Evidence Supporting a Role of Alternative Splicing Participates in Melon (Cucumis melo L.) Fruit Ripening
Abstract
1. Introduction
2. Results
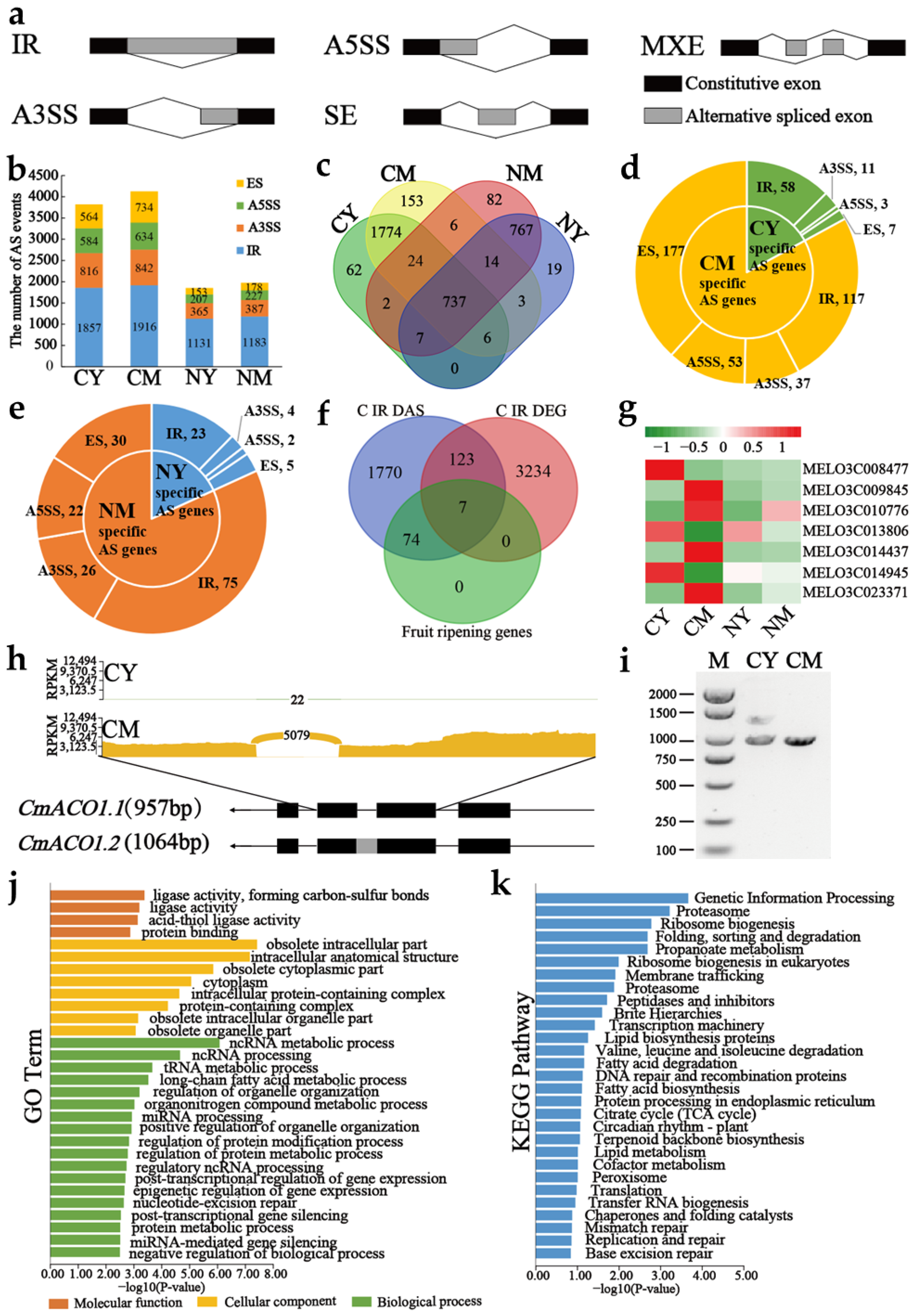
2.1. Alternative Splicing Landscape Changes in Young and Mature Stages
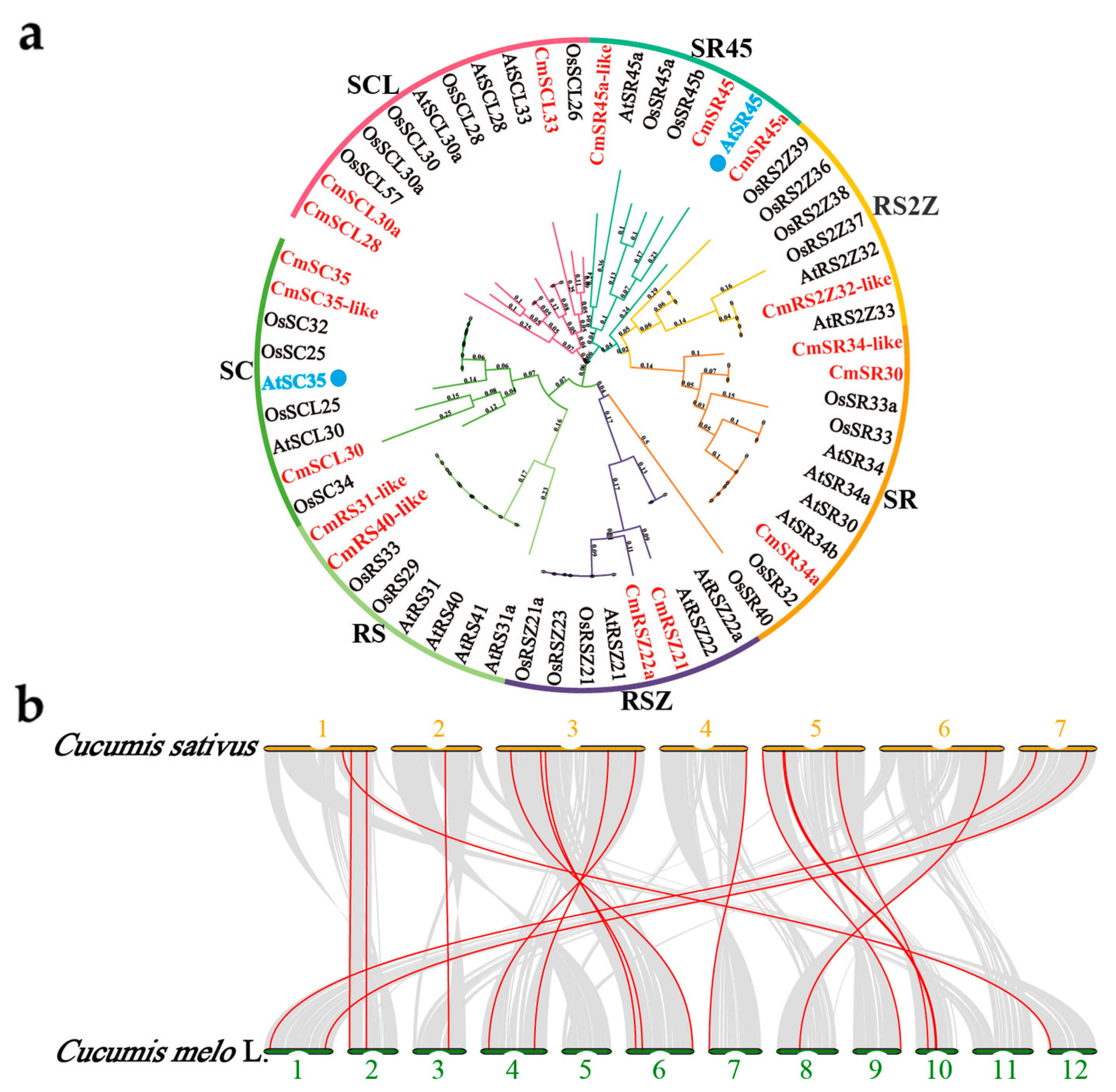
2.2. Identification of the CmSR Family
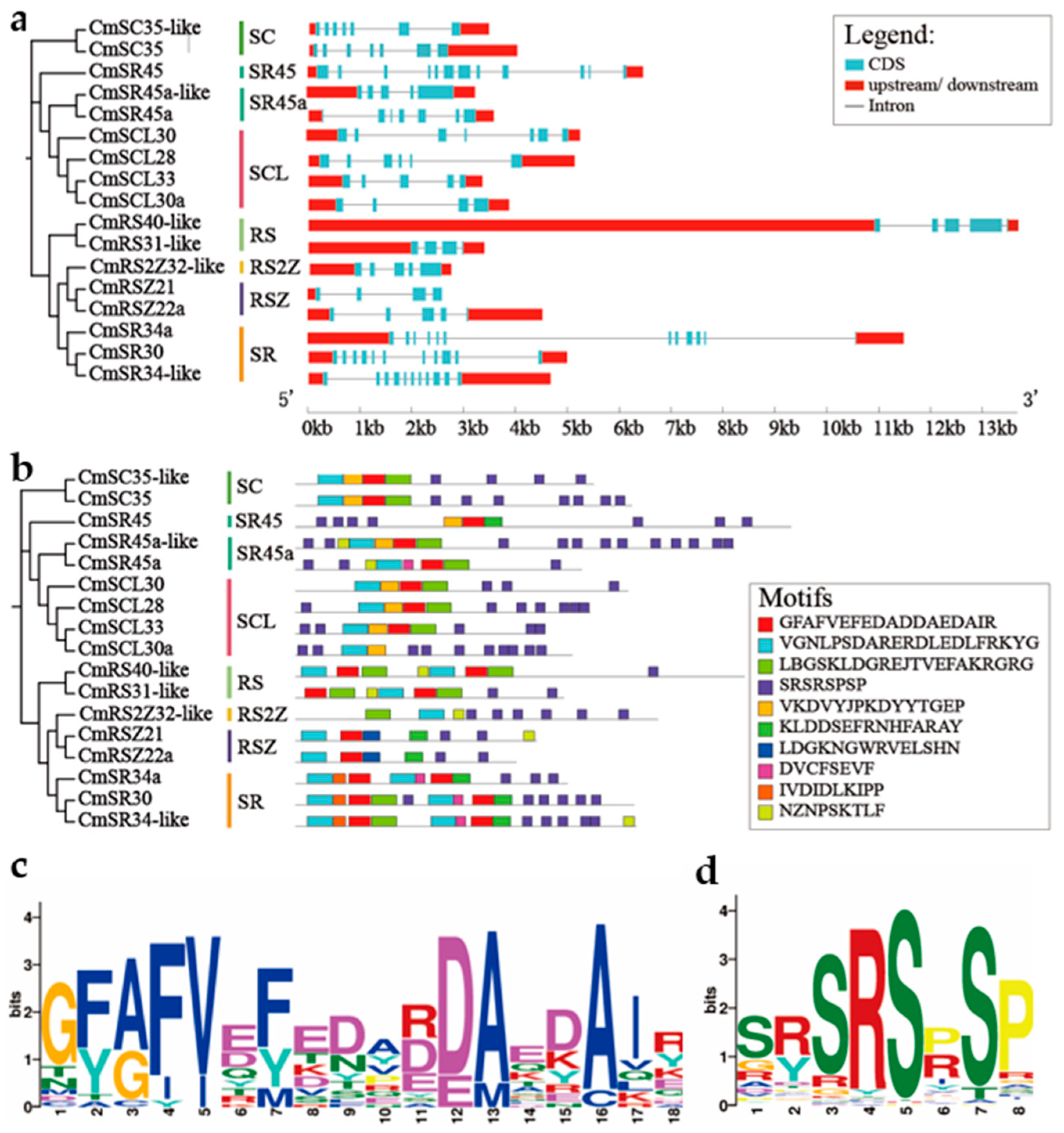
2.3. Phylogenetic Analysis, Syntenic Analysis, Gene Structure and Conserved Motifs of the CmSR Family
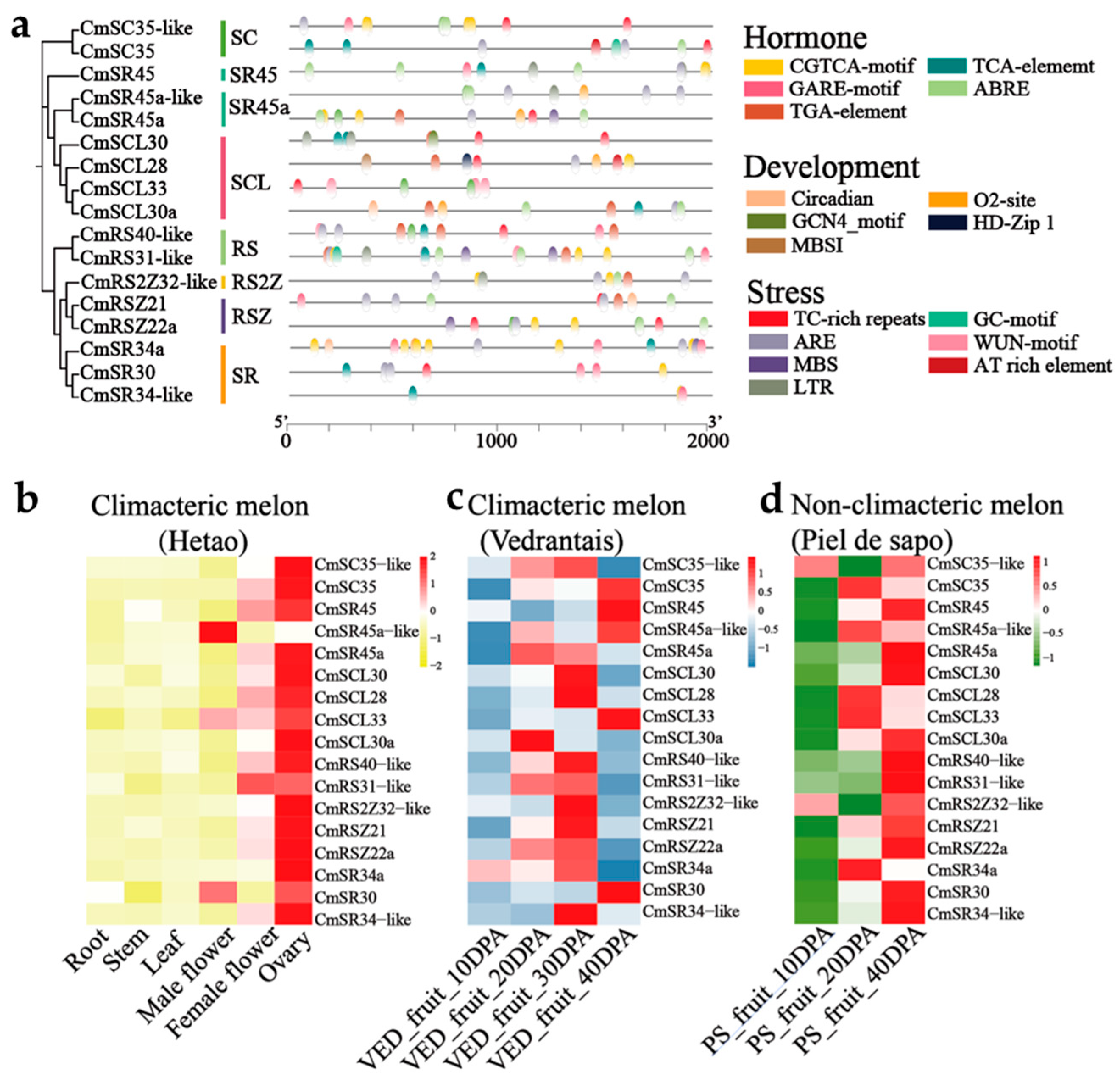
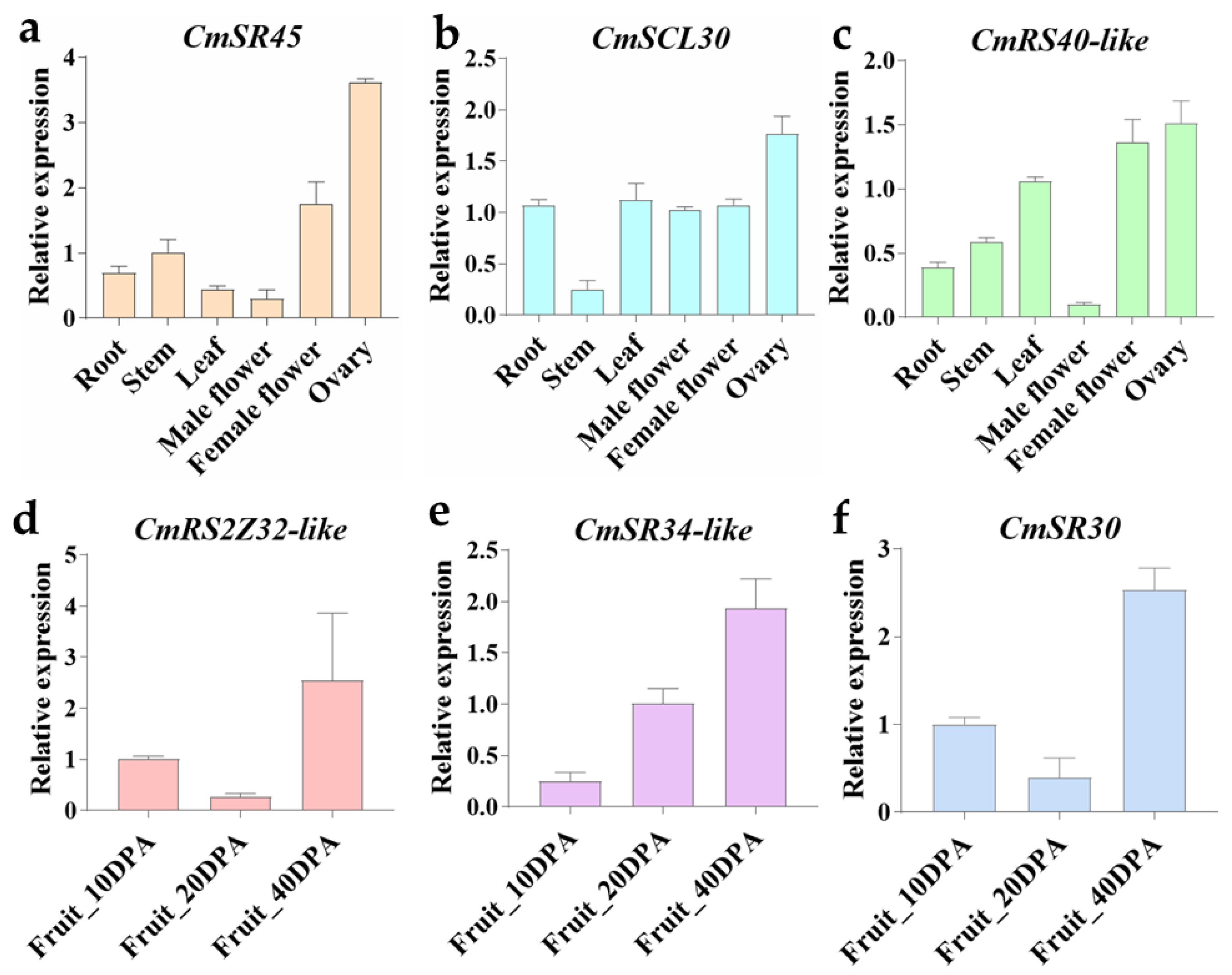
2.4. Cis-Elements in Promoter Regions and Gene Expression Analysis of SR Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Data Source
4.2. Identification of AS Genes and Events
4.3. Validation of AS Events
4.4. Identification of SR Genes in Cucumis melo L.
4.5. Phylogenetic Analysis of SR Family Members, Gene Duplication Analysis
4.6. Analysis of Gene Structure, Conserved Motifs, and cis-Acting Regulatory Elements
4.7. Preprocessing, Quality Control, and RNA-seq Data Analysis
4.8. QRT-PCR Analysis and Statistical Analysis
4.9. Analysis of Nucleotide Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shepard, P.J.; Hertel, K.J. The SR protein family. Genome Biol. 2009, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Cao, Y.; Ma, L. Alternative splicing in plant genes: A means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef] [PubMed]
- Szakonyi, D.; Duque, P. Alternative splicing as a regulator of early plant development. Front. Plant Sci. 2018, 9, 382146. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Mahfouz, M.M.; Zhou, S. Pre-mRNA alternative splicing as a modulator for heat stress response in plants. Trends Plant Sci. 2021, 26, 1153–1170. [Google Scholar] [CrossRef]
