GNN Codon Adjacency Tunes Protein Translation
Abstract
1. Introduction
2. Results and Discussion
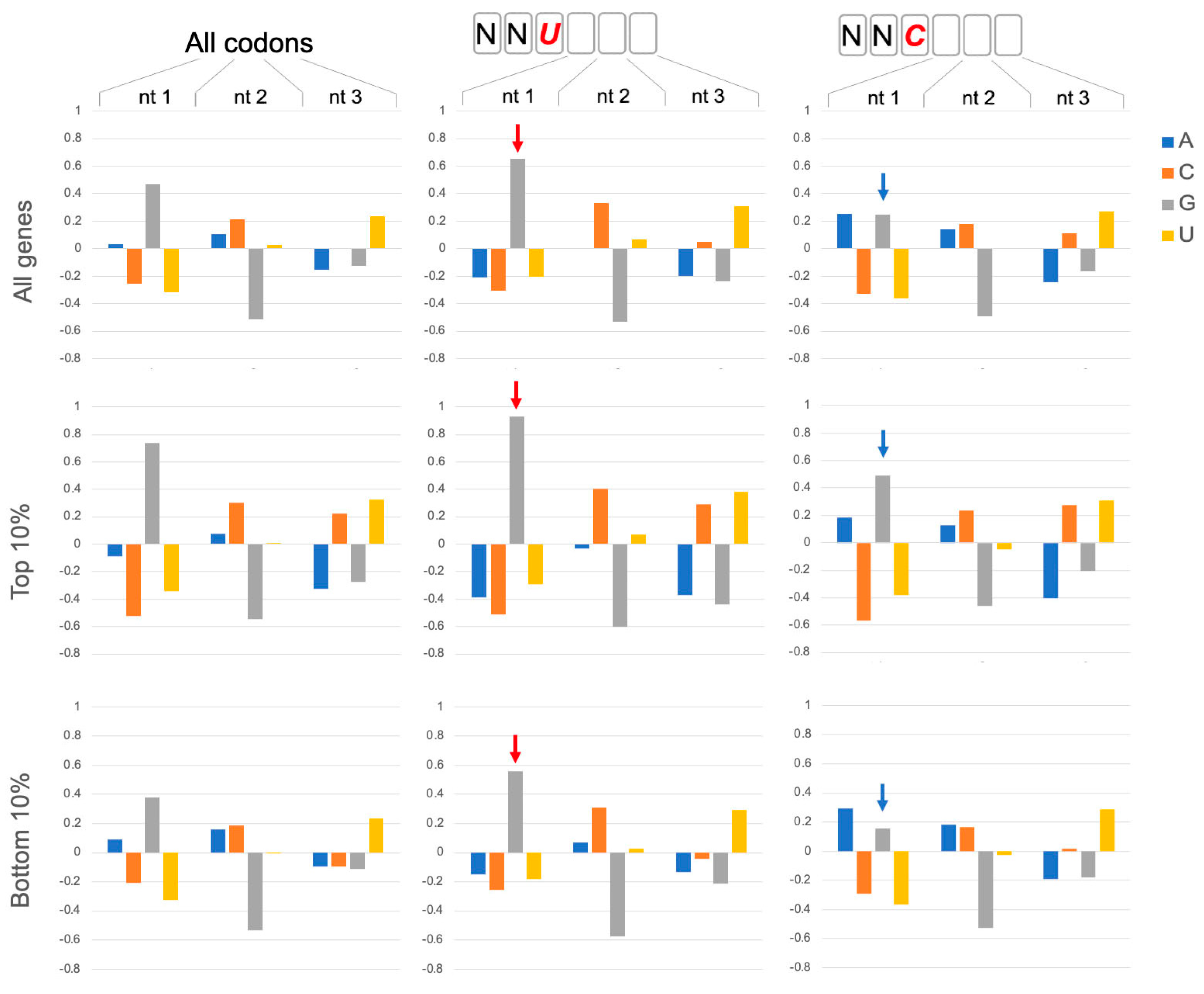
2.1. NNU Codons Have Distinctive Properties
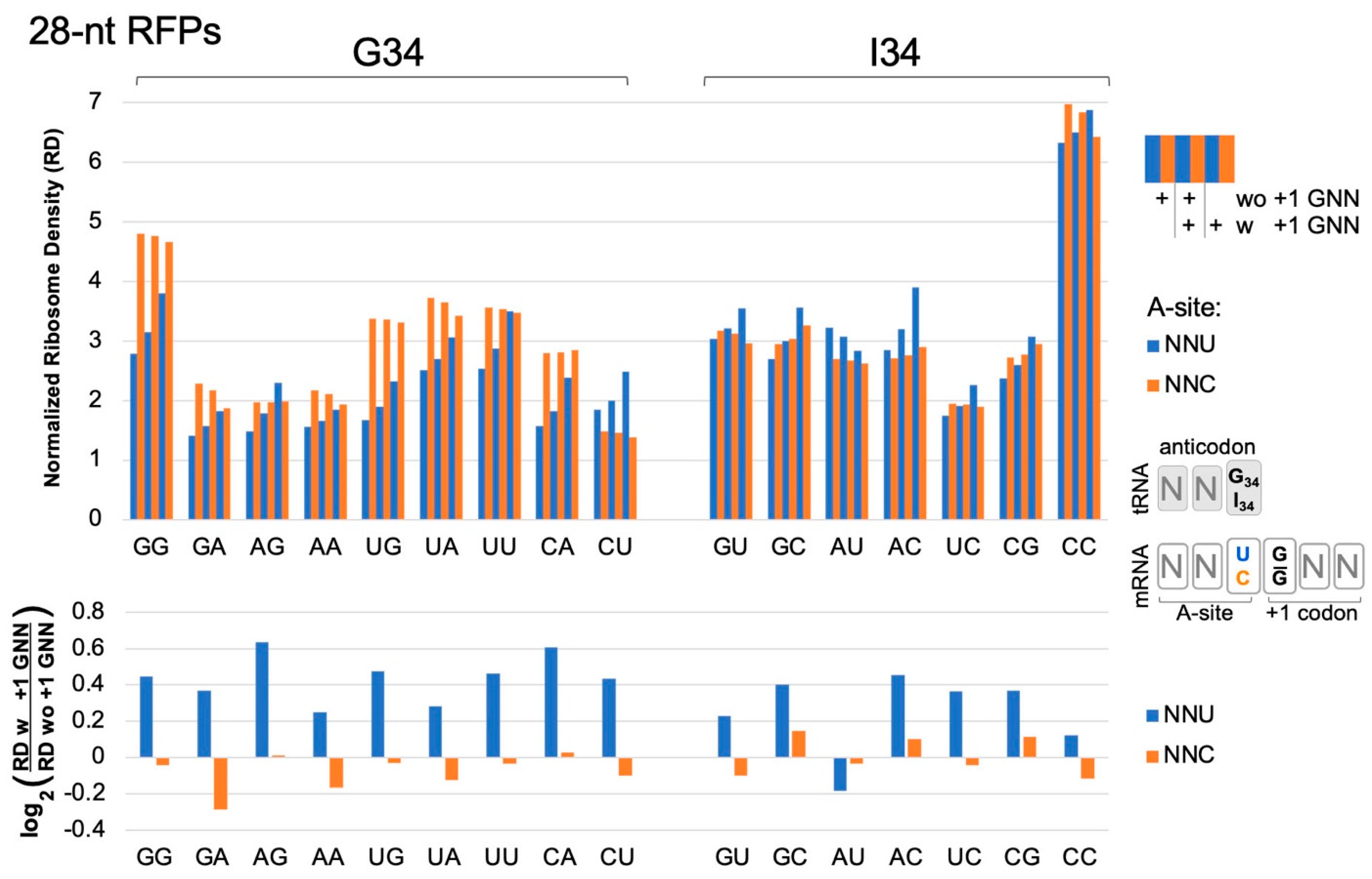
2.2. GNN Adjacency Slows Protein Translation
2.3. The CAR Surface Is Ideally Positioned to Mediate +1 GNN Regulation
2.4. A Sequence-Sensitive Ribosome Braking System
3. Materials and Methods
3.1. Ribosome Profiling Analysis
3.2. Molecular Dynamics Simulations
3.3. Codon Frequency and Information-Theoretic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barr, W.A.; Sheth, R.B.; Kwon, J.; Cho, J.; Glickman, J.W.; Hart, F.; Chatterji, O.K.; Scopino, K.; Voelkel-Meiman, K.; Krizanc, D.; et al. GCN sensitive protein translation in yeast. PLoS ONE 2020, 15, e0233197. [Google Scholar] [CrossRef] [PubMed]
- Curran, J.F.; Gross, B.L. Evidence that GHN phase bias does not constitute a framing code. J. Mol. Biol. 1994, 235, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Lagunez-Otero, J.; Trifonov, E.N. mRNA periodical infrastructure complementary to the proof-reading site in the ribosome. J. Biomol. Struct. Dyn. 1992, 10, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, L.; Mondragon, M.; Lagunez-Otero, J. Interaction of the 530 ribosomal site with regions of mRNA. Biosystems 1998, 46, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Trifonov, E.N. Translation framing code and frame-monitoring mechanism as suggested by the analysis of mRNA and 16 S rRNA nucleotide sequences. J. Mol. Biol. 1987, 194, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Abeyrathne, P.D.; Koh, C.S.; Grant, T.; Grigorieff, N.; Korostelev, A.A. Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome. Elife 2016, 5, e14874. [Google Scholar] [CrossRef] [PubMed]
