Monocytic Differentiation in Acute Myeloid Leukemia Cells: Diagnostic Criteria, Biological Heterogeneity, Mitochondrial Metabolism, Resistance to and Induction by Targeted Therapies
Abstract
1. Introduction
2. Monocytic FAB-M4/M5 AML Cell Differentiation: Morphological and Functional Characteristics, Molecular Markers, Genetic Heterogeneity and Minimal Prognostic Impact for Patients Receiving Conventional Chemotherapy
2.1. Monocytic Differentiation According to the FAB (French/American/British) Classification
2.2. The Genetic Heterogenety of Monocytic FAB-M4/M5 AML Patients
- Many patients with monocytic AML cells show normal or nonspecific cytogenetic abnormalities (e.g., +8 in FAB-M4 and t(8;16) in FAB-M5 [4]).
- The most common mutations observed for at least 10% of FAB-M4/M5 patients are NPM1 (38%), DNMT3A (37%), FLT3-ITD/TKD (32%), NRAS (11%) and RUNX (10%) [9].
- Several patients with NPM1 mutations fulfill the FAB criteria for monocytic differentiation, i.e., they can be classified as FAB-M4/M5 based on the morphological characteristics of their AML cells [14,15]. NPM1 mutations are common both for FAB-M4 and FAB-M5 patients [11] and seem to be (one of) the most common mutations in FAB-M4/M5 AML [9]. Patients with NPM1-mutated chemotherapy-related secondary AML show similar survival as patients with de novo NPM1-mutated AML [16], and this is better survival than other patients with therapy-associated AML who have an adverse prognosis [17,18,19]. The fusion oncogene NUP98:NSD1 shows a higher frequency in FAB-M4, and its detection at the time of first diagnosis is associated with an adverse prognosis [12].
- RAS mutations are significantly more frequent in FAB-M4/FAB-M5 and are associated with an increased risk of late relapse [13].
- PTPN11 mutations are more frequent in adults with monocytic AML and are independently associated with a lower complete remission rate and overall survival [10].
2.3. The Transcptomic Heterogeneity of Monocytic FAB-M4/M5 AML Cells
- A previous transcriptomic study identified eight different AML subsets based on the transcriptomic profile, and monocytic FAB-M4/M5 cells were classified in four of these eight subsets [20].
- A smaller transcriptomic study included a subset of monocytic AML patients, and the main difference was an increased expression of cells involved in cellular adhesion/communication for the monocytic cells compared to the other subsets [21].
- A previous study was based on the expression of 93 monocyte-specific genes, and based on this expression profile for a group of AML patients, they could classify the patients into a monocytic (144 patients) and a majority of 381 non-monocytic patients [22]. However, the monocytic cluster included 118 FAB-M4/M5 patients, whereas the non-monocytic cluster included 92 FAB-M4/M5 patients.
- The heterogeneity was also observed in a study of 27 patients with FAB-M5 AML; these authors did mRNA profiling based on the expression of 85 genes encoding histone-modifying proteins [23].
- A transcriptomic analysis of leukemic cells classified as mixed-lineage acute leukemia showed that even a subset of these patients was characterized by the high expression of monocyte genes [24].
2.4. Expression of Costimulatory Molecules and Checkpoint Ligands by AML Cells: High CD86 and PD-L1 Expression by FAB-M4/M5 AML Cells
2.5. Increased Constitutive Extracellular Release of Soluble Mediators by Monocytic AML Cells
2.6. The FAB Classification of AML Has No or Only Very Limited Prognostic Impact for Patients Receiving Intensive Conventional Antileukemic Chemotherapy
2.7. The Role of Nonleukemic Bone Marrow Cells in AML: The Importance of Leukemia-Supporting Stromal Cells and the Prognostic Impact of Monocyte Infiltration
2.8. Concluding Comment: The Limited Monocytic AML Cell Differentiation Is Not as Sufficient to Cause a Similar Prognostic Impact as the Bone Marrow Infiltration of Normal Monocytes/Macrophages
3. Variations in the Mitochondrial Energy Metabolism of AML Cells: Possible Associations between Metabolic Differences and Prognosis after Conventional Intensive Therapy
- 4-hydroxyphenylpyruvate dioxygenase like (HPDL, chromosome 1). This mitochondrial protein seems to function as a 4-hydroxyphenylpyruvate dioxygenase.
- Carnitine palmitoyltransferase 1A (CPT1A, chromosome 11). Mitochondrial oxidation of long-chain fatty acids is initiated by carnitine palmitoyltransferases and carnitine-dependent transport across the mitochondrial inner membrane.
- Isocitrate dehydrogenase (NAD(+)) 3 catalytic subunit alpha (IDH3A, chromosome 15). Isocitrate dehydrogenases catalyze the oxidative decarboxylation of isocitrate to 2-oxoglutarate. This NAD-dependent enzyme is localized to the mitochondrial matrix, and NAD-dependent isocitrate dehydrogenases catalyze the rate-limiting step of the tricarboxylic acid cycle. The encoded protein is a subunit of one of the NAD-dependent isocitrate dehydrogenases.
- Electron transfer flavoprotein subunit beta (ETFB, chromosome 19). This electron transfer flavoprotein shuttles electrons between primary flavoprotein dehydrogenases involved in mitochondrial fatty acid and amino acid catabolism and the membrane-bound electron transfer flavoprotein ubiquinone oxidoreductase.
- Enoyl-CoA hydratase, short chain 1 (ECHS1, chromosome 10). This mitochondrial matrix protein functions in the second step of the fatty acid-beta oxidation pathway.
- NADH:ubiquinone oxidoreductase subunit A1 (NDUFA1, chromosome X). This protein is an essential component of complex I of the respiratory chain that consists of at least 43 subunits, with 7 of them being encoded by the mitochondrial genome. This protein functions as an anchor to the inner mitochondrial membrane.
- NADH:ubiquinone oxidoreductase core subunit S2 (NDUFS2, chromosome 1). The protein is a core subunit of the mitochondrial respiratory chain complex I.
- Succinate dehydrogenase complex flavoprotein subunit A (SDHA, chromosome 5). This gene encodes a major catalytic subunit of succinate-ubiquinone oxidoreductase in complex III of the mitochondrial respiratory chain.
- Succinate-CoA ligase GDP/ADP-forming subunit alpha (SUCLG1, chromosome 2). This gene encodes a subunit of succinate coenzyme A ligase that catalyzes the conversion of succinyl CoA and ADP or GDP to a succinate and ATP or GTP.
4. Variations in the Mitochondrial Energy Metabolism of AML Cells: The Associations between Metabolic Differences and Monocytic FAB-M4/M5 AML Differentiation Is a Molecular Background for Resistance to Targeted Therapy
4.1. A Brief Overview of the Cellular Energy Metabolism
- Cytoplasmic glycolysis. The glycolytic pathway starts with the formation of glucose-6-phosphate; this is followed by a multistep process, finally generating pyruvate that can be transported into mitochondria to the tricarboxylic acid cycle, be further metabolized to lactate or be used for the synthesis of alanine. The 3-phosphoglycerate formed in the glycolysis can also be used for amino acid synthesis. The conversion of glucose to lactate provides 2 ATP molecules per glucose molecule, whereas complete glucose oxidation can provide up to 36 ATP molecules.
- Pentose phosphate pathway. This alternative cytoplasmic pathway utilizes glucose-6-phosphate; the final products include NADPH and ribose-5-phosphate, which are used in the synthesis of nucleotides and nucleic acids. NADPH is an important reducing agent in many cellular reactions (including lipid synthesis), and it protects against oxidative stress by regenerating reduced glutathione [84].
- The tricarboxylic acid cycle. After mitochondrial entry, pyruvate can be metabolized to acetyl-coenzyme A (CoA) that enters the mitochondrial tricarboxylic acid cycle. Acetyl-CoA can also be derived from fatty acid and amino acid metabolism.
- Citrate from the tricarboxylic acid cycle can be used for cytoplasmic fatty acid synthesis.
- The respiratory chain/oxidative phosphorylation. The chemical energy from the tricarboxylic acid cycle is converted into a mitochondrial electrochemical gradient that consists of five molecular complexes called complex 1/NADH:ubiquinone oxidoreductase, complex II/succinate dehydrogenase, complex III/ubiquinol:cytochrome c oxidoreductase, complex IV/cytochrome c oxidase and the final complex V/ATP synthase that converts ADP to ATP. Complex II/succinate dehydrogenase is also part of the tricarboxylic acid cycle. All complex II components are encoded by nuclear DNA, whereas the other four complexes also include proteins encoded by the mitochondrial genome.
- Lipid metabolism. Mitochondrial fatty acid oxidation breaks down fatty acids to acetyl-CoA that can enter the tricarboxylic acid cycle, whereas fatty acid synthesis takes place in the cytosol [84].
4.2. AML Cells Show a Mitochondrial Dysfunction but Also Heterogeneity with Regards to the Regulation of Energy Metabolism: Increased Reactive Oxygen Species in Monocytic FAB-M4/M5 Cells
4.3. Mutations in the Electron Chain Component Genes: No Association with FAB-M4/M5
5. Dual Functions of Mitochondrial Mediators: Their Regulation of Both Energy Metabolism and Cell Survival and Regulation of Mediator Expression by Differentiation
5.1. Mitochondrial Dynamics: Effects of Bcl-2 Family Members Depend on the Biological Context
5.2. Cytochrome C Oxidase: A Proapoptotic Mediator Essential for Cellular Energy Metabolism
5.3. Bcl-2: An Antiapoptotic Regulator Targeting the Metabolic Functions of Cytochrome C
5.4. Mcl-1: An Antiapoptotic and Metabolic Regulator Controlled by Glucose Metabolism
5.5. Bcl-xL: An Antiapoptotic Mediator That Modulates Several Metabolic Steps
5.6. The Proapoptotic Bcl-2 Family Members Bax, Bad, Puma and Noxa Modulate Cell Metabolism
5.7. TP53: Maintenance of Mitochondrial Integrity and Multiple Effects on Metabolic Regulation
- Crosstalk with the metabolic sensor mTORC1 (mammalian target of rapamycin complex 1).
- Suppression of glycolysis through downregulation of the glucose transporters, inhibition of glycolytic enzymes and indirect transcriptional effects. TP53 is also a modulator of the balance between the glycolytic and the pentose phosphate pathway, a downregulator of lipid synthesis, a driver of the oxidative phosphorylation and a maintainer of mitochondrial integrity.
- TP53 can reduce/eliminate the deleterious effects of oxidative stress by limiting the cellular damage or by using these species to eliminate cells damaged beyond repair.
5.8. Sirtuins Represent Possible Links between Protein Acetylation and Mitochondrial Metabolism
5.9. The Prognostic Impact of Glycolytic Profiles in AML Patients Receiving Intensive Therapy
5.10. The Prognostic Impact of Mitochondrial Lipid Metabolism in AML Patients Receiving Intensive Conventional Chemotherapy
5.11. The Heterogeneity of Monocytic FAB-M4/M5 AML Cells: Differences in the Regulation of Oxidative Phosphorylation Are Associated with Differences in Mitochondrial Translation and the Risk of Relapse after Intensive Chemotherapy
6. Resistance to Bcl-2 Targeting Therapy Depends on Both Specific Molecular Mechanisms Associated with Monocytic AML Cell Differentiation, as Well as General Susceptibility Biomarkers Identified in Previous Studies of Conventional Anti-AML Therapy
6.1. Clinical Studies of Venetoclax-Based AML-Stabilizing Therapy: The Importance of Genetic Abnormalities Together with Monocytic Differentiation
6.2. Venetoclax Combined with Intensive Chemotherapy
6.3. Venetoclax for the Treatment of Relapse after Allotransplantation
6.4. AML Cell Differentiation, Energy Metabolism and Venetoclax Resistance
- Ex vivo drug responses. A small ex vivo drug screening study including 34 patients described a lower antileukemic effect of venetoclax for FAB-M4/M5 monocytic AML cells than for undifferentiated FAB-M0/M1 AML cells [8]. Resistance was then associated with a low Bcl-2/Mcl-1 ratio. Another experimental study also described an association between myelomonocytic differentiation and venetoclax resistance [148]. These last authors also described that myelomonocytic leukemia and upregulated Bcl-2A1/CLEC7A, as well as mutations of PTPN11 and KRAS, conferred resistance to venetoclax and venetoclax combinations. Finally, ribosomal protein S6 kinase alpha-1 (RPS6KA1) also seems to be involved in the development of venetoclax resistance in monocytic AML cells [150]. These ex vivo observation further support and extend the observations in clinical studies (see Table 1, Section 6.1).
- Monocytic AML stem cells. A recent study characterized AML stem cell subsets [158]. These authors described a patient heterogeneity; some patients only had primitive AML stem cells, other patients had monocytic stem cells characterized by the expression of monocytic protein/differentiation markers and monocytic-associated transcription factors and the third group had a mixture of these two stem cell phenotypes that, at least for some patients, corresponded to underlying mutational variations. The ability of the monocytic stem cells to recapitulate AML development was verified in patient xenograft models. Although relatively few patients were examined, the presence of monocytic AML stem cells before venetoclax/azacitidine and/or the development of disease progression with monocytic AML cells during treatment were associated with a shorter duration of responses. The venetoclax/azacitidine combination then seemed to inhibit the electron transport chain and thereby cause the in vivo selection of monocytic AML cells, including monocytic AML stem cells [7,88].
- Relapsed monocytic AML cells rely on Mcl-1 for metabolic regulation and survival. Studies of the total AML cell population have shown that the venetoclax/azacitidine combination decreases oxidative phosphorylation and energy production, inhibits the electron transport chain, increases cellular reactive oxygen species and decreases glutathione levels [88,89]. Clinical venetoclax resistance can then be due to relatively low Bcl-2 levels as part of monocytic differentiation, and relapsed monocytic cells instead seem to rely on Mcl-1 for oxidative phosphorylation and survival [7,8,88].
- Reactive oxygen species in FAB-M4/M5 AML. Patients with the FAB-M4/M5 variants of AML have higher basal levels of reactive oxygen species (see Section 4.2) [90]. The monocytic AML cells seem to be transcriptionally distinct from undifferentiated/primitive AML cells; they also show an upregulation of genes important for oxidative phosphorylation and a higher basal respiration rate [7]. However, further studies are needed to clarify whether or how/whether these metabolic mechanisms contribute to monocytic-associated venetoclax resistance.
- Patients with venetoclax resistance show a genetic heterogeneity. Various genetic abnormalities are associated with the susceptibility of AML cells to venetoclax (Section 6.1). First, venetoclax-based treatment is highly effective in NPM1-mutated AML, but it is not known whether this is true also for monocytic NPM1-Ins AML cells [14,140,152,153]. Second, PTPN11 and KRAS mutations are associated both with monocytic AML cell differentiation [13,154,155] and venetoclax resistance [148]. Finally, a recent clinical study described a similar decreased responsiveness compared to other AML subsets both for FAB-M4 and FAB-M5 patients [164]. Thus, venetoclax resistance is associated with a genetic heterogeneity and probably also different (degrees of) monocytic AML cell differentiation.
- Venetoclax resistance can be mediated by various mechanisms. A recent study combined transcriptomic and ex vivo venetoclax responses to identify individual genes which expression was associated with venetoclax resistance in primary AML cells [165]. They identified four different venetoclax resistance clusters. (i) Their first cluster was associated with monocytic AML cell differentiation; this cluster was also associated with the decreased expression of Bcl-2 together with the increased expression of MCL-1 and BCL-2A1; activation of several metabolic pathways (glycolysis, oxidative phosphorylation and fatty acid metabolism); activation of PI3K-mTOR signaling and susceptibility to the inhibition of cyclin-dependent kinases. In contrast, on the other hand, the three other venetoclax resistance clusters had in common gene enrichment consistent with more immature leukemic cells. (ii) Cluster 2 was associated with FAB-M2 morphology, adverse ELN classification and NRAS mutations; (iii) cluster 3 showed the enrichment of TP53 mutations and signs of erythroid differentiation, (iv) whereas cluster 4 was associated with favorable ELN classification. Taken together, these observations suggest that venetoclax resistance can be mediated by various mechanisms and that the mechanisms behind FAB-M4/M5-associated resistance differ from other patients. This hypothesis is further supported by another recent study [149] describing that certain resistance mechanisms in primary AML cells reflect a more general resistance to various pharmacological agents independent of the drug target, whereas certain mechanisms for resistance/susceptibility to venetoclax can be independent/different from this general responsiveness.