- Will, C.L.; Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef] [PubMed]
- Zahler, A.M.; Lane, W.S.; Stolk, J.A.; Roth, M.B. SR proteins: A conserved family of pre-mRNA splicing factors. Genes Dev. 1992, 6, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, N.; Yoshimura, K.; Kimura, A.; Yabuta, Y.; Shigeoka, S. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol. 2007, 48, 1036–1049. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, M.; Tsumoto, A.; Shimamoto, K. The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell 2006, 18, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.N.; Rogers, M.F.; Labadorf, A.; Ben-Hur, A.; Guo, H.; Paterson, A.H.; Reddy, A.S. Comparative analysis of serine/arginine-rich proteins across 27 eukaryotes: Insights into sub-family classification and extent of alternative splicing. PLoS ONE 2011, 6, e24542. [Google Scholar] [CrossRef]
- Chen, S.; Li, J.; Liu, Y.; Li, H. Genome-wide analysis of serine/arginine-rich protein family in wheat and Brachypodium distachyon. Plants 2019, 8, 188. [Google Scholar] [CrossRef]
- Rosenkranz, R.R.; Bachiri, S.; Simm, S.; Schleiff, E.; Fragkostefanakis, S. Identification and regulation of tomato serine/arginine-rich proteins under high temperatures. Front. Plant Sci. 2021, 12, 645689. [Google Scholar] [CrossRef]
- Gu, J.; Ma, S.; Zhang, Y.; Wang, D.; Cao, S.; Wang, Z.-Y. Genome-wide identification of cassava serine/arginine-rich proteins: Insights into alternative splicing of pre-mRNAs and response to abiotic stress. Plant Cell Physiol. 2020, 61, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Chen, P.; Jian, H.; Sun, L.; Lv, X.; Wei, H.; Wang, H.; Hu, T.; Ma, L.; Fu, X. A comprehensive identification and function analysis of Serine/Arginine-Rich (SR) proteins in cotton (Gossypium spp.). Int. J. Mol. Sci. 2022, 23, 4566. [Google Scholar] [CrossRef]
