From the Microbiome to the Electrome: Implications for the Microbiota–Gut–Brain Axis
Abstract
1. Introduction
2. Results
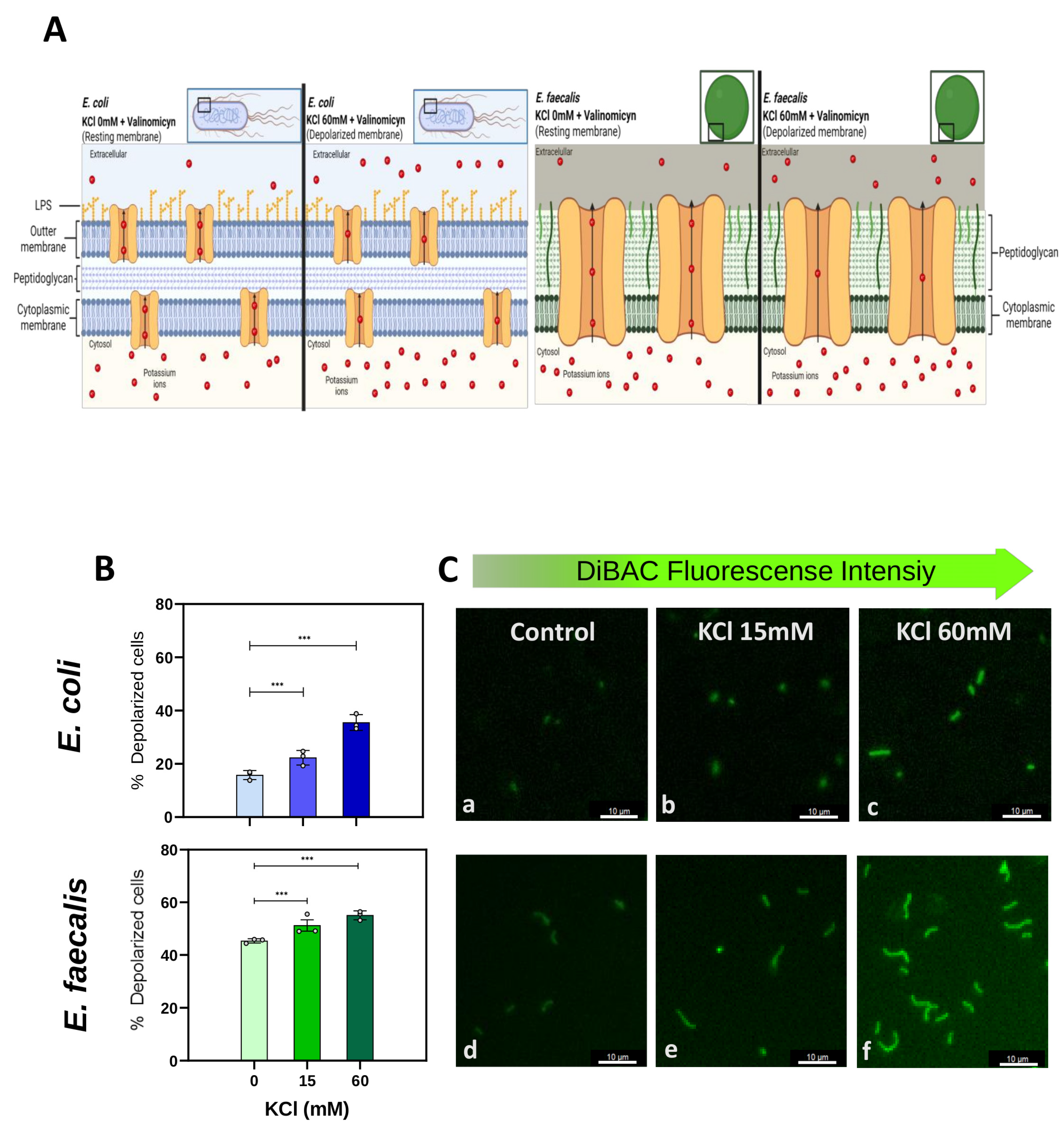
2.1. The Voltage-Sensitive Dye DiBAC Is a Reliable Marker for Detecting Membrane Potential (Vmem) Changes in E. coli and E. faecalis Cells
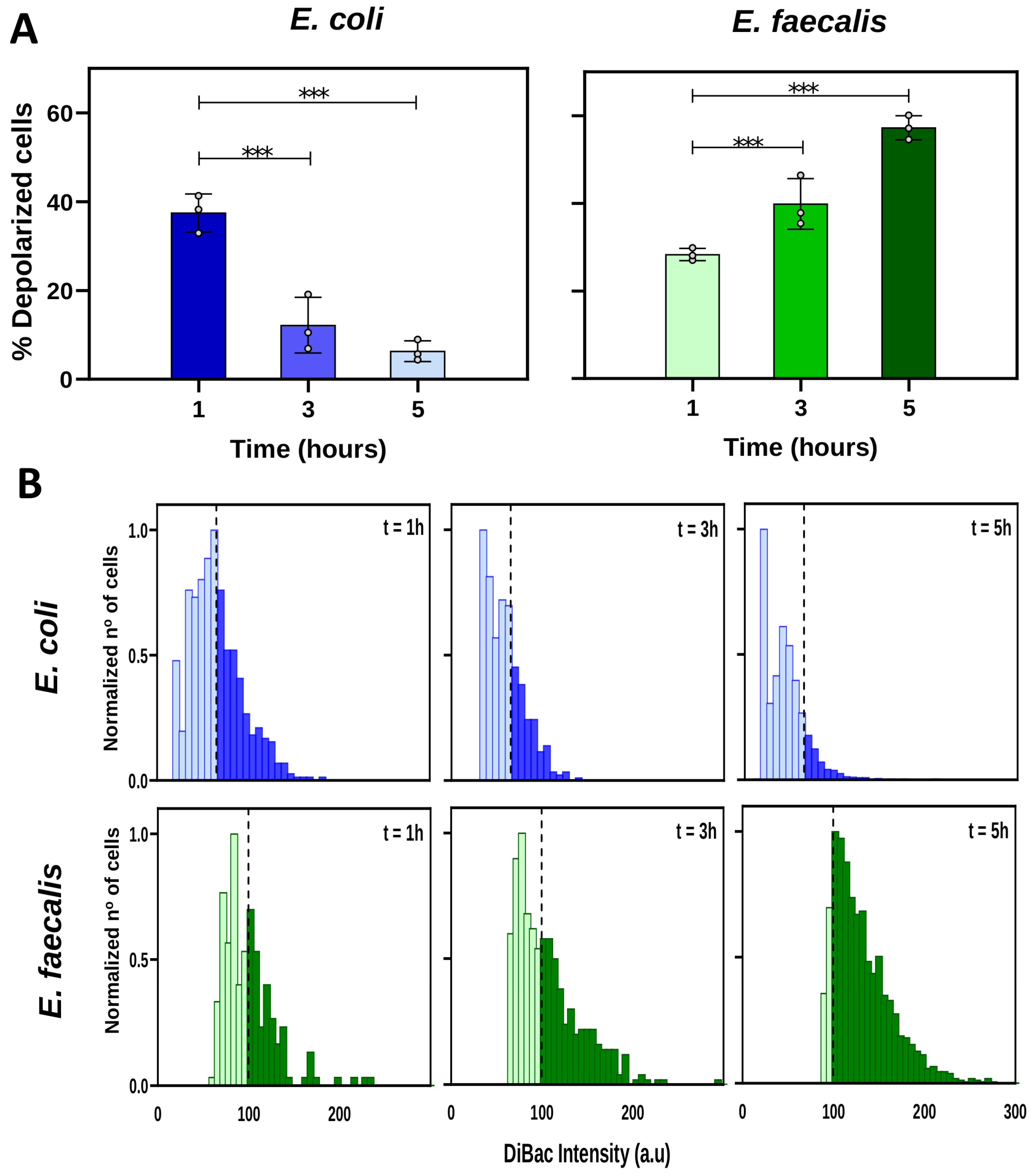
2.2. Changes in Vmem during Growth Phases Are Unique for Each Bacterial Strain
2.3. Glutamate and GABA Exposure Induces Vmem Changes in Bacteria
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Validation of (DiBAC4(3)), a Voltage-Sensitive Fluorescent Dye, in Bacterial Cultures
4.3. DiBAC4(3) Assay to Characterize Bioelectrical Profile during Growth
4.4. Effect of Neurotransmitters on E. coli and E. faecalis: Bioelectrical Profile, Growth, Cultivability, and Viability
4.5. Imaging and Image Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Hugon, P.; Dufour, J.C.; Colson, P.; Fournier, P.E.; Sallah, K.; Raoult, D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis. 2015, 15, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Parizadeh, M.; Arrieta, M.C. The global human gut microbiome: Genes, lifestyles, and diet. Trends Mol. Med. 2023, 29, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.O.; Lundgren, P.; Nath, K.; Thaiss, C.A. Metabolic control by the microbiome. Genome Med. 2022, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.; Syed, Y.A.; Khan, M.R. Understanding the Role of the Gut Microbiome in Brain Development and Its Association with Neurodevelopmental Psychiatric Disorders. Front. Cell Dev. Biol. 2022, 10, 880544. [Google Scholar] [CrossRef]
- Duvallet, C.; Gibbons, S.M.; Gurry, T.; Irizarry, R.A.; Alm, E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017, 8, 1784. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.G.; Goldenthal, A.R.; Uhlemann, A.C.; Mann, J.J.; Miller, J.M.; Sublette, M.E. Systematic Review of Gut Microbiota and Major Depression. Front. Psychiatry 2019, 10, 34. [Google Scholar] [CrossRef]
- Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018, 1693, 128–133. [Google Scholar] [CrossRef]
- Baj, A.; Moro, E.; Bistoletti, M.; Orlandi, V.; Crema, F.; Giaroni, C. Glutamatergic Signaling Along The Microbiota-Gut-Brain Axis. Int. J. Mol. Sci. 2019, 20, 1482. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Mao, Y.K.; Diorio, C.; Wang, L.; Huizinga, J.D.; Bienenstock, J.; Kunze, W. Lactobacillus reuteri ingestion and IK(Ca) channel blockade have similar effects on rat colon motility and myenteric neurones. Neurogastroenterol. Motil. 2010, 22, 98-e33. [Google Scholar] [CrossRef]