- Erythroid/megakaryocytic differentiation and venetoclax resistance. An experimental study described that AML cells with erythroid/megakaryocytic differentiation were dependent on antiapoptotic Bcl-xL (encoding the Bcl-2L2 molecule) rather than Bcl-2 or Mcl-1 for survival [156]. These cells were also susceptible to the Bcl-xL-selective inhibitors A-1331852 and navitoclax, whereas they were resistant to the Bcl-2 inhibitor venetoclax. Bcl-xL inhibition also caused extensive killing of such cells in AML xenograft models. Thus, venetoclax resistance due to an altered balance between Bcl-2 family members is not specific for monocytic AML cell differentiation.
7. AML Cell Differentiation and Targeted Therapies: Effect of Differentiation on the Antileukemic Efficiency and Induction of Differentiation by Targeted Therapies
7.1. Bromodomain Inhibitors: Increased Antileukemic Effect in Monocytic AML Cells
7.2. Histone Deacetylase Inhibitors: Epigenetic Modulation and Differentiation Induction
7.3. Lysine Demethylase 1 (LSD1/KDM1A) Inhibitors: Monocytic Differentiation in MLL-AML
7.4. Inhibition of DOT1-like Histone Lysine Methyltransferase: Differentiation Induction in MLL-Rearranged and DNMT3A-Mutated AML
7.5. Inhibition of the Scaffold Protein Menin: An Effective Strategy in Certain AML Subsets
- MLL-rearranged AML. Inhibition of the menin/MLL interaction is a possible strategy in MLL-rearranged AML. VTP50469 is a potent, highly selective and orally bioavailable inhibitor that displaces menin from protein complexes and inhibits its chromatin binding [191]. This loss of MLL binding leads to changes in gene expression, cellular differentiation and regulation of apoptosis, and these antileukemic effects have been observed even in AML xenograft models. Furthermore, the knockout/degradation of menin or treatment with the menin inhibitor SNDX-50469 reduce MLL fusion protein-induced AML cell differentiation and reduces AML cell viability, as well as the Bcl-2 and CDK6 levels, but despite the reduced Bcl-2 levels, the combination of SNDX-50469 with venetoclax or the CDK6 inhibitor abemaciclib has synergistic antileukemic effects for patient-derived AML cells harboring MLL1 rearrangements or NPM1-Ins [192].
- FLT3-ITD AML. Pharmacological inhibition of the menin-MLL complex caused specific changes in gene expression with downregulation of both the MEIS1 transcription factor and its target FLT3 [193]. Combined menin-MLL and FLT3 inhibition had synergistic antiproliferative and proapoptotic effects in models of human NPM1-mutated or MLL-rearranged AML with additional FLT3-ITD [194]. Importantly, AML cells from patients with both NPM1 and FLT3 mutations showed significantly better responses to combined menin/FLT3 inhibition than to single-drug treatment.
- NUP98 translocations. The menin-MLL1 interaction is essential in AML with translocations involving the NUP98 (nucleoporin 98 and 96 precursor) gene, and studies of NUP98 fusion leukemia in animal models have shown that inhibition of the menin-MLL1 interaction by VTP50469 has an antileukemic effect [195]. This inhibition upregulates various differentiation markers such as CD11b by removing MLL1 and NUP98 fusion proteins from chromatin sites at genes that are essential for the malignant phenotype. The antileukemic and differentiation-inducing effects were also observed in patient-derived xenografts of NUP98 fusion-driven AML.
- UBTF abnormalities. AML with UBTF (upstream binding transcription factor) tandem duplications (UBTF-TDs) has a transcriptional signature with similarities to NUP98-rearranged and NPM1-mutated AML; the primary cells from UBTF-TD AML are sensitive to the menin inhibitor SNDX-5613 that has antiproliferative and differentiation-inducing effects in these cells [196].
- MN1 translocations. Translocations involving meningioma-1 (MN1) occur in an AML subset and result in a high expression of either the full-length MN1 protein or a fusion protein including most of the N-terminus of MN1 [197]. Menin is essential for the self-renewal of MN1-driven AML, and pharmacological inhibition of the MLL-menin interaction also has antiproliferative and differentiation-inducing effects in this AML variant when tested in experimental in vitro and xenograft models.
7.6. Inhibitors of the Nuclear Exporter Exportin-1 (XPO1): Effects on NPM-Ins, as well as Other Leukemogenic Proteins
7.7. Ribosome Targeting Seems to Overcome Venetoclax Resistance
7.8. Proteasome Inhibition: Is the Antileukemic Effect Associated with Monocytic Differentiation?
7.9. Flt3 Inhibition: Differentiation Induction versus Cytotoxicity
7.10. The TRAIL Agonist Eftozanermin: Combined Treatment with Venetoclax
7.11. Aryl Hydrocarbon Receptor Agonists and Induction of Monocytic/Granulocytic Differentiation
7.12. IDH Mutations and IDH Inhibition: Venetoclax Sensitization and Differentiation Induction
7.13. Inhibition of Pyrimidine Biosynthesis and Purine Metabolism Induces Differentiation
7.14. Therapeutic Targeting of Energy Metabolism by Direct Inhibition of Metabolic Regulators or Inhibition of the Metabolic Sensor/Regulator PI3K-Akt-mTOR Pathway
7.15. The Possible Therapeutic Targeting of Monocytic Markers
- Targeting of Mcl-1. Monocytic AML cells rely on Mcl-1 rather than Bcl-2 for survival and metabolic regulation, and experimental studies suggest that Mcl-1 targeting is a possible strategy either as a monotherapy or in combination with conventional chemotherapy or Bcl-2 targeting [115,231,232,233,234,235,236]. Mcl-1 inhibition can be achieved either by direct inhibition [115,231,232,233,234] or indirectly through transcriptional or translational suppression [235,236]. However, even though this strategy seems to have an anticancer effect, the risk of cardiotoxicity may make it unfeasible [237].
- Integrin targeting. Blocking of integrin cell surface molecules by monoclonal antibodies is now used as a strategy for the inhibition of immunocompetent cell extravasation/chemotaxis [238]; the blocking of leukemia-supporting AML/endothelial cell interactions in their common bone marrow environment may similarly be used as an antileukemic strategy, either by using integrin-directed antibodies [238] or possibly soluble integrin molecules that compete for the binding on the integrin ligands [239,240]. The inhibition of β2 integrins by small molecular inhibitors may also be possible [241], although this strategy will not be specific for CD11b/CD18 and CD11c/CD18. Furthermore, one would expect the targeting of CD11b/CD18 and CD11c/CD18 to be associated with a risk of infection due to the inhibition of normal monocytes [242], and this may become particularly important for patients receiving AML therapy with hematological toxicity [3]. Finally, the effects of integrins on downstream intracellular signaling are very complex, and many integrins modulate downstream pathways that are also shared with other cell surface molecules/receptors [238].
- Targeting of intracellular signaling. The monocyte markers include several receptor molecules; these markers may be targeted by blocking of their downstream signaling by small molecule inhibitors. This strategy may be possible for PI3K-Akt-mTOR (CD7, CD56 and CD117) [243,244]; NF-κB (CD14) [245]; CD117/c-kit [246]; TIMP1 (CD63) [247,248] and FAK (CD63, CD117 and ITGB1) [249] signaling. It should be emphasized that these pathways are common for various upstream receptors; this approach will thus also block signaling not only from the corresponding monocyte marker/receptor but also other receptors/mediators upstream from the target.
- Targeting of cellular function. Several monocyte markers seem to affect intracellular transport/trafficking, and the targeting of these processes may also be possible [250,251,252]. However, this strategy should also be regarded as nonspecific and will not only affect signaling from/functions of specific monocytic markers.
- Immunotherapy. To the best of our knowledge, there is no firm evidence for an association between the effect of immunotherapy and monocytic AML differentiation.
8. Coagulopathy and Risk of Thrombosis in AML: A Risk Associated with Differentiation or Differentiation Induction?
9. Discussion
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef]
- Arber, D.A.; Brunning, R.D.; Orazi, A.; Porwit, A.; Peterson, L.C.; Thiele, J.; Le Beau, M.M.; Hasserjian, R.P. Acute myeloid leukemia, NOS. In WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Eds.; International Agency for Research on Cancer: Lyon, France, 2017; pp. 156–166. [Google Scholar]
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.R.; Sultan, C. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br. J. Haematol. 1976, 33, 451–458. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Arber, D.A.; Hasserjian, R.P.; LeBeau, M.M.; et al. WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues; International Agency for Research in Cancer: Lyon, France, 2017. [Google Scholar]
- Pei, S.; Pollyea, D.A.; Gustafson, A.; Stevens, B.M.; Minhajuddin, M.; Fu, R.; Riemondy, K.A.; Gillen, A.E.; Sheridan, R.M.; Kim, J.; et al. Monocytic Subclones Confer Resistance to Venetoclax-Based Therapy in Patients with Acute Myeloid Leukemia. Cancer Discov. 2020, 10, 536–551. [Google Scholar] [CrossRef]
- Kuusanmäki, H.; Leppä, A.M.; Pölönen, P.; Kontro, M.; Dufva, O.; Deb, D.; Yadav, B.; Brück, O.; Kumar, A.; Everaus, H.; et al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica 2020, 105, 708–720. [Google Scholar] [CrossRef]
- Cheng, Z.; Hu, K.; Tian, L.; Dai, Y.; Pang, Y.; Cui, W.; Zhao, H.; Qin, T.; Han, Y.; Hu, N.; et al. Clinical and biological implications of mutational spectrum in acute myeloid leukemia of FAB subtypes M4 and M5. Cancer Gene Ther. 2018, 25, 77–83. [Google Scholar] [CrossRef]
- Alfayez, M.; Issa, G.C.; Patel, K.P.; Wang, F.; Wang, X.; Short, N.J.; Cortes, J.E.; Kadia, T.; Ravandi, F.; Pierce, S.; et al. The Clinical impact of PTPN11 mutations in adults with acute myeloid leukemia. Leukemia 2021, 35, 691–700. [Google Scholar] [CrossRef]
- Canaani, J.; Beohou, E.; Labopin, M.; Socié, G.; Huynh, A.; Volin, L.; Cornelissen, J.; Milpied, N.; Gedde-Dahl, T.; Deconinck, E.; et al. Impact of FAB classification on predicting outcome in acute myeloid leukemia, not otherwise specified, patients undergoing allogeneic stem cell transplantation in CR1: An analysis of 1690 patients from the acute leukemia working party of EBMT. Am. J. Hematol. 2017, 92, 344–350. [Google Scholar] [CrossRef]
- Miyajima, T.; Onozawa, M.; Yoshida, S.; Miyashita, N.; Kimura, H.; Takahashi, S.; Yokoyama, S.; Matsukawa, T.; Goto, H.; Sugita, J.; et al. Clinical implications of NUP98::NSD1 fusion at diagnosis in adult FLT3-ITD positive AML. Eur. J. Haematol. 2023, 111, 620–627. [Google Scholar] [CrossRef]
- Sano, H.; Shimada, A.; Taki, T.; Murata, C.; Park, M.J.; Sotomatsu, M.; Tabuchi, K.; Tawa, A.; Kobayashi, R.; Horibe, K.; et al. RAS mutations are frequent in FAB type M4 and M5 of acute myeloid leukemia, and related to late relapse: A study of the Japanese Childhood AML Cooperative Study Group. Int. J. Hematol. 2012, 95, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Tsykunova, G.; Reikvam, H.; Hovland, R.; Bruserud, Ø. The surface molecule signature of primary human acute myeloid leukemia (AML) cells is highly associated with NPM1 mutation status. Leukemia 2012, 26, 557–559. [Google Scholar] [CrossRef] [PubMed]
- Walter, R.B.; Othus, M.; Burnett, A.K.; Löwenberg, B.; Kantarjian, H.M.; Ossenkoppele, G.J.; Hills, R.K.; van Montfort, K.G.; Ravandi, F.; Evans, A.; et al. Significance of FAB subclassification of “acute myeloid leukemia, NOS” in the 2008 WHO classification: Analysis of 5848 newly diagnosed patients. Blood 2013, 121, 2424–2431. [Google Scholar] [CrossRef] [PubMed]
- Othman, J.; Meggendorfer, M.; Tiacci, E.; Thiede, C.; Schlenk, R.; Dillon, R.; Stasik, S.; Venanzi, A.; Bertoli, S.; Delabesse, E.; et al. Overlapping features of therapy-related and de novo NPM1-mutated AML. Blood 2023, 141, 1846–1857. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cuadrón, D.; Megías-Vericat, J.E.; Serrano, J.; Martínez-Sánchez, P.; Rodríguez-Arbolí, E.; Gil, C.; Aguiar, E.; Bergua, J.; López-Lorenzo, J.L.; Bernal, T.; et al. Treatment patterns and outcomes of 2310 patients with secondary acute myeloid leukemia: A PETHEMA registry study. Blood Adv. 2022, 6, 1278–1295. [Google Scholar] [CrossRef] [PubMed]
- Granfeldt Østgård, L.S.; Medeiros, B.C.; Sengeløv, H.; Nørgaard, M.; Andersen, M.K.; Dufva, I.H.; Friis, L.S.; Kjeldsen, E.; Marcher, C.W.; Preiss, B.; et al. Epidemiology and Clinical Significance of Secondary and Therapy-Related Acute Myeloid Leukemia: A National Population-Based Cohort Study. J. Clin. Oncol. 2015, 33, 3641–3649. [Google Scholar] [CrossRef] [PubMed]