- Xie, M.; Zuo, R.; Bai, Z.; Zhao, C.; Huang, J.; Tong, C.; Liu, S. Genome-wide characterization of Serine/Arginine-rich gene family and its genetic effects on agronomic traits of Brassica napus. Front. Plant Sci. 2022, 13, 829668. [Google Scholar] [CrossRef]
- Chen, X.; Huang, S.; Jiang, M.; Chen, Y.; XuHan, X.; Zhang, Z.; Lin, Y.; Lai, Z. Genome-wide identification and expression analysis of the SR gene family in longan (Dimocarpus longan Lour.). PLoS ONE 2020, 15, e0238032. [Google Scholar] [CrossRef] [PubMed]
- Barta, A.; Kalyna, M.; Reddy, A.S. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 2010, 22, 2926–2929. [Google Scholar] [CrossRef]
- Kalyna, M.; Barta, A. A Plethora of Plant Serine/arginine-Rich Proteins: Redundancy or Evolution of Novel Gene Functions? Portland Press Ltd.: London, UK, 2004. [Google Scholar]
- Manley, J.L.; Krainer, A.R. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010, 24, 1073–1074. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.S.; Palusa, S.G.; Golovkin, M.; Prasad, J.; Manley, J.L.; Reddy, A.S. Regulation of plant developmental processes by a novel splicing factor. PLoS ONE 2007, 2, e471. [Google Scholar] [CrossRef]
- Sapra, A.K.; Änkö, M.-L.; Grishina, I.; Lorenz, M.; Pabis, M.; Poser, I.; Rollins, J.; Weiland, E.-M.; Neugebauer, K.M. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol. Cell 2009, 34, 179–190. [Google Scholar] [CrossRef]
- Zhang, W.; Du, B.; Liu, D.; Qi, X. Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 2014, 455, 312–317. [Google Scholar] [CrossRef]
- Chen, T.; Cui, P.; Chen, H.; Ali, S.; Zhang, S.; Xiong, L. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 2013, 9, e1003875. [Google Scholar] [CrossRef]