- Lagomarsino, V.N.; Kostic, A.D.; Chiu, I.M. Mechanisms of microbial-neuronal interactions in pain and nociception. Neurobiol. Pain. 2021, 9, 100056. [Google Scholar] [CrossRef] [PubMed]
- De Loof, A. The cell’s self-generated “electrome”: The biophysical essence of the immaterial dimension of Life? Commun. Integr. Biol. 2016, 9, e1197446. [Google Scholar] [CrossRef]
- Petroff, O.A. GABA and glutamate in the human brain. Neuroscientist 2002, 8, 562–573. [Google Scholar] [CrossRef]
- Benarroch, J.M.; Asally, M. The Microbiologist’s Guide to Membrane Potential Dynamics. Trends Microbiol. 2020, 28, 304–314. [Google Scholar] [CrossRef]
- Levin, M. Bioelectric networks: The cognitive glue enabling evolutionary scaling from physiology to mind. Anim. Cogn. 2023, 26, 1865–1891. [Google Scholar] [CrossRef]
- Mathews, J.; Levin, M. Cancer’s unique bioelectric properties: From cells to body-wide networks: Comment on: “The distinguishing electrical properties of cancer cells” by Elisabetta Di Gregorio, Simone Israel, Michael Staelens, Gabriella Tankel, Karthik Shankar, and Jack A. Tuszynski (this issue). Phys. Life Rev. 2023, 47, 113–115. [Google Scholar] [CrossRef] [PubMed]
- de Souza-Guerreiro, T.C.; Asally, M. Seeking Insights into Aging Through Yeast Mitochondrial Electrophysiology. Bioelectricity 2021, 3, 111–115. [Google Scholar] [CrossRef]
- Silic, M.R.; Zhang, G. Bioelectricity in Developmental Patterning and Size Control: Evidence and Genetically Encoded Tools in the Zebrafish Model. Cells 2023, 12, 1148. [Google Scholar] [CrossRef]
- Whited, J.L.; Levin, M. Bioelectrical controls of morphogenesis: From ancient mechanisms of cell coordination to biomedical opportunities. Curr. Opin. Genet. Dev. 2019, 57, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Levin, M. Molecular bioelectricity in developmental biology: New tools and recent discoveries: Control of cell behavior and pattern formation by transmembrane potential gradients. Bioessays 2012, 34, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, T.; Kema, I.P.; Levin, M. Serotonin signaling is a very early step in patterning of the left-right axis in chick and frog embryos. Curr. Biol. 2005, 15, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, L.S.; Duerstock, B.S.; Borgens, R.B. Reduction of the current of injury leaving the amputation inhibits limb regeneration in the red spotted newt. Dev. Biol. 1996, 178, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Hotary, K.B.; Robinson, K.R. Evidence of a role for endogenous electrical fields in chick embryo development. Development 1992, 114, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Brackenbury, W.J. Membrane potential and cancer progression. Front. Physiol. 2013, 4, 185. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.B.; Koshland, D.E., Jr. Sensory electrophysiology of bacteria: Relationship of the membrane potential to motility and chemotaxis in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1977, 74, 4752–4756. [Google Scholar] [CrossRef]