- Lalayanni, C.; Gavriilaki, E.; Athanasiadou, A.; Iskas, M.; Papathanasiou, M.; Marvaki, A.; Mpesikli, S.; Papaioannou, G.; Mallouri, D.; Batsis, I.; et al. Secondary Acute Myeloid Leukemia (sAML): Similarly Dismal Outcomes of AML After an Antecedent Hematologic Disorder and Therapy Related AML. Clin. Lymphoma Myeloma Leuk. 2022, 22, e233–e240. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.Y.; Li, J.F.; Zhu, Y.M.; Lin, X.J.; Wen, L.J.; Zhang, F.; Zhang, Y.L.; Zhao, M.; Fang, H.; Wang, S.Y.; et al. Transcriptome-based molecular subtypes and differentiation hierarchies improve the classification framework of acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2022, 119, e2211429119. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, N.C.; López-Pérez, R.; Hernández, J.M.; Isidro, I.; González, B.; Delgado, M.; Fermiñán, E.; García, J.L.; Vázquez, L.; González, M.; et al. Gene expression profile reveals deregulation of genes with relevant functions in the different subclasses of acute myeloid leukemia. Leukemia 2005, 19, 402–409. [Google Scholar] [CrossRef]
- Klamer, S.E.; Nota, B.; Moorhouse, M.; Voermans, C.; Schoot, C.E. Gene-expression-based monocyte-specific clustering of acute myeloid leukemias reveals novel associations. Leuk. Lymphoma 2017, 58, 1721–1725. [Google Scholar] [CrossRef]
- Xiao, P.F.; Tao, Y.F.; Hu, S.Y.; Cao, L.; Lu, J.; Wang, J.; Feng, X.; Pan, J.; Chai, Y.H. mRNA expression profiling of histone modifying enzymes in pediatric acute monoblastic leukemia. Pharmazie 2017, 72, 177–186. [Google Scholar] [PubMed]
- Wang, Q.; Cai, W.Z.; Wang, Q.R.; Zhu, M.Q.; Yan, L.Z.; Yu, Y.; Bao, X.B.; Shen, H.J.; Yao, H.; Xie, J.D.; et al. Integrative genomic and transcriptomic profiling reveals distinct molecular subsets in adult mixed phenotype acute leukemia. Am. J. Hematol. 2023, 98, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Tamura, H.; Dan, K.; Tamada, K.; Nakamura, K.; Shioi, Y.; Hyodo, H.; Wang, S.D.; Dong, H.; Chen, L.; Ogata, K. Expression of functional B7-H2 and B7.2 costimulatory molecules and their prognostic implications in de novo acute myeloid leukemia. Clin. Cancer Res. 2005, 11, 5708–5717. [Google Scholar] [CrossRef] [PubMed]
- Graf, M.; Reif, S.; Hecht, K.; Pelka-Fleischer, R.; Kroell, T.; Pfister, K.; Schmetzer, H. High expression of costimulatory molecules correlates with low relapse-free survival probability in acute myeloid leukemia (AML). Ann. Hematol. 2005, 84, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ma, L.; Zhang, X.; Huang, L.; Wei, J. Targeting PD-1/PD-L1 pathway in myelodysplastic syndromes and acute myeloid leukemia. Exp. Hematol. Oncol. 2022, 11, 11. [Google Scholar] [CrossRef]
- Zhang, L.; Gajewski, T.F.; Kline, J. PD-1/PD-L1 interactions inhibit antitumor immune responses in a murine acute myeloid leukemia model. Blood 2009, 114, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Munger, M.E.; Veenstra, R.G.; Weigel, B.J.; Hirashima, M.; Munn, D.H.; Murphy, W.J.; Azuma, M.; Anderson, A.C.; Kuchroo, V.K.; et al. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood 2011, 117, 4501–4510. [Google Scholar] [CrossRef]
- Zhou, Q.; Munger, M.E.; Highfill, S.L.; Tolar, J.; Weigel, B.J.; Riddle, M.; Sharpe, A.H.; Vallera, D.A.; Azuma, M.; Levine, B.L.; et al. Program death-1 signaling and regulatory T cells collaborate to resist the function of adoptively transferred cytotoxic T lymphocytes in advanced acute myeloid leukemia. Blood 2010, 116, 2484–2493. [Google Scholar] [CrossRef]
- Dong, Y.; Han, Y.; Huang, Y.; Jiang, S.; Huang, Z.; Chen, R.; Yu, Z.; Yu, K.; Zhang, S. PD-L1 Is Expressed and Promotes the Expansion of Regulatory T Cells in Acute Myeloid Leukemia. Front. Immunol. 2020, 11, 1710. [Google Scholar] [CrossRef]
- Chen, C.; Liang, C.; Wang, S.; Chio, C.L.; Zhang, Y.; Zeng, C.; Chen, S.; Wang, C.; Li, Y. Expression patterns of immune checkpoints in acute myeloid leukemia. J. Hematol. Oncol. 2020, 13, 28. [Google Scholar] [CrossRef]
- Krönig, H.; Kremmler, L.; Haller, B.; Englert, C.; Peschel, C.; Andreesen, R.; Blank, C.U. Interferon-induced programmed death-ligand 1 (PD-L1/B7-H1) expression increases on human acute myeloid leukemia blast cells during treatment. Eur. J. Haematol. 2014, 92, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Schnorfeil, F.M.; Lichtenegger, F.S.; Emmerig, K.; Schlueter, M.; Neitz, J.S.; Draenert, R.; Hiddemann, W.; Subklewe, M. T cells are functionally not impaired in AML: Increased PD-1 expression is only seen at time of relapse and correlates with a shift towards the memory T cell compartment. J. Hematol. Oncol. 2015, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, S.O.; Sughayer, M.; Khader, M.; Tbakhi, A.; Khudirat, S.; Hejazi, A.; AlRyalat, S.; Bustami, N.; Aladily, T.N. Programmed Death Ligand-1 is Frequently Expressed in Primary Acute Myeloid Leukemia and B-Acute Lymphoblastic Leukemia. Clin. Lab. 2022, 68, 748. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Basu, S.; Garcia-Manero, G.; Hourigan, C.S.; Oetjen, K.A.; Cortes, J.E.; Ravandi, F.; Jabbour, E.J.; Al-Hamal, Z.; Konopleva, M.; et al. The distribution of T-cell subsets and the expression of immune checkpoint receptors and ligands in patients with newly diagnosed and relapsed acute myeloid leukemia. Cancer 2019, 125, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Sallman, D.A.; McLemore, A.F.; Aldrich, A.L.; Komrokji, R.S.; McGraw, K.L.; Dhawan, A.; Geyer, S.; Hou, H.A.; Eksioglu, E.A.; Sullivan, A.; et al. TP53 mutations in myelodysplastic syndromes and secondary AML confer an immunosuppressive phenotype. Blood 2020, 136, 2812–2823. [Google Scholar] [CrossRef] [PubMed]
- Brodská, B.; Otevřelová, P.; Šálek, C.; Fuchs, O.; Gašová, Z.; Kuželová, K. High PD-L1 Expression Predicts for Worse Outcome of Leukemia Patients with Concomitant NPM1 and FLT3 Mutations. Int. J. Mol. Sci. 2019, 20, 2823. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Xing, M.; Han, L.; Gan, S.; Ma, J.; Wu, F.; Huang, Y.; Chen, Y.; Tian, W.; An, C.; et al. High PD-L1 expression drives glycolysis via an Akt/mTOR/HIF-1α axis in acute myeloid leukemia. Oncol. Rep. 2020, 43, 999–1009. [Google Scholar] [CrossRef]
- Wang, F.; Yang, L.; Xiao, M.; Zhang, Z.; Shen, J.; Anuchapreeda, S.; Tima, S.; Chiampanichayakul, S.; Xiao, Z. PD-L1 regulates cell proliferation and apoptosis in acute myeloid leukemia by activating PI3K-AKT signaling pathway. Sci. Rep. 2022, 12, 11444. [Google Scholar] [CrossRef]
- Soltani, M.; Ghanadian, M.; Ghezelbash, B.; Shokouhi, A.; Zamyatnin, A.A., Jr.; Bazhin, A.V.; Ganjalikhani-Hakemi, M. PD-L1 stimulation can promote proliferation and survival of leukemic cells by influencing glucose and fatty acid metabolism in acute myeloid leukemia. BMC Cancer 2023, 23, 447. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Yu, Z.; Huang, J.; Chen, Y.; Huang, S.; Yao, D.; Xu, L.; Lu, Y.; Chen, S.; Li, Y. Increased PD-1+Tim-3+ exhausted T cells in bone marrow may influence the clinical outcome of patients with AML. Biomark. Res. 2020, 8, 6. [Google Scholar] [CrossRef]
- Gonçalves Silva, I.; Yasinska, I.M.; Sakhnevych, S.S.; Fiedler, W.; Wellbrock, J.; Bardelli, M.; Varani, L.; Hussain, R.; Siligardi, G.; Ceccone, G.; et al. The Tim-3-galectin-9 Secretory Pathway is Involved in the Immune Escape of Human Acute Myeloid Leukemia Cells. EBioMedicine 2017, 22, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Chajuwan, T.; Kansuwan, P.; Kobbuaklee, S.; Chanswangphuwana, C. Characteristics and clinical correlation of TIM-3 and PD-1/PD-L1 expressions in leukemic cells and tumor microenvironment in newly diagnosed acute myeloid leukemia. Leuk. Lymphoma 2022, 63, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Yoyen-Ermis, D.; Tunali, G.; Tavukcuoglu, E.; Horzum, U.; Ozkazanc, D.; Sutlu, T.; Buyukasik, Y.; Esendagli, G. Myeloid maturation potentiates STAT3-mediated atypical IFN-γ signaling and upregulation of PD-1 ligands in AML and MDS. Sci. Rep. 2019, 9, 11697. [Google Scholar] [CrossRef] [PubMed]
- Berthon, C.; Driss, V.; Liu, J.; Kuranda, K.; Leleu, X.; Jouy, N.; Hetuin, D.; Quesnel, B. In acute myeloid leukemia, B7-H1 (PD-L1) protection of blasts from cytotoxic T cells is induced by TLR ligands and interferon-gamma and can be reversed using MEK inhibitors. Cancer Immunol. Immunother. 2010, 59, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.X.; Mei, Z.Y.; Zhou, J.H.; Yao, Y.S.; Li, Y.H.; Xu, Y.H.; Li, J.X.; Gao, X.N.; Zhou, M.H.; Jiang, M.M.; et al. Low dose decitabine treatment induces CD80 expression in cancer cells and stimulates tumor specific cytotoxic T lymphocyte responses. PLoS ONE 2013, 8, e62924. [Google Scholar] [CrossRef] [PubMed]
- Graf, M.; Hecht, K.; Reif, S.; Pelka-Fleischer, R.; Pfister, K.; Schmetzer, H. Expression and prognostic value of hemopoietic cytokine receptors in acute myeloid leukemia (AML): Implications for future therapeutical strategies. Eur. J. Haematol. 2004, 72, 89–106. [Google Scholar] [CrossRef] [PubMed]
- Brenner, A.K.; Tvedt, T.H.; Nepstad, I.; Rye, K.P.; Hagen, K.M.; Reikvam, H.; Bruserud, Ø. Patients with acute myeloid leukemia can be subclassified based on the constitutive cytokine release of the leukemic cells; the possible clinical relevance and the importance of cellular iron metabolism. Expert Opin. Ther. Targets 2017, 21, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Ryningen, A.; Olsnes, A.M.; Stordrange, L.; Øyan, A.M.; Kalland, K.H.; Gjertsen, B.T. Subclassification of patients with acute myelogenous leukemia based on chemokine responsiveness and constitutive chemokine release by their leukemic cells. Haematologica 2007, 92, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Honnemyr, M.; Bruserud, Ø.; Brenner, A.K. The constitutive protease release by primary human acute myeloid leukemia cells. J. Cancer Res. Clin. Oncol. 2017, 143, 1985–1998. [Google Scholar] [CrossRef]
- Ghannadan, M.; Wimazal, F.; Simonitsch, I.; Sperr, W.R.; Mayerhofer, M.; Sillaber, C.; Hauswirth, A.W.; Gadner, H.; Chott, A.; Horny, H.P.; et al. Immunohistochemical detection of VEGF in the bone marrow of patients with acute myeloid leukemia. Correlation between VEGF expression and the FAB category. Am. J. Clin. Pathol. 2003, 119, 663–671. [Google Scholar] [CrossRef]
- Cimino, G.; Sgadari, C.; Amadori, S.; Magliocca, V.; Poti, G.P.; Cimino, G.; Mandelli, F. High serum interleukin-2 levels in acute myeloid leukaemia (AML) are associated with FAB M4 and M5 subtypes. Br. J. Haematol. 1989, 73, 431. [Google Scholar] [CrossRef]
- Vinante, F.; Rigo, A.; Tecchio, C.; Patuzzo, C.; Ricetti, M.M.; Morosato, L.; Cassatella, M.A.; Chilosi, M.; Pizzolo, G. Preferential release of high amounts of interleukin-8 by myeloid blasts showing monocytic differentiation. Haematologica 1996, 81, 195–200. [Google Scholar]
- Bruserud, Ø.; Ulvestad, E. Expression and release of adhesion molecules by human acute myelogenous leukemia blasts. Leuk. Res. 1999, 23, 149–157. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Ulvestad, E. Soluble Fas/Apo-1 (CD95) levels during T cell activation in the presence of acute myelogenous leukemia accessory cells; contributions from local release and variations in systemic levels. Cancer Immunol. Immunother. 2000, 49, 377–387. [Google Scholar] [CrossRef]
- Aasebø, E.; Brenner, A.K.; Birkeland, E.; Tvedt, T.H.A.; Selheim, F.; Berven, F.S.; Bruserud, Ø. The Constitutive Extracellular Protein Release by Acute Myeloid Leukemia Cells-A Proteomic Study of Patient Heterogeneity and Its Modulation by Mesenchymal Stromal Cells. Cancers 2021, 13, 1509. [Google Scholar] [CrossRef]
- Bostrom, B.; Brunning, R.D.; McGlave, P.; Ramsay, N.; Nesbit, M., Jr.; Woods, W.G.; Hurd, D.; Krivit, W.; Kim, T.; Goldman, A.; et al. Bone marrow transplantation for acute nonlymphocytic leukemia in first remission: Analysis of prognostic factors. Blood 1985, 65, 1191–1196. [Google Scholar] [CrossRef]
- Saultz, J.N.; Tyner, J.W. Chasing leukemia differentiation through induction therapy, relapse and transplantation. Blood Rev. 2023, 57, 101000. [Google Scholar] [CrossRef]
- Nørgaard, J.M.; Olesen, L.H.; Olesen, G.; Meyer, K.; Kristensen, J.S.; Bendix, K.; Pedersen, B.; Kjeldsen, E.; Hokland, P. FAB M4 and high CD14 surface expression is associated with high cellular resistance to Ara-C and daunorubicin: Implications for clinical outcome in acute myeloid leukaemia. Eur. J. Haematol. 2001, 67, 221–229. [Google Scholar] [CrossRef]
- Miari, K.E.; Guzman, M.L.; Wheadon, H.; Williams, M.T.S. Macrophages in Acute Myeloid Leukaemia: Significant Players in Therapy Resistance and Patient Outcomes. Front. Cell Dev. Biol. 2021, 9, 692800. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Vo, A.K.; Rekvam, H. Hematopoiesis, Inflammation and Aging-The Biological Background and Clinical Impact of Anemia and Increased C-Reactive Protein Levels on Elderly Individuals. J. Clin. Med. 2022, 11, 706. [Google Scholar] [CrossRef]
- Mussai, F.; De Santo, C.; Abu-Dayyeh, I.; Booth, S.; Quek, L.; McEwen-Smith, R.M.; Qureshi, A.; Dazzi, F.; Vyas, P.; Cerundolo, V. Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood 2013, 122, 749–758. [Google Scholar] [CrossRef]