- Zhao, X.; Tan, L.; Wang, S.; Shen, Y.; Guo, L.; Ye, X.; Liu, S.; Feng, Y.; Wu, W. The SR splicing factors: Providing perspectives on their evolution, expression, alternative splicing, and function in Populus trichocarpa. Int. J. Mol. Sci. 2021, 22, 11369. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, L.; Zhang, Y.; Ouyang, Z.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 2014, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Liu, X.; Liu, C.; Liu, G.; Li, S.; Wang, L. Integrating omics and alternative splicing reveals insights into grape response to high temperature. Plant Physiol. 2017, 173, 1502–1518. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wu, T.; Shi, L.; Yang, L.; Zhang, C. Alternative splicing control of light and temperature stress responses and its prospects in vegetable crops. Veg. Res. 2023, 3, 17. [Google Scholar] [CrossRef]
- Yan, Q.; Xia, X.; Sun, Z.; Fang, Y. Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genet. 2017, 13, e1006663. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Zhang, D.; Li, Z.; Liang, H.; Deng, R.; Su, X.; Jiang, Y.; Duan, X. Alternative splicing of MaMYB16L regulates starch degradation in banana fruit during ripening. J. Integr. Plant Biol. 2021, 63, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, C.; Shao, G.; Wu, S.; Liu, P.; Cao, P.; Jiang, P.; Wang, S.; Zhu, H.; Lin, X. The reference genome and full-length transcriptome of pakchoi provide insights into cuticle formation and heat adaption. Hortic. Res. 2022, 9, uhac123. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Zeng, N.; Qu, G.; Fu, D.; Zhu, B.; Luo, Y.; Ostersetzer-Biran, O.; Zhu, H. Glycine-rich RNA-binding cofactor RZ1AL is associated with tomato ripening and development. Hortic. Res. 2022, 9, uhac134. [Google Scholar] [CrossRef]