- Dalgarno, C.; Scopino, K.; Raval, M.; Nachmanoff, C.; Sakkas, E.D.; Krizanc, D.; Thayer, K.M.; Weir, M.P. The CAR-mRNA Interaction Surface Is a Zipper Extension of the Ribosome A Site. Int. J. Mol. Sci. 2022, 23, 1417. [Google Scholar] [CrossRef] [PubMed]
- Scopino, K.; Dalgarno, C.; Nachmanoff, C.; Krizanc, D.; Thayer, K.M.; Weir, M.P. Arginine Methylation Regulates Ribosome CAR Function. Int. J. Mol. Sci. 2021, 22, 1335. [Google Scholar] [CrossRef] [PubMed]
- Scopino, K.; Williams, E.; Elsayed, A.; Barr, W.A.; Krizanc, D.; Thayer, K.M.; Weir, M.P. A Ribosome Interaction Surface Sensitive to mRNA GCN Periodicity. Biomolecules 2020, 10, 849. [Google Scholar] [CrossRef]
- Ghaemmaghami, S.; Huh, W.K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Burton, Z.F. “Superwobbling” and tRNA-34 Wobble and tRNA-37 Anticodon Loop Modifications in Evolution and Devolution of the Genetic Code. Life 2022, 12, 252. [Google Scholar] [CrossRef] [PubMed]
- Gamble, C.E.; Brule, C.E.; Dean, K.M.; Fields, S.; Grayhack, E.J. Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast. Cell 2016, 166, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Letzring, D.P.; Dean, K.M.; Grayhack, E.J. Control of translation efficiency in yeast by codon-anticodon interactions. Rna 2010, 16, 2516–2528. [Google Scholar] [CrossRef] [PubMed]
- Tesina, P.; Lessen, L.N.; Buschauer, R.; Cheng, J.; Wu, C.C.; Berninghausen, O.; Buskirk, A.R.; Becker, T.; Beckmann, R.; Green, R. Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J. 2020, 39, e103365. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Zinshteyn, B.; Wehner, K.A.; Green, R. High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol. Cell 2019, 73, 959–970 e955. [Google Scholar] [CrossRef] [PubMed]