- Al-Matary, Y.S.; Botezatu, L.; Opalka, B.; Hönes, J.M.; Lams, R.F.; Thivakaran, A.; Schütte, J.; Köster, R.; Lennartz, K.; Schroeder, T.; et al. Acute myeloid leukemia cells polarize macrophages towards a leukemia supporting state in a Growth factor independence 1 dependent manner. Haematologica 2016, 101, 1216–1227. [Google Scholar] [CrossRef]
- Zhai, Y.; Zhang, J.; Wang, H.; Lu, W.; Liu, S.; Yu, Y.; Weng, W.; Ding, Z.; Zhu, Q.; Shi, J. Growth differentiation factor 15 contributes to cancer-associated fibroblasts-mediated chemo-protection of AML cells. J. Exp. Clin. Cancer Res. 2016, 35, 147. [Google Scholar] [CrossRef]
- Sauerer, T.; Velázquez, G.F.; Schmid, C. Relapse of acute myeloid leukemia after allogeneic stem cell transplantation: Immune escape mechanisms and current implications for therapy. Mol. Cancer 2023, 22, 180. [Google Scholar] [CrossRef]
- Damiani, D.; Tiribelli, M. Checkpoint Inhibitors in Acute Myeloid Leukemia. Biomedicines 2023, 11, 1724. [Google Scholar] [CrossRef]
- Chen, E.C.; Garcia, J.S. Immunotherapy for acute myeloid leukemia: Current trends, challenges, and strategies. Acta Haematol. 2023, 147, 198–218. [Google Scholar] [CrossRef]
- Bruserud, O.; Ehninger, G.; Hamann, W.; Pawelec, G. Secretion of IL-2, IL-3, IL-4, IL-6 and GM-CSF by CD4+ and CD8+ TCR alpha beta+ T-cell clones derived early after allogeneic bone marrow transplantation. Scand. J. Immunol. 1993, 38, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Zhang, X.; Chen, Q.; Kantawong, F.; Wan, S.; Liu, J.; Li, H.; Zhou, J.; Lu, B.; Wu, J. Identification of a Mitochondria-Related Gene Signature to Predict the Prognosis in AML. Front. Oncol. 2022, 12, 823831. [Google Scholar] [CrossRef]
- Tong, X.; Zhou, F. Integrated bioinformatic analysis of mitochondrial metabolism-related genes in acute myeloid leukemia. Front. Immunol. 2023, 14, 1120670. [Google Scholar] [CrossRef]
- Bruserud, O.; Ulvestad, E. Acute myelogenous leukemia blasts as accessory cells during in vitro T lymphocyte activation. Cell Immunol. 2000, 206, 36–50. [Google Scholar] [CrossRef]
- Xu, Z.J.; Gu, Y.; Wang, C.Z.; Jin, Y.; Wen, X.M.; Ma, J.C.; Tang, L.J.; Mao, Z.W.; Qian, J.; Lin, J. The M2 macrophage marker CD206: A novel prognostic indicator for acute myeloid leukemia. Oncoimmunology 2019, 9, 1683347. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Lü, M.; Cao, F.; Wu, G.; Gao, F.; Pang, H.; Li, Y.; Zhang, Y.; Xing, H.; Liang, C.; et al. Single-cell map of diverse immune phenotypes in the acute myeloid leukemia microenvironment. Biomark. Res. 2021, 9, 15. [Google Scholar] [CrossRef]
- van Galen, P.; Hovestadt, V.; Wadsworth, M.H., II.; Hughes, T.K.; Griffin, G.K.; Battaglia, S.; Verga, J.A.; Stephansky, J.; Pastika, T.J.; Lombardi Story, J.; et al. Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell 2019, 176, 1265–1281. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Feng, W.; Wang, R.; Yang, F.; Wang, L.; Chen, S.; Ru, Y.; Cheng, T.; Zheng, G. Repolarizing heterogeneous leukemia-associated macrophages with more M1 characteristics eliminates their pro-leukemic effects. Oncoimmunology 2017, 7, e1412910. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.L.; Kornbluth, S. The tangled circuitry of metabolism and apoptosis. Mol. Cell 2013, 49, 399–410. [Google Scholar] [CrossRef]
- Li, C.; Zhang, G.; Zhao, L.; Ma, Z.; Chen, H. Metabolic reprogramming in cancer cells: Glycolysis, glutaminolysis, and Bcl-2 proteins as novel therapeutic targets for cancer. World J. Surg. Oncol. 2016, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Sousa, J.S.; D’Imprima, E.; Vonck, J. Mitochondrial Respiratory Chain Complexes. Subcell. Biochem. 2018, 87, 167–227. [Google Scholar] [PubMed]
- Bezawork-Geleta, A.; Rohlena, J.; Dong, L.; Pacak, K.; Neuzil, J. Mitochondrial Complex II: At the Crossroads. Trends Biochem. Sci. 2017, 42, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Papa, S.; Martino, P.L.; Capitanio, G.; Gaballo, A.; De Rasmo, D.; Signorile, A.; Petruzzella, V. The oxidative phosphorylation system in mammalian mitochondria. Adv. Exp. Med. Biol. 2012, 942, 3–37. [Google Scholar]
- Nolfi-Donegan, D.; Braganza, A.; Shiva, S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674. [Google Scholar] [CrossRef]
- Kalyanaraman, B.; Cheng, G.; Hardy, M.; Ouari, O.; Lopez, M.; Joseph, J.; Zielonka, J.; Dwinell, M.B. A review of the basics of mitochondrial bioenergetics, metabolism, and related signaling pathways in cancer cells: Therapeutic targeting of tumor mitochondria with lipophilic cationic compounds. Redox Biol. 2018, 14, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Berkers, C.R.; Maddocks, O.D.; Cheung, E.C.; Mor, I.; Vousden, K.H. Metabolic regulation by p53 family members. Cell Metab. 2013, 18, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Sriskanthadevan, S.; Jeyaraju, D.V.; Chung, T.E.; Prabha, S.; Xu, W.; Skrtic, M.; Jhas, B.; Hurren, R.; Gronda, M.; Wang, X.; et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood 2015, 125, 2120–2130. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.L.; Hege, K.; Kalpage, H.A.; Edwards, H.; Hüttemann, M.; Taub, J.W.; Ge, Y. Targeting mitochondrial respiration for the treatment of acute myeloid leukemia. Biochem. Pharmacol. 2020, 182, 114253. [Google Scholar] [CrossRef] [PubMed]
- Lagadinou, E.D.; Sach, A.; Callahan, K.; Rossi, R.M.; Neering, S.J.; Minhajuddin, M.; Ashton, J.M.; Pei, S.; Grose, V.; O’Dwyer, K.M.; et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013, 12, 329–341. [Google Scholar] [CrossRef]
- Pollyea, D.A.; Stevens, B.M.; Jones, C.L.; Winters, A.; Pei, S.; Minhajuddin, M.; D’Alessandro, A.; Culp-Hill, R.; Riemondy, K.A.; Gillen, A.E.; et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med. 2018, 24, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.M.; Thomas, D.; Corces-Zimmerman, M.R.; Xavy, S.; Rastogi, S.; Hong, W.J.; Zhao, F.; Medeiros, B.C.; Tyvoll, D.A.; Majeti, R. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat. Med. 2015, 21, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Mondet, J.; Presti, C.L.; Garrel, C.; Skaare, K.; Mariette, C.; Carras, S.; Park, S.; Carré, M.; Bulabois, C.E.; Molina, L.; et al. Adult patients with de novo acute myeloid leukemia show a functional deregulation of redox balance at diagnosis which is correlated with molecular subtypes and overall survival. Haematologica 2019, 104, e393–e397. [Google Scholar] [CrossRef]
- Wu, S.; Akhtari, M.; Alachkar, H. Characterization of Mutations in the Mitochondrial Encoded Electron Transport Chain Complexes in Acute Myeloid Leukemia. Sci. Rep. 2018, 8, 13301. [Google Scholar] [CrossRef]
- Ashton, T.M.; McKenna, W.G.; Kunz-Schughart, L.A.; Higgins, G.S. Oxidative Phosphorylation as an Emerging Target in Cancer Therapy. Clin. Cancer Res. 2018, 24, 2482–2490. [Google Scholar] [CrossRef]
- Damm, F.; Bunke, T.; Thol, F.; Markus, B.; Wagner, K.; Göhring, G.; Schlegelberger, B.; Heil, G.; Reuter, C.W.; Püllmann, K.; et al. Prognostic implications and molecular associations of NADH dehydrogenase subunit 4 (ND4) mutations in acute myeloid leukemia. Leukemia 2012, 26, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.A.; Markus, B.; Hermkens, M.; Damm, F.; Reinhardt, D.; Zimmermann, M.; Thol, F.; Bunke, T.; Bogoeva, D.; Reuter, C.W.; et al. NADH dehydrogenase subunit 4 variant sequences in childhood acute myeloid leukaemia. Br. J. Haematol. 2013, 161, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Silkjaer, T.; Nyvold, C.G.; Juhl-Christensen, C.; Hokland, P.; Nørgaard, J.M. Mitochondrial cytochrome c oxidase subunit II variations predict adverse prognosis in cytogenetically normal acute myeloid leukaemia. Eur. J. Haematol. 2013, 91, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, B.M.; Banik, B.K.; Borah, P.; Jain, A. Reactive Oxygen Species (ROS): Key Components in Cancer Therapies. Anticancer Agents Med. Chem. 2022, 22, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003, 552 Pt 2, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Sillar, J.R.; Germon, Z.P.; DeIuliis, G.N.; Dun, M.D. The Role of Reactive Oxygen Species in Acute Myeloid Leukaemia. Int. J. Mol. Sci. 2019, 20, 6003. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Shen, Q.; Claret, F.X. Novel roles of reactive oxygen species in the pathogenesis of acute myeloid leukemia. J. Leukoc. Biol. 2013, 94, 423–429. [Google Scholar] [CrossRef]
- Martinou, J.C.; Youle, R.J. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev. Cell 2011, 21, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C.; Delivani, P.; Cullen, S.P.; Martin, S.J. Bax- or Bak-induced mitochondrial fission can be uncoupled from cytochrome C release. Mol. Cell 2008, 31, 570–585. [Google Scholar] [CrossRef] [PubMed]
- Delivani, P.; Adrain, C.; Taylor, R.C.; Duriez, P.J.; Martin, S.J. Role for CED-9 and Egl-1 as regulators of mitochondrial fission and fusion dynamics. Mol. Cell 2006, 21, 761–773. [Google Scholar] [CrossRef]
- Perciavalle, R.M.; Stewart, D.P.; Koss, B.; Lynch, J.; Milasta, S.; Bathina, M.; Temirov, J.; Cleland, M.M.; Pelletier, S.; Schuetz, J.D.; et al. Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat. Cell Biol. 2012, 14, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, A.E.; Deshmukh, M. Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nat. Cell Biol. 2008, 10, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.C.; Borutaite, V. Regulation of apoptosis by the redox state of cytochrome c. Biochim. Biophys. Acta 2008, 1777, 877–881. [Google Scholar] [CrossRef]
- Brischigliaro, M.; Zeviani, M. Cytochrome c oxidase deficiency. Biochim. Biophys. Acta Bioenerg. 2021, 1862, 148335. [Google Scholar] [CrossRef] [PubMed]
- Hirai, I.; Wang, H.G. Survival-factor-induced phosphorylation of Bad results in its dissociation from Bcl-x(L) but not Bcl-2. Biochem. J. 2001, 359, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.H.; Thompson, C.B. The role of the Bcl-2 family in the regulation of outer mitochondrial membrane permeability. Cell Death Differ. 2000, 7, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Clément, M.V.; Hirpara, J.L.; Pervaiz, S. Decrease in intracellular superoxide sensitizes Bcl-2-overexpressing tumor cells to receptor and drug-induced apoptosis independent of the mitochondria. Cell Death Differ. 2003, 10, 1273–1285. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.X.; Pervaiz, S. Bcl-2 induces pro-oxidant state by engaging mitochondrial respiration in tumor cells. Cell Death Differ. 2007, 14, 1617–1627. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.X.; Pervaiz, S. Involvement of cytochrome c oxidase subunits Va and Vb in the regulation of cancer cell metabolism by Bcl-2. Cell Death Differ. 2010, 17, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Maurer, U.; Charvet, C.; Wagman, A.S.; Dejardin, E.; Green, D.R. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol. Cell 2006, 21, 749–760. [Google Scholar] [CrossRef]
- Zhao, Y.; Altman, B.J.; Coloff, J.L.; Herman, C.E.; Jacobs, S.R.; Wieman, H.L.; Wofford, J.A.; Dimascio, L.N.; Ilkayeva, O.; Kelekar, A.; et al. Glycogen synthase kinase 3alpha and 3beta mediate a glucose-sensitive antiapoptotic signaling pathway to stabilize Mcl-1. Mol. Cell Biol. 2007, 27, 4328–4339. [Google Scholar] [CrossRef] [PubMed]
- Coloff, J.L.; Macintyre, A.N.; Nichols, A.G.; Liu, T.; Gallo, C.A.; Plas, D.R.; Rathmell, J.C. Akt-dependent glucose metabolism promotes Mcl-1 synthesis to maintain cell survival and resistance to Bcl-2 inhibition. Cancer Res. 2011, 71, 5204–5213. [Google Scholar] [CrossRef] [PubMed]
- Carter, B.Z.; Mak, P.Y.; Tao, W.; Warmoes, M.; Lorenzi, P.L.; Mak, D.; Ruvolo, V.; Tan, L.; Cidado, J.; Drew, L.; et al. Targeting MCL-1 dysregulates cell metabolism and leukemia-stroma interactions and resensitizes acute myeloid leukemia to BCL-2 inhibition. Haematologica 2022, 107, 58–76. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.H.; Pan, H.; Seebacher, J.; Jang, I.H.; Hyberts, S.G.; Heffron, G.J.; Vander Heiden, M.G.; Yang, R.; Li, F.; Locasale, J.W.; et al. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell 2011, 146, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Alavian, K.N.; Li, H.; Collis, L.; Bonanni, L.; Zeng, L.; Sacchetti, S.; Lazrove, E.; Nabili, P.; Flaherty, B.; Graham, M.; et al. Bcl-xL regulates metabolic efficiency of neurons through interaction with the mitochondrial F1FO ATP synthase. Nat. Cell Biol. 2011, 13, 1224–1233. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.; Hayashi, T.; Wolozny, D.; Yin, B.; Su, T.C.; Betenbaugh, M.J.; Su, T.P. The non-apoptotic action of Bcl-xL: Regulating Ca(2+) signaling and bioenergetics at the ER-mitochondrion interface. J. Bioenerg. Biomembr. 2016, 48, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, R.A.; Franklin, J.L. Bax affects production of reactive oxygen by the mitochondria of non-apoptotic neurons. Exp. Neurol. 2007, 204, 458–461. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jones, R.G.; Bui, T.; White, C.; Madesh, M.; Krawczyk, C.M.; Lindsten, T.; Hawkins, B.J.; Kubek, S.; Frauwirth, K.A.; Wang, Y.L.; et al. The proapoptotic factors Bax and Bak regulate T Cell proliferation through control of endoplasmic reticulum Ca(2+) homeostasis. Immunity 2007, 27, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Colombini, M. Ceramide channels and their role in mitochondria-mediated apoptosis. Biochim. Biophys. Acta 2010, 1797, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