- Humphries, J.; Xiong, L.; Liu, J.; Prindle, A.; Yuan, F.; Arjes, H.A.; Tsimring, L.; Suel, G.M. Species-Independent Attraction to Biofilms through Electrical Signaling. Cell 2017, 168, 200–209.e212. [Google Scholar] [CrossRef] [PubMed]
- Strahl, H.; Hamoen, L.W. Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. USA 2010, 107, 12281–12286. [Google Scholar] [CrossRef]
- Kikuchi, K.; Galera-Laporta, L.; Weatherwax, C.; Lam, J.Y.; Moon, E.C.; Theodorakis, E.A.; Garcia-Ojalvo, J.; Süel, G.M. Electrochemical potential enables dormant spores to integrate environmental signals. Science 2022, 378, 43–49. [Google Scholar] [CrossRef]
- Muñoz-Rodríguez, D.; Bourqqia-Ramzi, M.; García-Esteban, M.T.; Murciano-Cespedosa, A.; Vian, A.; Lombardo-Hernández, J.; García-Pérez, P.; Conejero, F.; Mateos González, Á.; Geuna, S.; et al. Bioelectrical State of Bacteria Is Linked to Growth Dynamics and Response to Neurotransmitters: Perspectives for the Investigation of the Microbiota-Brain Axis. Int. J. Mol. Sci. 2023, 24, 13394. [Google Scholar] [CrossRef] [PubMed]
- Ansari, F.; Neshat, M.; Pourjafar, H.; Jafari, S.M.; Samakkhah, S.A.; Mirzakhani, E. The role of probiotics and prebiotics in modulating of the gut-brain axis. Front. Nutr. 2023, 10, 1173660. [Google Scholar] [CrossRef] [PubMed]
- Reissbrodt, R.; Hammes, W.P.; dal Bello, F.; Prager, R.; Fruth, A.; Hantke, K.; Rakin, A.; Starcic-Erjavec, M.; Williams, P.H. Inhibition of growth of Shiga toxin-producing Escherichia coli by nonpathogenic Escherichia coli. FEMS Microbiol. Lett. 2009, 290, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Rund, S.A.; Rohde, H.; Sonnenborn, U.; Oelschlaeger, T.A. Antagonistic effects of probiotic Escherichia coli Nissle 1917 on EHEC strains of serotype O104:H4 and O157:H7. Int. J. Med. Microbiol. 2013, 303, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wassenaar, T.M. Insights from 100 Years of Research with Probiotic E. coli. Eur. J. Microbiol. Immunol. (Bp) 2016, 6, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus spp.-Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Kong, M.S.; Lai, M.W.; Chao, H.C.; Chang, K.W.; Chen, S.Y.; Huang, Y.C.; Chiu, C.H.; Li, W.C.; Lin, P.Y.; et al. Probiotics have clinical, microbiologic, and immunologic efficacy in acute infectious diarrhea. Pediatr. Infect. Dis. J. 2010, 29, 135–138. [Google Scholar] [CrossRef]