- Liu, B.; Santo Domingo, M.; Mayobre, C.; Martín-Hernández, A.M.; Pujol, M.; Garcia-Mas, J. Knock-out of CmNAC-NOR affects melon climacteric fruit ripening. Front. Plant Sci. 2022, 13, 878037. [Google Scholar] [CrossRef]
- Bartlett, A.; O’Malley, R.C.; Huang, S.-s.C.; Galli, M.; Nery, J.R.; Gallavotti, A.; Ecker, J.R. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 2017, 12, 1659–1672. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Estrada, A.D.; Blakley, I.; Reid, R.; Patel, K.; Meyer, M.D.; Andersen, S.U.; Brown, A.F.; Lila, M.A.; Loraine, A.E. RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. Gigascience 2015, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Bai, D.; Song, H.; Lin, K.; Pang, E. Alternative splicing during fruit development among fleshy fruits. BMC Genom. 2021, 22, 762. [Google Scholar] [CrossRef] [PubMed]
- Zarid, M.; Esteras, C.; Gemma Sifres, A.; Cañizares, X.; Esteva, J.; Picó, B.; Monforte, A.; Fernández Trujillo, J. Gene Expression and Volatile Production during Melon Ripening. In Proceedings of the 6th Workshop on Agri-Food Research, Murcia, Spain, 8–9 May 2017; Artés-Hernández, F., Egea-Cortines, M., Fernández-Hernández, J.A., Calatrava, J., Aguayo, E., Alarcón, J.J., Cos, J.E., Eds.; CRAI Biblioteca. Universidad Politécnica de Cartagena: Murcia, Spain, 2017; pp. 27–30. [Google Scholar]
- Shi, M.; Ali, M.M.; Sun, K.; Gull, S.; Hu, X.; Kayima, V.; Cai, S.; Hou, Y.; Chen, F. Changes in fruit anthocyanins, their biosynthesis-related enzymes and related genes during fruit development of purple and yellow passion fruits. Fruit Res. 2023, 3, 17. [Google Scholar] [CrossRef]
- Li, Z.; Chen, C.; Zou, D.; Li, J.; Huang, Y.; Zheng, X.; Tan, B.; Cheng, J.; Wang, W.; Zhang, L. Ethylene accelerates grape ripening via increasing VvERF75-induced ethylene synthesis and chlorophyll degradation. Fruit Res. 2023, 3, 1–9. [Google Scholar] [CrossRef]