- Van Brocklyn, J.R.; Williams, J.B. The control of the balance between ceramide and sphingosine-1-phosphate by sphingosine kinase: Oxidative stress and the seesaw of cell survival and death. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 163, 26–36. [Google Scholar] [CrossRef]
- Danial, N.N.; Gramm, C.F.; Scorrano, L.; Zhang, C.Y.; Krauss, S.; Ranger, A.M.; Datta, S.R.; Greenberg, M.E.; Licklider, L.J.; Lowell, B.B.; et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 2003, 424, 952–956. [Google Scholar] [CrossRef]
- Lowman, X.H.; McDonnell, M.A.; Kosloske, A.; Odumade, O.A.; Jenness, C.; Karim, C.B.; Jemmerson, R.; Kelekar, A. The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Mol. Cell 2010, 40, 823–833. [Google Scholar] [CrossRef]
- Gottlieb, E.; Vousden, K.H. p53 regulation of metabolic pathways. Cold Spring Harb. Perspect. Biol. 2010, 2, a001040. [Google Scholar] [CrossRef]
- Yaseen, A.; Chen, S.; Hock, S.; Rosato, R.; Dent, P.; Dai, Y.; Grant, S. Resveratrol sensitizes acute myelogenous leukemia cells to histone deacetylase inhibitors through reactive oxygen species-mediated activation of the extrinsic apoptotic pathway. Mol. Pharmacol. 2012, 82, 1030–1041. [Google Scholar] [CrossRef] [PubMed]
- Sasca, D.; Hähnel, P.S.; Szybinski, J.; Khawaja, K.; Kriege, O.; Pante, S.V.; Bullinger, L.; Strand, S.; Strand, D.; Theobald, M.; et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood 2014, 124, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Bhatia, R. Role of SIRT1 in the growth and regulation of normal hematopoietic and leukemia stem cells. Curr. Opin. Hematol. 2015, 22, 324–329. [Google Scholar] [CrossRef]
- O’Brien, C.; Ling, T.; Berman, J.M.; Culp-Hill, R.; Reisz, J.A.; Rondeau, V.; Jahangiri, S.; St-Germain, J.; Macwan, V.; Astori, A.; et al. Simultaneous inhibition of Sirtuin 3 and cholesterol homeostasis targets acute myeloid leukemia stem cells by perturbing fatty acid β-oxidation and inducing lipotoxicity. Haematologica 2023, 108, 2343–2357. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, Y.; Wei, W.; Wang, W.; Jiang, D.; Ren, Y.; Peng, Z.; Fan, Q.; Cheng, J.; Ma, J. Dysregulation of SIRT3 SUMOylation Confers AML Chemoresistance via Controlling HES1-Dependent Fatty Acid Oxidation. Int. J. Mol. Sci. 2022, 23, 8282. [Google Scholar] [CrossRef] [PubMed]
- Herst, P.M.; Howman, R.A.; Neeson, P.J.; Berridge, M.V.; Ritchie, D.S. The level of glycolytic metabolism in acute myeloid leukemia blasts at diagnosis is prognostic for clinical outcome. J. Leukoc. Biol. 2011, 89, 51–55. [Google Scholar] [CrossRef]
- Catalano, G.; Zaza, A.; Banella, C.; Pelosi, E.; Castelli, G.; de Marinis, E.; Smigliani, A.; Travaglini, S.; Ottone, T.; Divona, M.; et al. MCL1 regulates AML cells metabolism via direct interaction with HK2. Metabolic signature at onset predicts overall survival in AMLs’ patients. Leukemia 2023, 37, 1600–1610. [Google Scholar] [CrossRef]
- Chen, M.; Tao, Y.; Yue, P.; Guo, F.; Yan, X. Construction and validation of a fatty acid metabolism risk signature for predicting prognosis in acute myeloid leukemia. BMC Genom. Data 2022, 23, 85. [Google Scholar] [CrossRef]
- Selheim, F.; Aasebø, E.; Bruserud, Ø.; Hernandez-Valladares, M. High Mitochondrial Protein Expression as a Potential Predictor of Relapse Risk in Acute Myeloid Leukemia Patients with the Monocytic FAB Subtypes M4 and M5. Cancers 2024, 16, 8. [Google Scholar] [CrossRef]
- Selheim, F.; Aasebø, E.; Reikvam, H.; Bruserud, Ø.; Hernandez-Valladares, M. Monocytic differentiation of human acute myeloid leukemia cells: A proteomic and phosphoproteomic comparison of FAB-M4/M5 patients with and without nucleophosmin 1 mutations. Int. J. Mol. Sci. 2024, 25, 5080. [Google Scholar] [CrossRef]
- Shimony, S.; Stone, R.M.; Stahl, M. Venetoclax combination therapy in acute myeloid leukemia and myelodysplastic syndromes. Curr. Opin. Hematol. 2022, 29, 63–73. [Google Scholar] [CrossRef]
- Jonas, B.A.; Pollyea, D.A. How we use venetoclax with hypomethylating agents for the treatment of newly diagnosed patients with acute myeloid leukemia. Leukemia 2019, 33, 2795–2804. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Jonas, B.A.; Pullarkat, V.; Thirman, M.J.; Garcia, J.S.; Wei, A.H.; Konopleva, M.; Döhner, H.; Letai, A.; Fenaux, P.; et al. Azacitidine and Venetoclax in Previously Untreated Acute Myeloid Leukemia. N. Engl. J. Med. 2020, 383, 617–629. [Google Scholar] [CrossRef]
- Gangat, N.; Johnson, I.; McCullough, K.; Farrukh, F.; Al-Kali, A.; Alkhateeb, H.; Begna, K.; Mangaonkar, A.; Litzow, M.; Hogan, W.; et al. Molecular predictors of response to venetoclax plus hypomethylating agent in treatment-naïve acute myeloid leukemia. Haematologica 2022, 107, 2501–2505. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Tiong, I.S.; Quaglieri, A.; MacRaild, S.; Loghavi, S.; Brown, F.C.; Thijssen, R.; Pomilio, G.; Ivey, A.; Salmon, J.M.; et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 2020, 135, 791–803. [Google Scholar] [CrossRef]
- Aldoss, I.; Yang, D.; Pillai, R.; Sanchez, J.F.; Mei, M.; Aribi, A.; Ali, H.; Sandhu, K.; Al Malki, M.M.; Salhotra, A.; et al. Association of leukemia genetics with response to venetoclax and hypomethylating agents in relapsed/refractory acute myeloid leukemia. Am. J. Hematol. 2019, 94, E253–E255. [Google Scholar] [CrossRef]
- Ucciero, A.; Pagnoni, F.; Scotti, L.; Pisterna, A.; Barone-Adesi, F.; Gaidano, G.; Patriarca, A.; Lunghi, M. Venetoclax with Hypomethylating Agents in Newly Diagnosed Acute Myeloid Leukemia: A Systematic Review and Meta-Analysis of Survival Data from Real-World Studies. Cancers 2023, 15, 4618. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Lachowiez, C.A.; Takahashi, K.; Loghavi, S.; Xiao, L.; Kadia, T.; Daver, N.; Adeoti, M.; Short, N.J.; Sasaki, K.; et al. Venetoclax Combined With FLAG-IDA Induction and Consolidation in Newly Diagnosed and Relapsed or Refractory Acute Myeloid Leukemia. J. Clin. Oncol. 2021, 39, 2768–2778. [Google Scholar] [CrossRef]
- Wang, Y.W.; Tsai, C.H.; Lin, C.C.; Tien, F.M.; Chen, Y.W.; Lin, H.Y.; Yao, M.; Lin, Y.C.; Lin, C.T.; Cheng, C.L.; et al. Cytogenetics and mutations could predict outcome in relapsed and refractory acute myeloid leukemia patients receiving BCL-2 inhibitor venetoclax. Ann. Hematol. 2020, 99, 501–511. [Google Scholar] [CrossRef]
- Byrne, M.; Danielson, N.; Sengsayadeth, S.; Rasche, A.; Culos, K.; Gatwood, K.; Wyatt, H.; Chinratanalab, W.; Dholaria, B.; Ferrell, P.B.; et al. The use of venetoclax-based salvage therapy for post-hematopoietic cell transplantation relapse of acute myeloid leukemia. Am. J. Hematol. 2020, 95, 1006–1014. [Google Scholar] [CrossRef]
- Schuler, E.; Wagner-Drouet, E.M.; Ajib, S.; Bug, G.; Crysandt, M.; Dressler, S.; Hausmann, A.; Heidenreich, D.; Hirschbühl, K.; Hoepting, M.; et al. Treatment of myeloid malignancies relapsing after allogeneic hematopoietic stem cell transplantation with venetoclax and hypomethylating agents-a retrospective multicenter analysis on behalf of the German Cooperative Transplant Study Group. Ann. Hematol. 2021, 100, 959–968. [Google Scholar] [CrossRef]
- Amit, O.; On, Y.B.; Perez, G.; Shargian-Alon, L.; Yeshurun, M.; Ram, R. Venetoclax and donor lymphocyte infusion for early relapsed acute myeloid leukemia after allogeneic hematopoietic cell transplantation. A retrospective multicenter trial. Ann. Hematol. 2021, 100, 817–824. [Google Scholar] [CrossRef]
- Zhang, H.; Nakauchi, Y.; Köhnke, T.; Stafford, M.; Bottomly, D.; Thomas, R.; Wilmot, B.; McWeeney, S.K.; Majeti, R.; Tyner, J.W. Integrated analysis of patient samples identifies biomarkers for venetoclax efficacy and combination strategies in acute myeloid leukemia. Nat. Cancer 2020, 1, 826–839. [Google Scholar] [CrossRef]
- White, B.S.; Khan, S.A.; Mason, M.J.; Ammad-Ud-Din, M.; Potdar, S.; Malani, D.; Kuusanmäki, H.; Druker, B.J.; Heckman, C.; Kallioniemi, O.; et al. Bayesian multi-source regression and monocyte-associated gene expression predict BCL-2 inhibitor resistance in acute myeloid leukemia. NPJ Precis. Oncol. 2021, 5, 71. [Google Scholar] [CrossRef]
- Weidenauer, K.; Schmidt, C.; Rohde, C.; Pauli, C.; Blank, M.F.; Heid, D.; Waclawiczek, A.; Corbacioglu, A.; Göllner, S.; Lotze, M.; et al. The ribosomal protein S6 kinase alpha-1 (RPS6KA1) induces resistance to venetoclax/azacitidine in acute myeloid leukemia. Leukemia 2023, 37, 1611–1625. [Google Scholar] [CrossRef]
- Cherry, E.M.; Abbott, D.; Amaya, M.; McMahon, C.; Schwartz, M.; Rosser, J.; Sato, A.; Schowinsky, J.; Inguva, A.; Minhajuddin, M.; et al. Venetoclax and azacitidine compared with induction chemotherapy for newly diagnosed patients with acute myeloid leukemia. Blood Adv. 2021, 5, 5565–5573. [Google Scholar] [CrossRef]
- Tiong, I.S.; Dillon, R.; Ivey, A.; The, T.C.; Nguyen, P.; Cummings, N.; Taussig, D.C.; Latif, A.L.; Potter, N.E.; Runglall, M.; et al. Venetoclax induces rapid elimination of NPM1 mutant measurable residual disease in combination with low-intensity chemotherapy in acute myeloid leukaemia. Br. J. Haematol. 2021, 192, 1026–1030. [Google Scholar] [CrossRef]
- Falini, B.; Brunetti, L.; Sportoletti, P.; Martelli, M.P. NPM1-mutated acute myeloid leukemia: From bench to bedside. Blood 2020, 136, 1707–1721. [Google Scholar] [CrossRef]
- Tartaglia, M.; Martinelli, S.; Cazzaniga, G.; Cordeddu, V.; Iavarone, I.; Spinelli, M.; Palmi, C.; Carta, C.; Pession, A.; Aricò, M.; et al. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood 2004, 104, 307–313. [Google Scholar] [CrossRef]
- Tartaglia, M.; Martinelli, S.; Iavarone, I.; Cazzaniga, G.; Spinelli, M.; Giarin, E.; Petrangeli, V.; Carta, C.; Masetti, R.; Aricò, M.; et al. Somatic PTPN11 mutations in childhood acute myeloid leukaemia. Br. J. Haematol. 2005, 129, 333–339. [Google Scholar] [CrossRef]
- Kuusanmäki, H.; Dufva, O.; Vähä-Koskela, M.; Leppä, A.M.; Huuhtanen, J.; Vänttinen, I.; Nygren, P.; Klievink, J.; Bouhlal, J.; Pölönen, P.; et al. Erythroid/megakaryocytic differentiation confers BCL-XL dependency and venetoclax resistance in acute myeloid leukemia. Blood 2023, 141, 1610–1625. [Google Scholar] [CrossRef]
- Oyogoa, E.; Traer, E.; Tyner, J.; Lachowiez, C. Building on Foundations: Venetoclax-Based Combinations in the Treatment of Acute Myeloid Leukemia. Cancers 2023, 15, 3589. [Google Scholar] [CrossRef]
- Pei, S.; Shelton, I.T.; Gillen, A.E.; Stevens, B.M.; Gasparetto, M.; Wang, Y.; Liu, L.; Liu, J.; Brunetti, T.M.; Engel, K.; et al. A Novel Type of Monocytic Leukemia Stem Cell Revealed by the Clinical Use of Venetoclax-Based Therapy. Cancer Discov. 2023, 13, 2032–2049. [Google Scholar] [CrossRef]
- Griffioen, M.S.; de Leeuw, D.C.; Janssen, J.J.W.M.; Smit, L. Targeting Acute Myeloid Leukemia with Venetoclax; Biomarkers for Sensitivity and Rationale for Venetoclax-Based Combination Therapies. Cancers 2022, 14, 3456. [Google Scholar] [CrossRef]
- Waclawiczek, A.; Leppä, A.M.; Renders, S.; Stumpf, K.; Reyneri, C.; Betz, B.; Janssen, M.; Shahswar, R.; Donato, E.; Karpova, D.; et al. Combinatorial BCL2 Family Expression in Acute Myeloid Leukemia Stem Cells Predicts Clinical Response to Azacitidine/Venetoclax. Cancer Discov. 2023, 13, 1408–1427. [Google Scholar] [CrossRef]
- Rahmani, M.; Nkwocha, J.; Hawkins, E.; Pei, X.; Parker, R.E.; Kmieciak, M.; Leverson, J.D.; Sampath, D.; Ferreira-Gonzalez, A.; Grant, S. Cotargeting BCL-2 and PI3K Induces BAX-Dependent Mitochondrial Apoptosis in AML Cells. Cancer Res. 2018, 78, 3075–3086. [Google Scholar] [CrossRef]
- Emadi, A.; Kapadia, B.; Bollino, D.; Bhandary, B.; Baer, M.R.; Niyongere, S.; Strovel, E.T.; Kaizer, H.; Chang, E.; Choi, E.Y.; et al. Venetoclax and pegcrisantaspase for complex karyotype acute myeloid leukemia. Leukemia 2021, 35, 1907–1924. [Google Scholar] [CrossRef]
- Eide, C.A.; Kurtz, S.E.; Kaempf, A.; Long, N.; Joshi, S.K.; Nechiporuk, T.; Huang, A.; Dibb, C.A.; Taylor, A.; Bottomly, D.; et al. Clinical Correlates of Venetoclax-Based Combination Sensitivities to Augment Acute Myeloid Leukemia Therapy. Blood Cancer Discov. 2023, 4, 452–467. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yang, J.; Chen, M.; Xiang, X.; Ma, H.; Niu, T.; Gong, Y.; Chen, X.; Liu, J.; Wu, Y. Myelomonocytic and monocytic acute myeloid leukemia demonstrate comparable poor outcomes with venetoclax-based treatment: A monocentric real-world study. Ann. Hematol. 2024, 103, 1197–1209. [Google Scholar] [CrossRef]
- Mohanty, V.; Baran, N.; Huang, Y.; Ramage, C.L.; Cooper, L.M.; He, S.; Iqbal, R.; Daher, M.; Tyner, J.W.; Mills, G.B.; et al. Transcriptional and phenotypic heterogeneity underpinning venetoclax resistance in AML. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.; Xie, J.; Xu, T.; Chen, M.; Guo, Y.; Wang, Y.; Qiu, H.; Wu, D.; Zeng, Z.; Chen, S.; et al. Identification of a venetoclax-resistance prognostic signature base on 6-senescence genes and its clinical significance for acute myeloid leukemia. Front. Oncol. 2023, 13, 1302356. [Google Scholar] [CrossRef]