- Herrera-Rincon, C.; Pare, J.F.; Martyniuk, C.J.; Jannetty, S.K.; Harrison, C.; Fischer, A.; Dinis, A.; Keshari, V.; Novak, R.; Levin, M. An in vivo brain-bacteria interface: The developing brain as a key regulator of innate immunity. NPJ Regen. Med. 2020, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Jameson, K.G.; Olson, C.A.; Kazmi, S.A.; Hsiao, E.Y. Toward Understanding Microbiome-Neuronal Signaling. Mol. Cell 2020, 78, 577–583. [Google Scholar] [CrossRef]
- Murciano-Brea, J.; Garcia-Montes, M.; Geuna, S.; Herrera-Rincon, C. Gut Microbiota and Neuroplasticity. Cells 2021, 10, 2084. [Google Scholar] [CrossRef]
- Herrera-Rincon, C.; Murciano-Brea, J.; Geuna, S. Can we promote neural regeneration through microbiota-targeted strategies? Introducing the new concept of neurobiotics. Neural Regen. Res. 2022, 17, 1965–1966. [Google Scholar] [CrossRef] [PubMed]
- Levin, M. Bioelectric signaling: Reprogrammable circuits underlying embryogenesis, regeneration, and cancer. Cell 2021, 184, 1971–1989. [Google Scholar] [CrossRef] [PubMed]
- McMillen, P.; Oudin, M.J.; Levin, M.; Payne, S.L. Beyond Neurons: Long Distance Communication in Development and Cancer. Front. Cell Dev. Biol. 2021, 9, 739024. [Google Scholar] [CrossRef]
- Manicka, S.; Pai, V.P.; Levin, M. Information integration during bioelectric regulation of morphogenesis of the embryonic frog brain. iScience 2023, 26, 108398. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Rincon, C.; Pai, V.P.; Moran, K.M.; Lemire, J.M.; Levin, M. The brain is required for normal muscle and nerve patterning during early Xenopus development. Nat. Commun. 2017, 8, 587. [Google Scholar] [CrossRef]
- Levin, M.; Martyniuk, C.J. The bioelectric code: An ancient computational medium for dynamic control of growth and form. Biosystems 2018, 164, 76–93. [Google Scholar] [CrossRef]
- Levin, M.; Selberg, J.; Rolandi, M. Endogenous Bioelectrics in Development, Cancer, and Regeneration: Drugs and Bioelectronic Devices as Electroceuticals for Regenerative Medicine. iScience 2019, 22, 519–533. [Google Scholar] [CrossRef] [PubMed]
- Kralj, J.M.; Hochbaum, D.R.; Douglass, A.D.; Cohen, A.E. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science 2011, 333, 345–348. [Google Scholar] [CrossRef]
- Bruni, G.N.; Weekley, R.A.; Dodd, B.J.T.; Kralj, J.M. Voltage-gated calcium flux mediates Escherichia coli mechanosensation. Proc. Natl. Acad. Sci. USA 2017, 114, 9445–9450. [Google Scholar] [CrossRef]
- Stratford, J.P.; Edwards, C.L.A.; Ghanshyam, M.J.; Malyshev, D.; Delise, M.A.; Hayashi, Y.; Asally, M. Electrically induced bacterial membrane-potential dynamics correspond to cellular proliferation capacity. Proc. Natl. Acad. Sci. USA 2019, 116, 9552–9557. [Google Scholar] [CrossRef]