- Stadler, M.; Fire, A. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA 2011, 17, 2063–2073. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef]
- Lin, Y.; May, G.E.; Kready, H.; Nazzaro, L.; Mao, M.; Spealman, P.; Creeger, Y.; McManus, C.J. Impacts of uORF codon identity and position on translation regulation. Nucleic Acids Res. 2019, 47, 9358–9367. [Google Scholar] [CrossRef]
- Komar, A.A.; Samatova, E.; Rodnina, M.V. Translation Rates and Protein Folding. J. Mol. Biol. 2023, 168384. [Google Scholar] [CrossRef]
- Liu, Y. A code within the genetic code: Codon usage regulates co-translational protein folding. Cell Commun. Signal 2020, 18, 145. [Google Scholar] [CrossRef] [PubMed]
- Waudby, C.A.; Dobson, C.M.; Christodoulou, J. Nature and Regulation of Protein Folding on the Ribosome. Trends Biochem. Sci. 2019, 44, 914–926. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 2020, 182, 404–416 e414. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef]
- Endres, L.; Rose, R.E.; Doyle, F.; Rahn, T.; Lee, B.; Seaman, J.; McIntyre, W.D.; Fabris, D. 2′-O-ribose methylation of transfer RNA promotes recovery from oxidative stress in Saccharomyces cerevisiae. PLoS ONE 2020, 15, e0229103. [Google Scholar] [CrossRef]
- Gu, C.; Begley, T.J.; Dedon, P.C. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014, 588, 4287–4296. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.M.; Leonardi, A.; Dedon, P.C.; Begley, T.J. The Versatile Roles of the tRNA Epitranscriptome during Cellular Responses to Toxic Exposures and Environmental Stress. Toxics 2019, 7, 17. [Google Scholar] [CrossRef]