- Ramsey, H.E.; Fischer, M.A.; Lee, T.; Gorska, A.E.; Arrate, M.P.; Fuller, L.; Boyd, K.L.; Strickland, S.A.; Sensintaffar, J.; Hogdal, L.J.; et al. A Novel MCL1 Inhibitor Combined with Venetoclax Rescues Venetoclax-Resistant Acute Myelogenous Leukemia. Cancer Discov. 2018, 8, 1566–1581. [Google Scholar] [CrossRef]
- Zuber, J.; Shi, J.; Wang, E.; Rappaport, A.R.; Herrmann, H.; Sison, E.A.; Magoon, D.; Qi, J.; Blatt, K.; Wunderlich, M.; et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011, 478, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Romine, K.A.; Nechiporuk, T.; Bottomly, D.; Jeng, S.; McWeeney, S.K.; Kaempf, A.; Corces, M.R.; Majeti, R.; Tyner, J.W. Monocytic differentiation and AHR signaling as Primary Nodes of BET Inhibitor Response in Acute Myeloid Leukemia. Blood Cancer Discov. 2021, 2, 518–531. [Google Scholar] [CrossRef]
- Morabito, F.; Voso, M.T.; Hohaus, S.; Gentile, M.; Vigna, E.; Recchia, A.G.; Iovino, L.; Benedetti, E.; Lo-Coco, F.; Galimberti, S. Panobinostat for the treatment of acute myelogenous leukemia. Expert Opin. Investig. Drugs 2016, 25, 1117–1131. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, X.; Yu, L. Roles of Histone Deacetylases in Acute Myeloid Leukemia with Fusion Proteins. Front. Oncol. 2021, 11, 741746. [Google Scholar] [CrossRef]
- Bots, M.; Verbrugge, I.; Martin, B.P.; Salmon, J.M.; Ghisi, M.; Baker, A.; Stanley, K.; Shortt, J.; Ossenkoppele, G.J.; Zuber, J.; et al. Differentiation therapy for the treatment of t(8;21) acute myeloid leukemia using histone deacetylase inhibitors. Blood 2014, 123, 1341–1352. [Google Scholar] [CrossRef]
- Petruccelli, L.A.; Pettersson, F.; Del Rincón, S.V.; Guilbert, C.; Licht, J.D.; Miller, W.H., Jr. Expression of leukemia-associated fusion proteins increases sensitivity to histone deacetylase inhibitor-induced DNA damage and apoptosis. Mol. Cancer Ther. 2013, 12, 1591–1604. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Maru, Y.; Tanaka, J. Action mechanisms of histone deacetylase inhibitors in the treatment of hematological malignancies. Cancer Sci. 2016, 107, 1543–1549. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Tsykunova, G.; Hernandez-Valladares, M.; Reikvam, H.; Tvedt, T.H.A. Therapeutic Use of Valproic Acid and All-Trans Retinoic Acid in Acute Myeloid Leukemia-Literature Review and Discussion of Possible Use in Relapse after Allogeneic Stem Cell Transplantation. Pharmaceuticals 2021, 14, 423. [Google Scholar] [CrossRef] [PubMed]
- Fredly, H.; Ersvær, E.; Kittang, A.O.; Tsykunova, G.; Gjertsen, B.T.; Bruserud, Ø. The combination of valproic acid, all-trans retinoic acid and low-dose cytarabine as disease-stabilizing treatment in acute myeloid leukemia. Clin. Epigenet. 2013, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Rücker, F.G.; Lang, K.M.; Fütterer, M.; Komarica, V.; Schmid, M.; Döhner, H.; Schlenk, R.F.; Döhner, K.; Knudsen, S.; Bullinger, L. Molecular dissection of valproic acid effects in acute myeloid leukemia identifies predictive networks. Epigenetics 2016, 11, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Li, G.; Cui, Z.; Chen, P.; Wang, J.; Hu, Z.; Zhang, L.; Wei, L. The Histone Deacetylase Inhibitor I1 Induces Differentiation of Acute Leukemia Cells with MLL Gene Rearrangements via Epigenetic Modification. Front. Pharmacol. 2022, 13, 876076. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Duan, Y.; Wang, J.; Liu, Y.; Zhao, Y.; Wang, H.; Zhang, L.; Chen, Z.S.; Hu, Z.; Wei, L. Histone Deacetylase Inhibitor I3 Induces the Differentiation of Acute Myeloid Leukemia Cells with t(8; 21) or MLL Gene Translocation and Leukemic Stem-Like cells. J. Oncol. 2022, 2022, 3345536. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhao, M.; Wu, Z.X.; Yao, J.; Zhang, L.; Wang, J.; Hu, Z.; Wie, L.; Chen, Z.S. The Histone Deacetylase Inhibitor I13 Induces Differentiation of M2, M3 and M5 Subtypes of Acute Myeloid Leukemia Cells and Leukemic Stem-Like Cells. Front. Oncol. 2022, 12, 855570. [Google Scholar] [CrossRef] [PubMed]
- Maiques-Diaz, A.; Spencer, G.J.; Lynch, J.T.; Ciceri, F.; Williams, E.L.; Amaral, F.M.R.; Wiseman, D.H.; Harris, W.J.; Li, Y.; Sahoo, S.; et al. Enhancer Activation by Pharmacologic Displacement of LSD1 from GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell Rep. 2018, 22, 3641–3659. [Google Scholar] [CrossRef]
- Harris, W.J.; Huang, X.; Lynch, J.T.; Spencer, G.J.; Hitchin, J.R.; Li, Y.; Ciceri, F.; Blaser, J.G.; Greystoke, B.F.; Jordan, A.M.; et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012, 21, 473–487. [Google Scholar] [CrossRef]
- Smitheman, K.N.; Severson, T.M.; Rajapurkar, S.R.; McCabe, M.T.; Karpinich, N.; Foley, J.; Pappalardi, M.B.; Hughes, A.; Halsey, W.; Thomas, E.; et al. Lysine specific demethylase 1 inactivation enhances differentiation and promotes cytotoxic response when combined with all-trans retinoic acid in acute myeloid leukemia across subtypes. Haematologica 2019, 104, 1156–1167. [Google Scholar] [CrossRef]
- Noce, B.; Di Bello, E.; Fioravanti, R.; Mai, A. LSD1 inhibitors for cancer treatment: Focus on multi-target agents and compounds in clinical trials. Front. Pharmacol. 2023, 14, 1120911. [Google Scholar] [CrossRef]
- Salamero, O.; Montesinos, P.; Willekens, C.; Pérez-Simón, J.A.; Pigneux, A.; Récher, C.; Popat, R.; Carpio, C.; Molinero, C.; Mascaró, C.; et al. First-in-Human Phase I Study of Iadademstat (ORY-1001): A First-in-Class Lysine-Specific Histone Demethylase 1A Inhibitor, in Relapsed or Refractory Acute Myeloid Leukemia. J. Clin. Oncol. 2020, 38, 4260–4273. [Google Scholar] [CrossRef]
- Brzezinka, K.; Nevedomskaya, E.; Lesche, R.; Steckel, M.; Eheim, A.L.; Haegebarth, A.; Stresemann, C. Functional diversity of inhibitors tackling the differentiation blockage of MLL-rearranged leukemia. J. Hematol. Oncol. 2019, 12, 66. [Google Scholar] [CrossRef]
- Rau, R.E.; Rodriguez, B.A.; Luo, M.; Jeong, M.; Rosen, A.; Rogers, J.H.; Campbell, C.T.; Daigle, S.R.; Deng, L.; Song, Y.; et al. DOT1L as a therapeutic target for the treatment of DNMT3A-mutant acute myeloid leukemia. Blood 2016, 128, 971–981. [Google Scholar] [CrossRef]
- Dafflon, C.; Craig, V.J.; Méreau, H.; Gräsel, J.; Schacher Engstler, B.; Hoffman, G.; Nigsch, F.; Gaulis, S.; Barys, L.; Ito, M.; et al. Complementary activities of DOT1L and Menin inhibitors in MLL-rearranged leukemia. Leukemia 2017, 31, 1269–1277. [Google Scholar] [CrossRef]
- Stein, E.M.; Garcia-Manero, G.; Rizzieri, D.A.; Tibes, R.; Berdeja, J.G.; Savona, M.R.; Jongen-Lavrenic, M.; Altman, J.K.; Thomson, B.; Blakemore, S.J.; et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 2018, 131, 2661–2669. [Google Scholar] [CrossRef]
- Dempke, W.C.M.; Desole, M.; Chiusolo, P.; Sica, S.; Schmidt-Hieber, M. Targeting the undruggable: Menin inhibitors ante portas. J. Cancer Res. Clin. Oncol. 2023, 149, 9451–9459. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Evans, K.; Gadrey, J.Y.; Eschle, B.K.; Hatton, C.; Uckelmann, H.J.; Ross, K.N.; Perner, F.; Olsen, S.N.; Pritchard, T.; et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer Cell 2019, 36, 660–673. [Google Scholar] [CrossRef]
- Fiskus, W.; Boettcher, S.; Daver, N.; Mill, C.P.; Sasaki, K.; Birdwell, C.E.; Davis, J.A.; Takahashi, K.; Kadia, T.M.; DiNardo, C.D.; et al. Effective Menin inhibitor-based combinations against AML with MLL rearrangement or NPM1 mutation (NPM1c). Blood Cancer J. 2022, 12, 5. [Google Scholar] [CrossRef]
- Dzama, M.M.; Steiner, M.; Rausch, J.; Sasca, D.; Schönfeld, J.; Kunz, K.; Taubert, M.C.; McGeehan, G.M.; Chen, C.W.; Mupo, A.; et al. Synergistic targeting of FLT3 mutations in AML via combined menin-MLL and FLT3 inhibition. Blood 2020, 136, 2442–2456. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.D.; Nathan, S.; Larson, M.; Hussain, M.J.; Katz, D.A.; Varma, A.; Miller, I.; Ustun, C. Erythroid differentiation of myeloblast induced by gilteritinib in relapsed FLT3-ITD-positive acute myeloid leukemia. Blood Adv. 2019, 3, 3709–3712. [Google Scholar] [CrossRef]
- Heikamp, E.B.; Henrich, J.A.; Perner, F.; Wong, E.M.; Hatton, C.; Wen, Y.; Barwe, S.P.; Gopalakrishnapillai, A.; Xu, H.; Uckelmann, H.J.; et al. The menin-MLL1 interaction is a molecular dependency in NUP98-rearranged AML. Blood 2022, 139, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Barajas, J.M.; Rasouli, M.; Umeda, M.; Hiltenbrand, R.L.; Abdelhamed, S.; Mohnani, R.; Arthur, B.; Westover, T.; Thomas, M.E., 3rd; Ashtiani, M.; et al. Acute myeloid leukemias with UBTF tandem duplications are sensitive to Menin inhibitors. Blood 2023, 143, 619–630. [Google Scholar] [CrossRef]
- Libbrecht, C.; Xie, H.M.; Kingsley, M.C.; Haladyna, J.N.; Riedel, S.S.; Alikarami, F.; Lenard, A.; McGeehan, G.M.; Ernst, P.; Bernt, K.M. Menin is necessary for long term maintenance of meningioma-1 driven leukemia. Leukemia 2021, 35, 1405–1417. [Google Scholar] [CrossRef]
- Brunetti, L.; Gundry, M.C.; Sorcini, D.; Guzman, A.G.; Huang, Y.H.; Ramabadran, R.; Gionfriddo, I.; Mezzasoma, F.; Milano, F.; Nabet, B.; et al. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell 2018, 34, 499–512. [Google Scholar] [CrossRef]
- Lin, K.H.; Rutter, J.C.; Xie, A.; Killarney, S.T.; Vaganay, C.; Benaksas, C.; Ling, F.; Sodaro, G.; Meslin, P.A.; Bassil, C.F.; et al. P2RY2-AKT activation is a therapeutically actionable consequence of XPO1 inhibition in acute myeloid leukemia. Nat. Cancer 2022, 3, 837–851. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wu, S.; Liu, S.; Li, X.; Gai, Y.; Lin, H.; Wang, Y.; Edwards, H.; Ge, Y.; Wang, G. Venetoclax enhances DNA damage induced by XPO1 inhibitors: A novel mechanism underlying the synergistic antileukaemic effect in acute myeloid leukaemia. J. Cell Mol. Med. 2022, 26, 2646–2657. [Google Scholar] [CrossRef]
- Garzon, R.; Savona, M.; Baz, R.; Andreeff, M.; Gabrail, N.; Gutierrez, M.; Savoie, L.; Mau-Sorensen, P.M.; Wagner-Johnston, N.; Yee, K.; et al. A phase 1 clinical trial of single-agent selinexor in acute myeloid leukemia. Blood 2017, 129, 3165–3174. [Google Scholar] [CrossRef]
- Bhatnagar, B.; Zhao, Q.; Mims, A.S.; Vasu, S.; Behbehani, G.K.; Larkin, K.; Blachly, J.S.; Badawi, M.A.; Hill, K.L.; Dzwigalski, K.R.; et al. Phase 1 study of selinexor in combination with salvage chemotherapy in Adults with relapsed or refractory Acute myeloid leukemia. Leuk. Lymphoma 2023, 64, 2091–2100. [Google Scholar] [CrossRef]
- Ranganathan, P.; Yu, X.; Santhanam, R.; Hofstetter, J.; Walker, A.; Walsh, K.; Bhatnagar, B.; Klisovic, R.; Vasu, S.; Phelps, M.A.; et al. Decitabine priming enhances the antileukemic effects of exportin 1 (XPO1) selective inhibitor selinexor in acute myeloid leukemia. Blood 2015, 125, 2689–2692. [Google Scholar] [CrossRef]
- Etchin, J.; Berezovskaya, A.; Conway, A.S.; Galinsky, I.A.; Stone, R.M.; Baloglu, E.; Senapedis, W.; Landesman, Y.; Kauffman, M.; Shacham, S.; et al. KPT-8602, a second-generation inhibitor of XPO1-mediated nuclear export, is well tolerated and highly active against AML blasts and leukemia-initiating cells. Leukemia 2017, 31, 143–150. [Google Scholar] [CrossRef]
- Sharon, D.; Chan, S.M. Repurposing ribosome-targeting antibiotics to overcome resistance to venetoclax in acute myeloid leukemia. Mol. Cell Oncol. 2020, 7, 1712182. [Google Scholar] [CrossRef] [PubMed]
- Riccioni, R.; Senese, M.; Diverio, D.; Riti, V.; Buffolino, S.; Mariani, G.; Boe, A.; Cedrone, M.; Lo-Coco, F.; Foà, R.; et al. M4 and M5 acute myeloid leukaemias display a high sensitivity to Bortezomib-mediated apoptosis. Br. J. Haematol. 2007, 139, 194–205. [Google Scholar] [CrossRef]
- Stapnes, C.; Døskeland, A.P.; Hatfield, K.; Ersvaer, E.; Ryningen, A.; Lorens, J.B.; Gjertsen, B.T.; Bruserud, O. The proteasome inhibitors bortezomib and PR-171 have antiproliferative and proapoptotic effects on primary human acute myeloid leukaemia cells. Br. J. Haematol. 2007, 136, 814–828. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, M.; Murphy, E.; Morrell, R.; Knapper, S.; O’Dwyer, M.; Samali, A.; Szegezdi, E. The Proteasome Inhibitor Bortezomib Sensitizes AML with Myelomonocytic Differentiation to TRAIL Mediated Apoptosis. Cancers 2011, 3, 1329–1350. [Google Scholar] [CrossRef]
- Arries, C.D.; Yohe, S.L. Monocytic Maturation Induced by FLT3 Inhibitor Therapy of Acute Myeloid Leukemia: Morphologic and Immunophenotypic Characteristics. Lab. Med. 2020, 51, 478–483. [Google Scholar] [CrossRef]
- Kondo, T.; Onozawa, M.; Fujisawa, S.; Harada, S.; Ogasawara, R.; Izumiyama, K.; Saito, M.; Morioka, M.; Mori, A.; Teshima, T. Myelomonocytic differentiation of leukemic blasts accompanied by differentiation syndrome in a case of FLT3-ITD-positive AML treated with gilteritinib. Hematology 2021, 26, 256–260. [Google Scholar] [CrossRef]