- Feder, A.; Jiao, C.; Galpaz, N.; Vrebalov, J.; Xu, Y.; Portnoy, V.; Tzuri, G.; Gonda, I.; Burger, J.; Gur, A. Melon ethylene-mediated transcriptome and methylome dynamics provide insights to volatile production. bioRxiv 2020, preprint. [Google Scholar] [CrossRef]
- Li, Y.; Qi, H.; Jin, Y.; Tian, X.; Sui, L.; Qiu, Y. Role of ethylene in biosynthetic pathway of related-aroma volatiles derived from amino acids in oriental sweet melons (Cucumis melo var. makuwa Makino). Sci. Hortic. 2016, 201, 24–35. [Google Scholar] [CrossRef]
- Boldt, R.; Edner, C.; Kolukisaoglu, U.; Hagemann, M.; Weckwerth, W.; Wienkoop, S.; Morgenthal, K.; Bauwe, H. D-GLYCERATE 3-KINASE, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell 2005, 17, 2413–2420. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.; Priest, H.D.; Megraw, M.; Mockler, T.C. Alternative splicing in plants: Directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol. 2015, 24, 125–135. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 1–4. [Google Scholar] [CrossRef]
- Sanjur, O.I.; Piperno, D.R.; Andres, T.C.; Wessel-Beaver, L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: Implications for crop plant evolution and areas of origin. Proc. Natl. Acad. Sci. USA 2002, 99, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Dai, Z.; Zeng, B.; Li, J.; Ouyang, J.; Kang, L.; Wang, W.; Jia, W. Autocatalytic biosynthesis of abscisic acid and its synergistic action with auxin to regulate strawberry fruit ripening. Hortic. Res. 2022, 9, uhab076. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Lian, Q.; Zhang, Z.; Fu, Q.; He, Y.; Ma, S.; Ruggieri, V.; Monforte, A.J.; Wang, P.; Julca, I. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 2019, 51, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhao, S.; Li, P.; Yin, Y.; Niu, Q.; Yan, J.; Huang, D. Plant buffering against the high-light stress-induced accumulation of CsGA2ox8 transcripts via alternative splicing to finely tune gibberellin levels and maintain hypocotyl elongation. Hortic. Res. 2021, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fanara, S.; Schloesser, M.; Joris, M.; De Franco, S.; Vandevenne, M.; Kerff, F.; Hanikenne, M.; Motte, P. The Arabidopsis SR45 splicing factor bridges the splicing machinery and the exon-exon junction complex. bioRxiv 2023, preprint. [Google Scholar] [CrossRef]
- Oudelaar, A.M.; Higgs, D.R. The relationship between genome structure and function. Nat. Rev. Genet. 2021, 22, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Sun, K.; Mackaluso, J.D.; Seddon, A.E.; Jin, R.; Thomashow, M.F.; Shiu, S.-H. Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 14992–14997. [Google Scholar] [CrossRef] [PubMed]
- Heldenbrand, J.; Ren, Y.; Asmann, Y.; Mainzer, L.S. Step-by-Step Guide for Downloading Very Large Datasets to a Supercomputer Using the SRA Toolkit. 2017. Available online: https://slack.protocols.io:8443/view/step-by-step-guide-for-downloading-very-large-data-kb6csre.html (accessed on 1 October 2022).
- Shen, S.; Park, J.W.; Lu, Z.-x.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.; Jiao, C.; Guo, S.; Zhao, K.; Blanca, J.; Zhang, Z.; Huang, S. Cucurbit Genomics Database (CuGenDB): A central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2019, 47, D1128–D1136. [Google Scholar] [CrossRef] [PubMed]