- Jenner, L.; Starosta, A.L.; Terry, D.S.; Mikolajka, A.; Filonava, L.; Yusupov, M.; Blanchard, S.C.; Wilson, D.N.; Yusupova, G. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3812–3816. [Google Scholar] [CrossRef] [PubMed]
- Budkevich, T.; Giesebrecht, J.; Altman, R.B.; Munro, J.B.; Mielke, T.; Nierhaus, K.H.; Blanchard, S.C.; Spahn, C.M. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell 2011, 44, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kingsford, C.; McManus, C.J. Using the Ribodeblur pipeline to recover A-sites from yeast ribosome profiling data. Methods 2018, 137, 67–70. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Hwang, P.; Sakkas, E.D.; Zhou, Y.; Perez, L.; Dave, I.; Kwon, J.B.; McMahon, A.E.; Wichman, M.; Raval, M.; et al. GNN Codon Adjacency Tunes Protein Translation. Int. J. Mol. Sci. 2024, 25, 5914. https://doi.org/10.3390/ijms25115914
Sun J, Hwang P, Sakkas ED, Zhou Y, Perez L, Dave I, Kwon JB, McMahon AE, Wichman M, Raval M, et al. GNN Codon Adjacency Tunes Protein Translation. International Journal of Molecular Sciences. 2024; 25(11):5914. https://doi.org/10.3390/ijms25115914
Chicago/Turabian StyleSun, Joyce, Pete Hwang, Eric D. Sakkas, Yancheng Zhou, Luis Perez, Ishani Dave, Jack B. Kwon, Audrey E. McMahon, Mia Wichman, Mitsu Raval, and et al. 2024. "GNN Codon Adjacency Tunes Protein Translation" International Journal of Molecular Sciences 25, no. 11: 5914. https://doi.org/10.3390/ijms25115914
APA StyleSun, J., Hwang, P., Sakkas, E. D., Zhou, Y., Perez, L., Dave, I., Kwon, J. B., McMahon, A. E., Wichman, M., Raval, M., Scopino, K., Krizanc, D., Thayer, K. M., & Weir, M. P. (2024). GNN Codon Adjacency Tunes Protein Translation. International Journal of Molecular Sciences, 25(11), 5914. https://doi.org/10.3390/ijms25115914