- McMahon, C.M.; Canaani, J.; Rea, B.; Sargent, R.L.; Qualtieri, J.N.; Watt, C.D.; Morrissette, J.J.D.; Carroll, M.; Perl, A.E. Gilteritinib induces differentiation in relapsed and refractory FLT3-mutated acute myeloid leukemia. Blood Adv. 2019, 3, 1581–1585. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Wei, A.H. How I treat acute myeloid leukemia in the era of new drugs. Blood 2020, 135, 85–96. [Google Scholar] [CrossRef]
- Bertoli, S.; Boutzen, H.; David, L.; Larrue, C.; Vergez, F.; Fernandez-Vidal, A.; Yuan, L.; Hospital, M.A.; Tamburini, J.; Demur, C.; et al. CDC25A governs proliferation and differentiation of FLT3-ITD acute myeloid leukemia. Oncotarget 2015, 6, 38061–38078. [Google Scholar] [CrossRef] [PubMed]
- Tahir, S.K.; Calvo, E.; Carneiro, B.A.; Yuda, J.; Shreenivas, A.; Jongen-Lavrencic, M.; Gort, E.; Ishizawa, K.; Morillo, D.; Biesdorf, C.; et al. Activity of eftozanermin alfa plus venetoclax in preclinical models and patients with acute myeloid leukemia. Blood 2023, 141, 2114–2126. [Google Scholar] [CrossRef]
- Han, H.; Shin, D.Y.; Kim, D.; Kim, H.; Lee, C.; Koh, Y.; Hong, J.; Yoon, S.S. Induction of leukemic stem cell differentiation by aryl hydrocarbon receptor agonist and synergy with gilteritinib in FLT3-ITD + acute myeloid leukemia. Leuk. Lymphoma 2020, 61, 1932–1942. [Google Scholar] [CrossRef]
- Hogg, G.; Severson, E.A.; Cai, L.; Hoffmann, H.M.; Holden, K.A.; Fitzgerald, K.; Kenyon, A.; Zeng, Q.; Mooney, M.; Gardner, S.; et al. Clinical characterization of the mutational landscape of 24,639 real-world samples from patients with myeloid malignancies. Cancer Genet. 2023, 278–279, 38–49. [Google Scholar] [CrossRef]
- Zeidner, J.F. Differentiating the Differentiation Syndrome Associated with IDH Inhibitors in AML. Clin. Cancer Res. 2020, 26, 4174–4176. [Google Scholar] [CrossRef] [PubMed]
- Norsworthy, K.J.; Mulkey, F.; Scott, E.C.; Ward, A.F.; Przepiorka, D.; Charlab, R.; Dorff, S.E.; Deisseroth, A.; Kazandjian, D.; Sridhara, R.; et al. Differentiation Syndrome with Ivosidenib and Enasidenib Treatment in Patients with Relapsed or Refractory IDH-Mutated AML: A U.S. Food and Drug Administration Systematic Analysis. Clin. Cancer Res. 2020, 26, 4280–4288. [Google Scholar] [CrossRef]
- Chen, X.; Xing, H.; Xie, X.; Kou, L.; Li, J.; Li, Y. Efficacy and safety of FDA-approved IDH inhibitors in the treatment of IDH mutated acute myeloid leukemia: A systematic review and meta-analysis. Clin. Epigenet. 2023, 15, 113. [Google Scholar] [CrossRef]
- Cathelin, S.; Sharon, D.; Subedi, A.; Cojocari, D.; Phillips, D.C.; Leverson, J.D.; MacBeth, K.J.; Nicolay, B.; Narayanaswamy, R.; Ronseaux, S.; et al. Enasidenib-induced differentiation promotes sensitivity to venetoclax in IDH2-mutated acute myeloid leukemia. Leukemia 2022, 36, 869–872. [Google Scholar] [CrossRef]
- Cao, L.; Weetall, M.; Trotta, C.; Cintron, K.; Ma, J.; Kim, M.J.; Furia, B.; Romfo, C.; Graci, J.D.; Li, W.; et al. Targeting of Hematologic Malignancies with PTC299, A Novel Potent Inhibitor of Dihydroorotate Dehydrogenase with Favorable Pharmaceutical Properties. Mol. Cancer Ther. 2019, 18, 3–16. [Google Scholar] [CrossRef]
- Jones, C.L.; Stevens, B.M.; D’Alessandro, A.; Reisz, J.A.; Culp-Hill, R.; Nemkov, T.; Pei, S.; Khan, N.; Adane, B.; Ye, H.; et al. Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer Cell 2018, 34, 724–740. [Google Scholar] [CrossRef]
- Zhou, J.; Yiying Quah, J.; Ng, Y.; Chooi, J.Y.; Hui-Min Toh, S.; Lin, B.; Zea Tan, T.; Hosoi, H.; Osato, M.; Seet, Q.; et al. ASLAN003, a potent dihydroorotate dehydrogenase inhibitor for differentiation of acute myeloid leukemia. Haematologica 2020, 105, 2286–2297. [Google Scholar] [CrossRef]
- So, J.; Lewis, A.C.; Smith, L.K.; Stanley, K.; Franich, R.; Yoannidis, D.; Pijpers, L.; Dominguez, P.; Hogg, S.J.; Vervoort, S.J.; et al. Inhibition of pyrimidine biosynthesis targets protein translation in acute myeloid leukemia. EMBO Mol. Med. 2022, 14, e15203. [Google Scholar] [CrossRef]
- Passaniti, A.; Kim, M.S.; Polster, B.M.; Shapiro, P. Targeting mitochondrial metabolism for metastatic cancer therapy. Mol. Carcinog. 2022, 61, 827–838. [Google Scholar] [CrossRef]
- Fiorillo, M.; Ózsvári, B.; Sotgia, F.; Lisanti, M.P. High ATP Production Fuels Cancer Drug Resistance and Metastasis: Implications for Mitochondrial ATP Depletion Therapy. Front. Oncol. 2021, 11, 740720. [Google Scholar] [CrossRef]
- Pardee, T.S.; Anderson, R.G.; Pladna, K.M.; Isom, S.; Ghiraldeli, L.P.; Miller, L.D.; Chou, J.W.; Jin, G.; Zhang, W.; Ellis, L.R.; et al. A Phase I Study of CPI-613 in Combination with High-Dose Cytarabine and Mitoxantrone for Relapsed or Refractory Acute Myeloid Leukemia. Clin. Cancer Res. 2018, 24, 2060–2073. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, T.; Ding, X.; Chang, Y.; Liu, C.; Zhang, Y.; Hao, S.; Yin, Q.; Jiang, B. Inhibition of mitochondrial complex III induces differentiation in acute myeloid leukemia. Biochem. Biophys. Res. Commun. 2021, 547, 162–168. [Google Scholar] [CrossRef]
- Zhang, H.; Fang, H.; Wang, K. Reactive oxygen species in eradicating acute myeloid leukemic stem cells. Stem Cell Investig. 2014, 1, 13. [Google Scholar]
- Liu, L.; Patnana, P.K.; Xie, X.; Frank, D.; Nimmagadda, S.C.; Rosemann, A.; Liebmann, M.; Klotz, L.; Opalka, B.; Khandanpour, C. High Metabolic Dependence on Oxidative Phosphorylation Drives Sensitivity to Metformin Treatment in MLL/AF9 Acute Myeloid Leukemia. Cancers 2022, 14, 486. [Google Scholar] [CrossRef]
- Wei, A.H.; Roberts, A.W.; Spencer, A.; Rosenberg, A.S.; Siegel, D.; Walter, R.B.; Caenepeel, S.; Hughes, P.; McIver, Z.; Mezzi, K.; et al. Targeting MCL-1 in hematologic malignancies: Rationale and progress. Blood Rev. 2020, 44, 100672. [Google Scholar] [CrossRef]
- Zhang, Q.; Riley-Gillis, B.; Han, L.; Jia, Y.; Lodi, A.; Zhang, H.; Ganesan, S.; Pan, R.; Konoplev, S.N.; Sweeney, S.R.; et al. Activation of RAS/MAPK pathway confers MCL-1 mediated acquired resistance to BCL-2 inhibitor venetoclax in acute myeloid leukemia. Signal Transduct. Target Ther. 2022, 7, 51. [Google Scholar] [CrossRef]
- Roberts, A.W.; Wei, A.H.; Huang, D.C.S. BCL2 and MCL1 inhibitors for hematologic malignancies. Blood 2021, 138, 1120–1136. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.-T.; Wu, Y.-Y.; Lee, Y.-C.; Chang, L.-S. BCL2L1 inhibitor A-1331852 inhibits MCL1 transcription and triggers apoptosis in acute myeloid leukemia cells. Biochem. Pharmacol. 2023, 215, 115738. [Google Scholar] [CrossRef]
- Tibes, R.; Bogenberger, J.M. Transcriptional Silencing of MCL-1 Through Cyclin-Dependent Kinase Inhibition in Acute Myeloid Leukemia. Front. Oncol. 2019, 9, 1205. [Google Scholar] [CrossRef]
- Lee, Y.; Shi, Y.; Wang, L.; Chiou, J.; Huang, C.; Chang, L. GSK3β suppression inhibits MCL1 protein synthesis in human acute myeloid leukemia cells. J. Cell. Physiol. 2021, 236, 570–586. [Google Scholar] [CrossRef] [PubMed]
- Yuda, J.; Will, C.; Phillips, D.C.; Abraham, L.; Alvey, C.; Avigdor, A.; Buck, W.; Besenhofer, L.; Boghaert, E.; Cheng, D.; et al. Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients. Commun. Med. 2023, 3, 154. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; He, X.; Qiu, Z.; Zhang, H.; Xie, R.; Liu, Z.; Gu, Y.; Zhao, N.; Xiang, Q.; Cui, Y. Targeting integrin pathways: Mechanisms and advances in therapy. Signal Transduct. Target. Ther. 2023, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Gomez, I.G.; Tang, J.; Wilson, C.L.; Yan, W.; Heinecke, J.W.; Harlan, J.M.; Raines, E.W. Metalloproteinase-mediated Shedding of Integrin β2 promotes macrophage efflux from inflammatory sites. J. Biol. Chem. 2012, 287, 4581–4589. [Google Scholar] [CrossRef]
- Itoh, Y. Modulation of Microenvironment Signals by Proteolytic Shedding of Cell Surface Extracellular Matrix Receptors. Front. Cell Dev. Biol. 2021, 9, 736735. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cronin, C.G.; Cao, Z.; Wang, C.; Ruan, J.; Pulikkot, S.; Hall, A.; Sun, H.; Groisman, A.; Chen, Y.; et al. Nexinhib20 Inhibits Neutrophil Adhesion and β2 Integrin Activation by Antagonizing Rac-1–Guanosine 5′-Triphosphate Interaction. J. Immunol. 2022, 209, 1574–1585. [Google Scholar] [CrossRef]
- Jawhara, S.; Pluskota, E.; Cao, W.; Plow, E.F.; Soloviev, D.A. Distinct Effects of Integrins αXβ2 and αMβ2 on Leukocyte Subpopulations during Inflammation and Antimicrobial Responses. Infect. Immun. 2016, 85, e00644-16. [Google Scholar]
- Nepstad, I.; Hatfield, K.J.; Grønningsæter, I.S.; Reikvam, H. The PI3K-Akt-mTOR Signaling Pathway in Human Acute Myeloid Leukemia (AML) Cells. Int. J. Mol. Sci. 2020, 21, 2907. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.K.D.C.; Machado, C.B.; Pessoa, F.M.C.D.P.; Barreto, I.V.; Gadelha, R.B.; Oliveira, D.D.S.; Ribeiro, R.M.; Lopes, G.S.; Filho, M.O.D.M.; DE Moraes, M.E.A.; et al. Development and Clinical Applications of PI3K/AKT/mTOR Pathway Inhibitors as a Therapeutic Option for Leukemias. Cancer Diagn. Progn. 2024, 4, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, B.; Verzella, D.; Capece, D.; Vecchiotti, D.; Nolfi, M.D.V.; Flati, I.; Cornice, J.; Di Padova, M.; Angelucci, A.; Alesse, E.; et al. NF-κB: A Druggable Target in Acute Myeloid Leukemia. Cancers 2022, 14, 3557. [Google Scholar] [CrossRef] [PubMed]
- Lennartsson, J.; Rönnstrand, L. Stem cell factor receptor/c-Kit: From basic science to clinical implications. Physiol. Rev. 2012, 92, 1619–1649. [Google Scholar] [CrossRef] [PubMed]
- Justo, B.L.; Jasiulionis, M.G. Characteristics of TIMP1, CD63, and β1-Integrin and the Functional Impact of Their Interaction in Cancer. Int. J. Mol. Sci. 2021, 22, 9319. [Google Scholar] [CrossRef] [PubMed]
- Chirco, R.; Liu, X.-W.; Jung, K.-K.; Kim, H.-R.C. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2006, 25, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Dawson, J.C.; Serrels, A.; Stupack, D.G.; Schlaepfer, D.D.; Frame, M.C. Targeting FAK in anticancer combination therapies. Nat. Rev. Cancer 2021, 21, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Bartaula-Brevik, S.; Leitch, C.; Hernandez-Valladares, M.; Aasebø, E.; Berven, F.S.; Selheim, F.; Brenner, A.K.; Rye, K.P.; Hagen, M.; Reikvam, H.; et al. Vacuolar ATPase Is a Possible Therapeutic Target in Acute Myeloid Leukemia: Focus on Patient Heterogeneity and Treatment Toxicity. J. Clin. Med. 2023, 12, 5546. [Google Scholar] [CrossRef] [PubMed]
- Aasebø, E.; Bartaula-Brevik, S.; Hernandez-Valladares, M.; Bruserud, Ø. Vacuolar ATPase as a possible therapeutic target in human acute myeloid leukemia. Expert Rev. Hematol. 2018, 11, 13–24. [Google Scholar] [CrossRef]
- Stransky, L.; Cotter, K.; Forgac, M.; van der Wijst, J.; Belge, H.; Bindels, R.J.M.; Devuyst, O. The Function of V-ATPases in Cancer. Physiol. Rev. 2016, 96, 1071–1091. [Google Scholar] [CrossRef]
- Libourel, E.J.; Klerk, C.P.W.; van Norden, Y.; de Maat, M.P.M.; Kruip, M.J.; Sonneveld, P.; Löwenberg, B.; Leebeek, F.W.G. Disseminated intravascular coagulation at diagnosis is a strong predictor for thrombosis in acute myeloid leukemia. Blood 2016, 128, 1854–1861. [Google Scholar] [CrossRef]
- Ku, G.H.; White, R.H.; Chew, H.K.; Harvey, D.J.; Zhou, H.; Wun, T. Venous thromboembolism in patients with acute leukemia: Incidence, risk factors, and effect on survival. Blood 2009, 113, 3911–3917. [Google Scholar] [CrossRef]
- De Stefano, V.; Sorà, F.; Rossi, E.; Chiusolo, P.; Laurenti, L.; Fianchi, L.; Zini, G.; Pagano, L.; Sica, S.; Leone, G. The risk of thrombosis in patients with acute leukemia: Occurrence of thrombosis at diagnosis and during treatment. J. Thromb. Haemost. 2005, 3, 1985–1992. [Google Scholar] [CrossRef] [PubMed]
- Olivi, M.; Di Biase, F.; Lanzarone, G.; Arrigo, G.; Martella, F.; Apolito, V.; Secreto, C.; Freilone, R.; Bruno, B.; Audisio, E.; et al. Thrombosis in Acute Myeloid Leukemia: Pathogenesis, Risk Factors and Therapeutic Challenges. Curr. Treat. Options Oncol. 2023, 24, 693–710. [Google Scholar] [CrossRef]
- Tobelem, G.; Jacquillat, C.; Chastang, C.; Auclerc, M.F.; Lechevallier, T.; Weil, M.; Daniel, M.T.; Flandrin, G.; Harrousseau, J.L.; Schaison, G.; et al. Acute monoblastic leukemia: A clinical and biologic study of 74 cases. Blood 1980, 55, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Mannan, A.; Muhsen, I.N.; Barragán, E.; Sanz, M.A.; Mohty, M.; Hashmi, S.K.; Aljurf, M. Genotypic and Phenotypic Characteristics of Acute Promyelocytic Leukemia Translocation Variants. Hematol. Oncol. Stem Cell Ther. 2020, 13, 189–201. [Google Scholar] [CrossRef]
- Sanz, M.A.; Fenaux, P.; Tallman, M.S.; Estey, E.H.; Löwenberg, B.; Naoe, T.; Lengfelder, E.; Döhner, H.; Burnett, A.K.; Chen, S.J.; et al. Management of acute promyelocytic leukemia: Updated recommendations from an expert panel of the European LeukemiaNet. Blood 2019, 133, 1630–1643. [Google Scholar] [CrossRef] [PubMed]
- Zhen, A.; Krutzik, S.R.; Levin, B.R.; Kasparian, S.; Zack, J.A.; Kitchen, S.G. CD4 ligation on human blood monocytes triggers macrophage differentiation and enhances HIV infection. J. Virol. 2014, 88, 9934–9946. [Google Scholar] [CrossRef]