- Jones, J.M.; Larkin, J.W. Toward Bacterial Bioelectric Signal Transduction. Bioelectricity 2021, 3, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Buttress, J.A.; Halte, M.; Te Winkel, J.D.; Erhardt, M.; Popp, P.F.; Strahl, H. A guide for membrane potential measurements in Gram-negative bacteria using voltage-sensitive dyes. Microbiology 2022, 168, 001227. [Google Scholar] [CrossRef] [PubMed]
- Hudson, M.A.; Siegele, D.A.; Lockless, S.W. Use of a Fluorescence-Based Assay To Measure Escherichia coli Membrane Potential Changes in High Throughput. Antimicrob. Agents Chemother. 2020, 64, e00910-20. [Google Scholar] [CrossRef]
- Han, X.; Foster, B.R.; Payne, C.K. Electrical Control of Escherichia coli Growth Measured with Simultaneous Modulation and Imaging. Bioelectricity 2020, 2, 221–228. [Google Scholar] [CrossRef]
- Leive, L. Studies on the permeability change produced in coliform bacteria by ethylenediaminetetraacetate. J. Biol. Chem. 1968, 243, 2373–2380. [Google Scholar] [CrossRef] [PubMed]
- Whatmore, A.M.; Chudek, J.A.; Reed, R.H. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 1990, 136, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.S.; Levin, M. Measuring resting membrane potential using the fluorescent voltage reporters DiBAC4(3) and CC2-DMPE. Cold Spring Harb. Protoc. 2012, 2012, 459–464. [Google Scholar] [CrossRef]
- Sträuber, H.; Müller, S. Viability states of bacteria--specific mechanisms of selected probes. Cytometry A 2010, 77, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Rojas, E.R.; Huang, K.C.; Theriot, J.A. Homeostatic Cell Growth Is Accomplished Mechanically through Membrane Tension Inhibition of Cell-Wall Synthesis. Cell Syst. 2017, 5, 578–590.e576. [Google Scholar] [CrossRef]
- Rojas, E.R.; Huang, K.C. Regulation of microbial growth by turgor pressure. Curr. Opin. Microbiol. 2018, 42, 62–70. [Google Scholar] [CrossRef]
- Dicks, L.M.T. Gut Bacteria and Neurotransmitters. Microorganisms 2022, 10, 1838. [Google Scholar] [CrossRef] [PubMed]
- Lauder, J.M. Neurotransmitters as morphogens. Prog. Brain Res. 1988, 73, 365–387. [Google Scholar] [CrossRef]
- Sullivan, K.G.; Levin, M. Neurotransmitter signaling pathways required for normal development in Xenopus laevis embryos: A pharmacological survey screen. J. Anat. 2016, 229, 483–502. [Google Scholar] [CrossRef]
- Zhou, Y.; Danbolt, N.C. Glutamate as a neurotransmitter in the healthy brain. J. Neural Transm. 2014, 121, 799–817. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Tymianski, M. Glutamate receptors, neurotoxicity and neurodegeneration. Pflug. Arch. 2010, 460, 525–542. [Google Scholar] [CrossRef] [PubMed]