- Duque, P. A role for SR proteins in plant stress responses. Plant Signal. Behav. 2011, 6, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; He, F.; Berkowitz, O.; Liu, J.; Cao, P.; Tang, M.; Shi, H.; Wang, W.; Li, Q.; Shen, Z. Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell 2018, 30, 2267–2285. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Seto, D.; Mahadevan, P.; Ackermann, H.-W.; Kropinski, A.M. Unifying classical and molecular taxonomic classification: Analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 2008, 159, 406–414. [Google Scholar] [CrossRef]
- Cazalla, D.; Zhu, J.; Manche, L.; Huber, E.; Krainer, A.R.; Cáceres, J.F. Nuclear export and retention signals in the RS domain of SR proteins. Mol. Cell. Biol. 2002, 22, 6871–6882. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Wei, Y.; Xu, Z.; Shen, C.; Li, A.; Guan, D.; Zhang, X.; Liu, B. Evidence Supporting a Role of Alternative Splicing Participates in Melon (Cucumis melo L.) Fruit Ripening. Int. J. Mol. Sci. 2024, 25, 5886. https://doi.org/10.3390/ijms25115886
Wang W, Wei Y, Xu Z, Shen C, Li A, Guan D, Zhang X, Liu B. Evidence Supporting a Role of Alternative Splicing Participates in Melon (Cucumis melo L.) Fruit Ripening. International Journal of Molecular Sciences. 2024; 25(11):5886. https://doi.org/10.3390/ijms25115886
Chicago/Turabian StyleWang, Wenjiao, Yuping Wei, Zhaoying Xu, Chengcheng Shen, Ang Li, Dailu Guan, Xuejun Zhang, and Bin Liu. 2024. "Evidence Supporting a Role of Alternative Splicing Participates in Melon (Cucumis melo L.) Fruit Ripening" International Journal of Molecular Sciences 25, no. 11: 5886. https://doi.org/10.3390/ijms25115886
APA StyleWang, W., Wei, Y., Xu, Z., Shen, C., Li, A., Guan, D., Zhang, X., & Liu, B. (2024). Evidence Supporting a Role of Alternative Splicing Participates in Melon (Cucumis melo L.) Fruit Ripening. International Journal of Molecular Sciences, 25(11), 5886. https://doi.org/10.3390/ijms25115886





