −4.624 | 0.0156 | cell wall | |
| F4KIM7_Q9C5N2 | Endomembrane family protein 70 | At5g25100 | M | −4.651 | 0.0223 | N/A | |
| F4J1V2_Q94AW8 | Chaperone protein dnaJ 3 | At3g44110 | M | −4.652 | 0.0881 | stress | |
| P22953 | Probable mediator of RNA polymerase II transcription subunit 37e (Heat Shock cognate Protein 70-1) | At5g02500 | S | −4.668 | 0.0276 | stress | |
| Q9XIE2 | ABC transporter G family member 36 (AtABCG36)(PEN3)(PDR8) | At1g | M | −4.718 | 0.2347 | transport | |
| P31414 | Pyrophosphate-energized vacuolar membrane proton pump 1 | At1g15690 | M | −4.742 | 0.2033 | transport | |
| Q9S791 | Putative uncharacterized protein | At1g70770 | O | −4.752 | 0.1353 | N/A | |
| Slowest | Q43348 | Acid beta-fructofuranosidase 3, vacuolar (Vacuolar invertase 3) | At1g62660 | S | −6.129 | 0.3853 | major CHO met |
| Q9C8Y9 | Beta-glucosidase 22 | At1g66280 | O | −6.150 | 0.2910 | CHO hydrolases | |
| P43297 | Cysteine proteinase RD21a | At1g47128 | M | −6.170 | 0.2050 | prot.degrad | |
| P25819 | Catalase-2 | At4g35090 | O | −6.176 | 0.4179 | redox | |
| Q9FF53 | Probable aquaporin PIP2-4 [Cleaved into: Probable aquaporin PIP2-4, N-terminally processed] | At5g60660 | M | −6.227 | 0.0228 | transport | |
| P46422 | Glutathione S-transferase F2 | At4g02520 | S | −6.244 | 0.0349 | GST | |
| A8MR01_F4JR94_O23179 | Patatin-like protein 1 (AtPLP1) | At4g37070 | M | −6.245 | 0.5113 | development | |
| Q9LHB9 | Peroxidase 32 | At3g32980 | M | −6.261 | 0.2944 | peroxidases | |
| Q9SIE7 | Putative uncharacterized protein (PLAT-plant-stress domain-containing protein) | At2g22170 | S | −6.314 | 0.0765 | N/A | |
| Q9LTQ5 | TRAF-like family protein | At3g20370 | O | −6.320 | 0.3226 | N/A | |
| Q9C8Y9 | Beta-glucosidase 22 | At1g66280 | M | −6.594 | 0.5007 | CHO hydrolases |
| ID b | Protein | AGI c | Fraction d | Turnover Rate e | SD f | Functional Category g | |
|---|---|---|---|---|---|---|---|
| Fastest | B9DG18_Q42547 | Catalase-3 | At1g20620 | S | −4.479 | 0.1605 | redox |
| Q9CA67 | Geranylgeranyl diphosphate reductase, chloroplastic | At1g74470 | M | −4.746 | 0.1219 | 2nd met | |
| Q9CA67 | Geranylgeranyl diphosphate reductase, chloroplastic | At1g74470 | O | −4.857 | 0.1659 | 2nd met | |
| P56761 | Photosystem II D2 protein | AtCg00270 | M | −4.979 | 0.1141 | PS.light | |
| P56761 | Photosystem II D2 protein | AtCg00270 | O | −4.986 | 0.0366 | PS.light | |
| P56778 | Photosystem II CP43 reaction center protein | AtCg00280 | M | −5.101 | 0.1626 | PS.light | |
| P56778 | Photosystem II CP43 reaction center protein | AtCg00280 | O | −5.127 | 0.0665 | PS.light | |
| P42761 | Glutathione S-transferase F10 (GST class-phi member 10) | At2g30870 | S | −5.168 | 0.3743 | GST | |
| Q9LKR3 | Mediator of RNA polymerase II transcription subunit 37a (Heat Shock Protein 70-11) | At5g28540 | M | −5.201 | 0.4357 | stress | |
| P27202 | Photosystem II 10 kDa polypeptide, chloroplastic | At1g79040 | M | −5.220 | 0.2322 | PS.light | |