- Prindle, A.; Liu, J.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Süel, G.M. Ion channels enable electrical communication in bacterial communities. Nature 2015, 527, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, R.; Salvador-Recatalà, V.; Dreyer, I. Electrical Wiring and Long-Distance Plant Communication. Trends Plant Sci. 2016, 21, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Strandwitz, P.; Kim, K.H.; Terekhova, D.; Liu, J.K.; Sharma, A.; Levering, J.; McDonald, D.; Dietrich, D.; Ramadhar, T.R.; Lekbua, A.; et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019, 4, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Dover, S.; Halpern, Y.S. Utilization of γ-Aminobutyric Acid as the Sole Carbon and Nitrogen Source by Escherichia coli K-12 Mutants. J. Bacteriol. 1972, 109, 835–843. [Google Scholar] [CrossRef]
- Te Winkel, J.D.; Gray, D.A.; Seistrup, K.H.; Hamoen, L.W.; Strahl, H. Analysis of Antimicrobial-Triggered Membrane Depolarization Using Voltage Sensitive Dyes. Front. Cell Dev. Biol. 2016, 4, 29. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bourqqia-Ramzi, M.; Mansilla-Guardiola, J.; Muñoz-Rodriguez, D.; Quarta, E.; Lombardo-Hernandez, J.; Murciano-Cespedosa, A.; Conejero-Meca, F.J.; Mateos González, Á.; Geuna, S.; Garcia-Esteban, M.T.; et al. From the Microbiome to the Electrome: Implications for the Microbiota–Gut–Brain Axis. Int. J. Mol. Sci. 2024, 25, 6233. https://doi.org/10.3390/ijms25116233
Bourqqia-Ramzi M, Mansilla-Guardiola J, Muñoz-Rodriguez D, Quarta E, Lombardo-Hernandez J, Murciano-Cespedosa A, Conejero-Meca FJ, Mateos González Á, Geuna S, Garcia-Esteban MT, et al. From the Microbiome to the Electrome: Implications for the Microbiota–Gut–Brain Axis. International Journal of Molecular Sciences. 2024; 25(11):6233. https://doi.org/10.3390/ijms25116233
Chicago/Turabian StyleBourqqia-Ramzi, Marwane, Jesús Mansilla-Guardiola, David Muñoz-Rodriguez, Elisa Quarta, Juan Lombardo-Hernandez, Antonio Murciano-Cespedosa, Francisco José Conejero-Meca, Álvaro Mateos González, Stefano Geuna, María Teresa Garcia-Esteban, and et al. 2024. "From the Microbiome to the Electrome: Implications for the Microbiota–Gut–Brain Axis" International Journal of Molecular Sciences 25, no. 11: 6233. https://doi.org/10.3390/ijms25116233
APA StyleBourqqia-Ramzi, M., Mansilla-Guardiola, J., Muñoz-Rodriguez, D., Quarta, E., Lombardo-Hernandez, J., Murciano-Cespedosa, A., Conejero-Meca, F. J., Mateos González, Á., Geuna, S., Garcia-Esteban, M. T., & Herrera-Rincon, C. (2024). From the Microbiome to the Electrome: Implications for the Microbiota–Gut–Brain Axis. International Journal of Molecular Sciences, 25(11), 6233. https://doi.org/10.3390/ijms25116233