- Wang, T.; Ge, Y.; Xiao, M.; Lopez-Coral, A.; Li, L.; Roesch, A.; Huang, C.; Alexander, P.; Vogt, T.; Xu, X.; et al. SECTM1 produced by tumor cells attracts human monocytes via CD7-mediated activation of the PI3K pathway. J. Investig. Dermatol. 2014, 134, 1108–1118. [Google Scholar] [CrossRef]
- Humphries, J.D.; Humphries, M.J. CD14 is a ligand for the integrin alpha4beta1. FEBS Lett. 2007, 581, 757–763. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Reikvam, H.; Brenner, A.K. Toll-like Receptor 4, Osteoblasts and Leukemogenesis; the Lesson from Acute Myeloid Leukemia. Molecules 2022, 27, 735. [Google Scholar] [CrossRef] [PubMed]
- Lubos, E.; Mahoney, C.E.; Leopold, J.A.; Zhang, Y.Y.; Loscalzo, J.; Handy, D.E. Glutathione peroxidase-1 modulates lipopolysaccharide-induced adhesion molecule expression in endothelial cells by altering CD14 expression. FASEB J. 2010, 24, 2525–2532. [Google Scholar] [CrossRef] [PubMed]
- Kindle, L.; Rothe, L.; Kriss, M.; Osdoby, P.; Collin-Osdoby, P. Human microvascular endothelial cell activation by IL-1 and TNF-alpha stimulates the adhesion and transendothelial migration of circulating human CD14+ monocytes that develop with RANKL into functional osteoclasts. J. Bone Miner. Res. 2006, 21, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Szlasa, W.; Wilk, K.; Knecht-Gurwin, K.; Gurwin, A.; Froń, A.; Sauer, N.; Krajewski, W.; Saczko, J.; Szydełko, T.; Kulbacka, J.; et al. Prognostic and Therapeutic Role of CD15 and CD15s in Cancer. Cancers 2022, 14, 2203. [Google Scholar] [CrossRef] [PubMed]
- Karawajczyk, M.; Douhan Håkansson, L.; Lipcsey, M.; Hultström, M.; Pauksens, K.; Frithiof, R.; Larsson, A. High expression of neutrophil and monocyte CD64 with simultaneous lack of upregulation of adhesion receptors CD11b, CD162, CD15, CD65 on neutrophils in severe COVID-19. Ther. Adv. Infect. Dis. 2021, 8, 20499361211034065. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, M.; Sato, N.; Sugimori, H.; Mori, K.; Oshimi, K. A minor E-selectin ligand, CD65, is critical for extravascular infiltration of acute myeloid leukemia cells. Leuk. Res. 2001, 25, 847–853. [Google Scholar] [CrossRef]
- Bordessoule, D.; Jones, M.; Gatter, K.C.; Mason, D.Y. Immunohistological patterns of myeloid antigens: Tissue distribution of CD13, CD14, CD16, CD31, CD36, CD65, CD66 and CD67. Br. J. Haematol. 1993, 83, 370–383. [Google Scholar] [CrossRef]
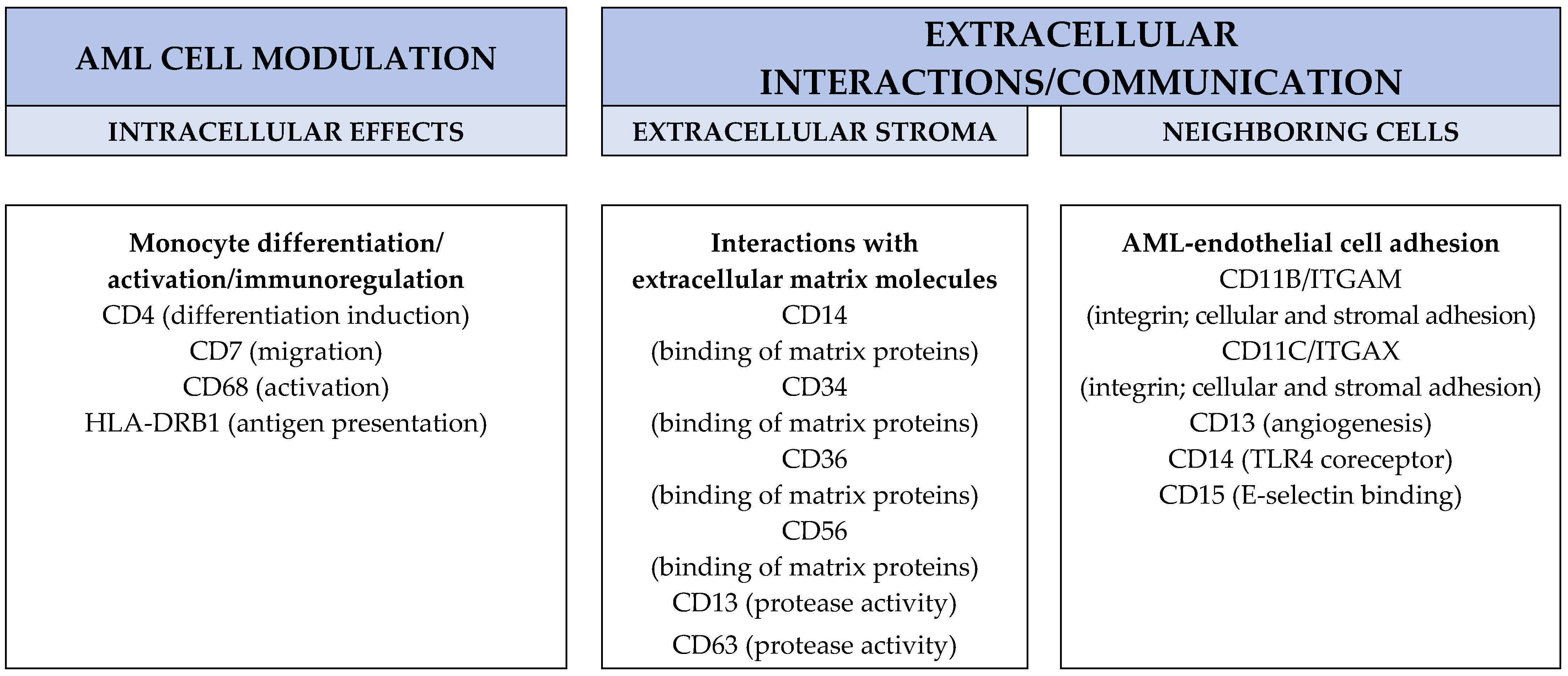
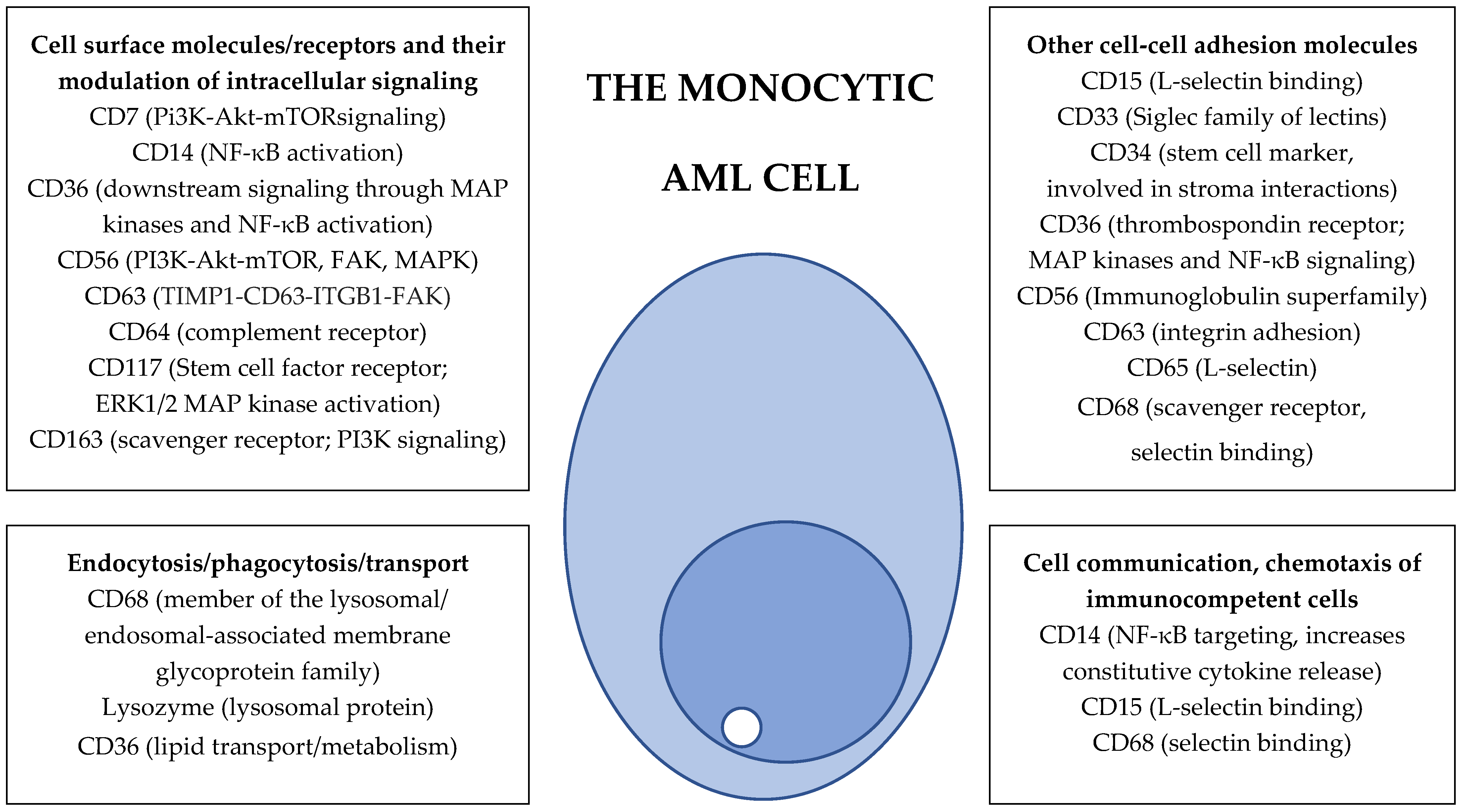
| Characteristic | FAB-M4; Acute Myelomonocytic Leukemia | FAB-M5; Acute Monoblastic and Monocytic Leukemia |
|---|---|---|
| <Morphology of the AML cells |
|
|
| Histochemistry (the AML cells) |
| Monocytes and promyelocytes are usually positive only for non-specific esterase but not myeloperoxidase |
| Immunophenotype of the AML cells |
|
|
| Protein levels of T-cell costimulatory and checkpoint molecules |
| ↑ CD86? |
| ↑ PD-L1? |
| Cell surface protein levels of cytokine receptors |
| ↑ GM-CSF receptor |
| ↑ Flt3 |
| ↓ SCF receptor/CD117 |
| Release of soluble protein mediators |
| ↑ Several CCL and CXCL chemokines |
| ↑ Several Interleukins |
| ↑ Several proteases and protease regulators |
| ↑ Several hematopoietic growth factors |
| Mediator | Effect of the Mediator on Cell Metabolism | Effect of Cell Metabolism on the Mediator |
|---|---|---|
| Bax |
| |
| Cytochrome C | Passing electrons from complex III to complex IV in the electron transport chain | Glucose-stimulated glutathione production inactivates, but reactive oxygen species activate cytochrome c |
| Bcl-2 |
| Metabolic stress modulates Bcl-2 activity |
| Mcl-1 |
| Targeted for degradation in the absence of glucose |
| Bcl-xL |
| |
| Bad | Regulates glucose metabolism | |
| Puma | Its expression is induced/increased by glucose deprivation | |
| Noxa |
| The levels are decreased by glucose |
| TP53 |
| There is a crosstalk between the metabolic sensor mTORC1 and TP53 |
| Sirtuins | These lysine deacetylases modulate cellular metabolism, e.g., lipid metabolism and urea cycle regulation | Protein acetylation is also influenced by availability of acetyl-CoA from the tricarboxylic acid cycle |
| Clinical studies of venetoclax resistance in AML | |
| Morphological differentiation | Monocytic differentiation is associated with venetoclax resistance [7,8,88,151,159]. Patients with newly diagnosed monocytic AML show significantly lower remission rate and overall survival compared to non-monocytic subsets and with no significant differences between the FAB-M4 and FAB-M5 subtypes [164]. Lower remission rate for monocytic variants was also observed for relapse/refractory AML patients, but the overall survival did not differ for these patients [164]. |
| Gene expression profiles | Gene expression profiling of primary AML cells detected four patient clusters associated with venetoclax resistance. (i) One cluster showed an enrichment of monocytic AML cells, as well as transcriptional activation of PI3K-AKT-mTOR signaling and energy metabolism pathways; these cells showed sensitivity to mTOR and CDK inhibition. (ii) The second cluster was enriched for NRAS mutations and associated with decreased HOX expression. (iii) The third was characterized by TP53 mutations and the expression of erythroid markers, and (iv) the last cluster showed overexpression of interferon signaling and high rates of DNMT3A mutations. A patient’s AML cell population may consist of cell subsets mimicking different resistance clusters [165]. |
| Bcl-2 family | The combinatorial ratio/score of the antiapoptotic mediators Bcl-2, Bcl-xL and MCL-1 expression in leukemic stem cells can predict the clinical responses to venetoclax [160]. |
| Senescence | Regulation of senescence together with PD1 signaling and NADP oxidase activity seems to be involved in the clinical venetoclax resistance associated with monocytic differentiation [166]. |
| AML stem cells | There seems to be a selective outgrowth of monocytic AML cells at the time of progression during venetoclax therapy [7]. This progression arises from leukemia stem cells that seem to differ from other primitive leukemic stem cells with regards to their immunophenotype (CD34−, CD4+, CD11b−, CD14− and CD36−); their reliance on purine metabolism and a different chemosensitivity profile. These monocytic stem cells may co-reside with other subsets of more primitive leukemic stem cells [158]. |
| Other factors than monocytic AML cell differentiation associated with venetoclax susceptibility/resistance | |
| Mutations | Sensitivity to venetoclax seems to be associated with NPM1, IDH1/2, TET2, RUNX1 and ASXL1 mutations, whereas venetoclax resistance is associated with FLT3-ITD, TP53, K/NRAS or PTPN11 mutations [139,140,148,159]. |
| Karyotype | Adverse karyotype according to the ELN classification is also associated with resistance [141]. |
| Clonal heterogeneity | Clonal heterogeneity is associated with venetoclax resistance [143,144,164]. |
| Experimental studies | |
| Differentiation | There is an association between venetoclax resistance and monocytic differentiation [163]. |
| Bcl-2 family | MCL-1 inhibition is an effective antileukemic strategy in venetoclax-resistant AML cells and patient-derived xenografts, and molecular profiling can be used to predict the efficiency of Mcl-1 versus Bcl-2 inhibition [167]. Resistance to venetoclax is associated with a low Bcl-2/Mcl-1 ratio [7]. Myelomonocytic, as well as upregulated, Bcl-2A1 and CLEC7A confer resistance [148]. |
| Mutations | Mutations of PTPN11 and KRAS also confer resistance to venetoclax-based therapy [148]. |
| Ribosome | The ribosomal protein S6 kinase alpha-1 mediates venetoclax resistance in monocytic AML [150]. |
| Metabolism | Metabolic effects of venetoclax involving oxidative phosphorylation seem to be important for its antileukemic effects in Bcl-2-dependent AML cells [87,88]. |
| Combination of venetoclax with other targeted therapies | |
| PI3K-mTOR | Combination of venetoclax with PI3K-mTOR inhibition has synergistic effects, and this increases sensitivity depends especially on BAX but also on BAK expression [161]. |
| Asparaginase | Combination of venetoclax and pegylated asparaginase has synergistic effects; this is possibly due to reduction of glutathione levels and thereby also reduced Mcl-1 expression [162]. |
| Mcl-1 | Combination of Bcl-2 and Mcl-1 inhibition has synergistic effects [167]. |
| Target | Effect of AML Cell Differentiation on Targeted Therapy or Effects of Targeted Therapy on AML Cell Differentiation |
|---|---|
| Epigenetic targeting/histone modification | |
| BET | Less effective in immature/undifferentiated AML cells, and monocytic markers correlates with sensitivity. The inhibitors induce monocytic differentiation. |
| Histone deacetylase | HDAC inhibition induces myeloid differentiation with induction of the transcription factors associated with differentiation and reduced expression of CD117, together with increased expression of CD11b/CD18. |
| Lysine demethylase 1 | LSD1 is important for the differentiation block in MLL variants of AML; LSD1 inhibitors induce morphological signs of monocytic differentiation; decreased CD34 expression and increased expression of CD14, CD36 and CD86. |
| DOT1L | In vitro studies have shown that DOT1L inhibitors induce both morphological and molecular signs of myeloid differentiation in MLL-rearranged and DNMT3A-mutated AML. |
| Menin | Menin inhibition is a possible strategy that modulates AML cell differentiation, especially in MLL-rearranged and NPM1-mutated AML but possibly also AML variants with FLT3-ITD, NUP98 translocations, UBTF tandem duplications and MN1 translocations. |
| Molecular transport | |
| Exportin 1 | Exportin 1 inhibition in patients with NPM1 mutation induces morphological signs of monocytic or granulocytic differentiation together with increased CD11b and CD14 expression. |
| Protein synthesis and modification | |
| Ribosome targeting | Combination of venetoclax (decreased susceptibility in FAB-M4/M5) with the ribosome-targeting tedizolid increases the antileukemic effects compared to monotherapy. |
| Proteasome | Increased antileukemic effect in monocytic AML cells. |
| Intracellular signaling | |
| FLT3 | Monocytic differentiation seems most common; even granulocytic and erythroid differentiation are described. |
| TRAIL agonist | Combination of venetoclax (decreased susceptibility in FAB-M4/M5) with the TRAIL agonist eftozanermin increases the antileukemic effects compared to monotherapy. |
| Aryl hydrocarbon receptor | Receptor agonists can inhibit AML stem cells and induce AML stem cell differentiation by the induction of monocyte and/or granulocyte markers in patients with Flt3-ITD characterized by increased levels of CD11b, CD14 and CD15, as well as morphological signs of differentiation. |
| Cellular metabolism | |
| IDH1/IDH2 | IDH mutations are associated with the FAB-M2 phenotype. Induction of monocytic and or granulocytic differentiation. |
| Pyrimidine biosynthesis | Inhibition of the rate-limiting step induces morphological AML cell differentiation with increased expression of CD11b, CD13, CD14, and CD33 and functional signs, as well as gene expression profile modulation consistent with neutrophil differentiation. |
| Energy metabolism | Inhibition of the mitochondrial electron transport chain complex III A inhibits proliferation and induces AML cell differentiation. The antileukemic effects of targeting antiapoptotic and metabolism-regulatory Bcl-2 family members depend on the differentiation status of the AML cells. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruserud, Ø.; Selheim, F.; Hernandez-Valladares, M.; Reikvam, H. Monocytic Differentiation in Acute Myeloid Leukemia Cells: Diagnostic Criteria, Biological Heterogeneity, Mitochondrial Metabolism, Resistance to and Induction by Targeted Therapies. Int. J. Mol. Sci. 2024, 25, 6356. https://doi.org/10.3390/ijms25126356
Bruserud Ø, Selheim F, Hernandez-Valladares M, Reikvam H. Monocytic Differentiation in Acute Myeloid Leukemia Cells: Diagnostic Criteria, Biological Heterogeneity, Mitochondrial Metabolism, Resistance to and Induction by Targeted Therapies. International Journal of Molecular Sciences. 2024; 25(12):6356. https://doi.org/10.3390/ijms25126356
Chicago/Turabian StyleBruserud, Øystein, Frode Selheim, Maria Hernandez-Valladares, and Håkon Reikvam. 2024. "Monocytic Differentiation in Acute Myeloid Leukemia Cells: Diagnostic Criteria, Biological Heterogeneity, Mitochondrial Metabolism, Resistance to and Induction by Targeted Therapies" International Journal of Molecular Sciences 25, no. 12: 6356. https://doi.org/10.3390/ijms25126356
APA StyleBruserud, Ø., Selheim, F., Hernandez-Valladares, M., & Reikvam, H. (2024). Monocytic Differentiation in Acute Myeloid Leukemia Cells: Diagnostic Criteria, Biological Heterogeneity, Mitochondrial Metabolism, Resistance to and Induction by Targeted Therapies. International Journal of Molecular Sciences, 25(12), 6356. https://doi.org/10.3390/ijms25126356