| Q9LJG3 | GDSL esterase/lipase ESM1 | At3g14210 | O | −5.261 | 0.0880 | 2nd met | |
| O80860 | ATP-dependent zinc metalloprotease FTSH 2, chloroplastic | At2g30950 | O | −5.307 | 0.1091 | prot.degrad | |
| O80860 | ATP-dependent zinc metalloprotease FTSH 2, chloroplastic | At2g30950 | M | −5.312 | 0.1564 | prot.degrad | |
| Q9SRV5 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 2 (AtMS2) | At3g03780 | S | −5.356 | 0.2480 | AA met | |
| Slowest | O80934 | Uncharacterized protein, chloroplastic | At2g37660 | S | −6.783 | 0.2293 | N/A |
| Q8LE52 | Glutathione S-transferase DHAR3, chloroplastic | At5g16710 | S | −6.816 | 0.1549 | redox | |
| P25857 | Glyceraldehyde-3-phosphate dehydrogenase GAPB, chloroplastic | At1g42970 | M | −6.861 | 0.1967 | PS.calvin cycle | |
| Q9XFT3-2 | Oxygen-evolving enhancer protein 3-1, chloroplastic (OEE3) | At4g21280 | M | −6.928 | 0.2714 | PS.light | |
| Q9SR37 | Beta-glucosidase 23 | At3g09260 | O | −7.200 | 0.2308 | CHO hydrolases | |
| Q9SR37 | Beta-glucosidase 23 | At3g09260 | M | −7.218 | 0.2027 | CHO hydrolases | |
| Q8W4H8 | Inactive GDSL esterase/lipase-like protein 23 (Probable myrosinase-associated protein GLL23) | At1g54010 | O | −7.438 | 0.1398 | 2nd met | |
| Q9SR37 | Beta-glucosidase 23 | At3g09260 | S | −7.684 | 0.6082 | CHO hydrolases | |
| Q9LXC9 | Soluble inorganic pyrophosphatase 6, chloroplastic (PPase 6) | At5g09650 | S | −7.774 | 1.5988 | nucleotide met | |
| Q9LTQ5 | TRAF-like family protein | At3g20370 | O | −7.976 | 0.2116 | N/A | |
| Q93Z83 | TRAF-like family protein | At5g26280 | O | −8.472 | 0.5887 | N/A | |
| F4IB98 | Jacalin-related lectin 11 | At1g52100 | O | −8.879 | 1.2147 | hormone met |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, K.-T.; Xu, Y.; Hegeman, A.D. Elevated Temperature Effects on Protein Turnover Dynamics in Arabidopsis thaliana Seedlings Revealed by 15N-Stable Isotope Labeling and ProteinTurnover Algorithm. Int. J. Mol. Sci. 2024, 25, 5882. https://doi.org/10.3390/ijms25115882
Fan K-T, Xu Y, Hegeman AD. Elevated Temperature Effects on Protein Turnover Dynamics in Arabidopsis thaliana Seedlings Revealed by 15N-Stable Isotope Labeling and ProteinTurnover Algorithm. International Journal of Molecular Sciences. 2024; 25(11):5882. https://doi.org/10.3390/ijms25115882
Chicago/Turabian StyleFan, Kai-Ting, Yuan Xu, and Adrian D. Hegeman. 2024. "Elevated Temperature Effects on Protein Turnover Dynamics in Arabidopsis thaliana Seedlings Revealed by 15N-Stable Isotope Labeling and ProteinTurnover Algorithm" International Journal of Molecular Sciences 25, no. 11: 5882. https://doi.org/10.3390/ijms25115882
APA StyleFan, K.-T., Xu, Y., & Hegeman, A. D. (2024). Elevated Temperature Effects on Protein Turnover Dynamics in Arabidopsis thaliana Seedlings Revealed by 15N-Stable Isotope Labeling and ProteinTurnover Algorithm. International Journal of Molecular Sciences, 25(11), 5882. https://doi.org/10.3390/ijms25115882







